Abstract
The primary role of DNA polymerases is to accurately and efficiently replicate the genome in order to ensure the maintenance of the genetic information and its faithful transmission through generations. This is not a simple task considering the size of the genome and its constant exposure to endogenous and environmental DNA damaging agents. Thus, a number of DNA repair pathways operate in cells to protect the integrity of the genome. In addition to their role in replication, DNA polymerases play a central role in most of these pathways. Given the multitude and the complexity of DNA transactions that depend on DNA polymerase activity, it is not surprising that cells in all organisms contain multiple highly specialized DNA polymerases, the majority of which have only recently been discovered. Five DNA polymerases are now recognized in Escherichia coli, 8 in Saccharomyces cerevisiae, and at least 15 in humans. While polymerases in bacteria, yeast and mammalian cells have been extensively studied much less is known about their counterparts in plants. For example, the plant model organism Arabidopsis thaliana is thought to contain 12 DNA polymerases, whose functions are mostly unknown. Here we review the properties and functions of DNA polymerases focusing on yeast and mammalian cells but paying special attention to the plant enzymes and the special circumstances of replication and repair in plant cells.
Keywords: DNA Polymerase, DNA repair, DNA replication, TLS, translesion synthesis, fidelity
I. Introduction - DNA Polymerase families
The first evidence of the existence of an enzymatic activity capable of synthesizing DNA came in 1958 with the discovery of E. coli Pol I by A. Kornberg and colleagues (Lehman, et al., 1958). The discovery of several other polymerase activities soon followed, and it was realized that they possessed significantly different properties. However, it was not until sequence information became readily available that the reasons behind those biochemical differences could begin to be understood. It became clear that polymerases, although sometimes clearly evolutionarily related, were nevertheless divergent, and the comparison of the features of their primary sequence led to a classification into families that is still current (families A, B, C and X; Ito and Braithwaite, 1991; Braithwaite and Ito, 1993; see Table 1). The development of massive sequencing projects resulted in a revolution in the polymerase field. In a brief amount of time, several novel DNA polymerase genes were identified (Goodman and Tippin, 2000). One of the main results was the identification of a novel family of DNA polymerases, family Y (Ohmori, et al., 2001), whose members are widely believed to conduct synthesis opposite template lesions in a process known as translesion synthesis (TLS; Prakash, et al., 2005). Thus DNA polymerases are generally classified into five families. However, many eukaryotic genomes encode one or more retrotransriptases. Among these is the enzyme telomerase, which appears to be essential for telomere maintenance (Autexier and Lue, 2006). In addition, a family of enzymes (family D) exists, composed of DNA polymerases that are only present in archaea (Cann and Ishino, 1999), and a novel class of DNA polymerases that are related to archaeal primases has recently been identified in bacteria (Pitcher, et al., 2005).
Table I.
Mammalian DNA Polymerases
| Polymerase | Family | Mol. Wt.(kDa) Cat. domaina | Subunit composition | Additional activities | Process/Functions |
|---|---|---|---|---|---|
| α (alpha) | B | 165 | p67, p58+p49 primase | primase | DNA replication, DSB repair: HR S-phase checkpoint |
| β (beta) | X | 39 | - | dRP lyase AP lyase | BER |
| γ (gamma) | A | 140 | p55 | 3′ Exonuclease dRP lyase | mitochondrial replication and repair |
| δ (delta) | B | 100 | p12 p58+p66 | 3′ Exonuclease | replication, repair: MMR, BER, NER, DSBs |
| ε (epsilon) | B | 225 | p17,p12, p59 | 3′ Exonuclease | replication, repair: BER, NER DSBs HR, S-phase checkpoint |
| ζ (zeta) | B | 353 | Rev7 | TLS, ICL repair somatic hypermutation | |
| η (eta) | Y | 78 | - | TLS, somatic hypermutation, HR | |
| θ (theta) | A | 290 | - | ATPase, Helicase motif | ICL repair?, somatic hypermutation, BER, TLS |
| Ι (iota) | Y | 80 | - | dRP lyase | TLS, BER? Specialized MMR? |
| κ (kappa) | Y | 76 | - | TLS, NER | |
| λ (lambda) | X | 66 | - | dRP lyase, | DSB repair: NHEJ, V(D)J recombination, BER |
| μ (mu) | X | 55 | - | TdT | DSB repair: NHEJ, V(D)J recombination |
| ν (nu) | A | 100 | - | ICL repair ? | |
| Rev1 | Y | 138 | - | TLS | |
| TdT | X | 56 | - | V(D)J recombination |
Molecular weight of human polymerases deduced from primary structure.
A. Family A
The prototype enzyme in this family is E. coli Pol I, discovered 50 years ago (Lehman, et al., 1958). It was the first DNA polymerase to be isolated and the first polymerase whose structure was solved (Ollis, et al., 1985). Although initially thought to be the main replicative polymerase in bacterial cells, it is now clear that its role is related to DNA repair and Okazaki fragment maturation (Kornberg and Baker, 1992). To assist in these roles, E. coli Pol I contains two additional activities besides DNA polymerization, a 3′-5′ and a 5′-3′ exonuclease. Of these, the 3′-5′ exonuclease activity is conserved in several other members of the family. This activity is termed proofreading activity because it can excise nucleotides misinserted by the polymerase. Interestingly, despite the fact that the bacterial members of the family only have a minor role in replication, members of this family belonging to other organisms do in fact carry out the bulk of genomic replication. This is the case of phage polymerases (such as T7; Doublie and Ellenberger, 1998) or, in eukaryotes, that of the mitochondrial replicative polymerase, Pol γ (Graziewicz, et al., 2006).
Besides Pol γ, mammalian cells contain two more polymerases of this family. Pol ν (Marini, et al., 2003) and Pol θ (Sharief, et al., 1999; Seki, et al., 2003). Both enzymes lack an associated proofreading activity and their role is still unclear. Pol θ has been shown to participate in the antigen variability generation process of somatic hypermutation (Masuda, et al., 2006) and is also thought to participate in DNA repair (Yoshimura, et al., 2006). Both enzymes have been suggested to play a role in TLS (Seki, et al., 2004; Takata, et al., 2006).
B. Family B
The main replicative enzymes in eukaryotes belong to family B (Garg and Burgers, 2005). Like most family A enzymes, most family B enzymes contain an associated 3′-5′ exonuclease activity. However, unlike members of other families, family B polymerases are multisubunit enzymes. It seems clear that Pols δ and ε share the monumental task of replicating the billions of base pairs in the genome of higher eukaryotes. Both are among the most faithful and processive enzymes in the presence of their accessory proteins (Shcherbakova, et al., 2003; Fortune, et al., 2005). Replication, however, is dependent on the dual activities of Pol α, which is a complex of a primase and a polymerase (Garg and Burgers, 2005). As is the case with family A, several bacteriophages (such as T4 (Benkovic, et al., 2001) or Phi29 (Blanco and Salas, 1996)) utilize a family B enzyme as their main replicase.
Besides Pols α, δ and ε, eukaryotes contain an additional family B protein: Pol ζ, whose role is not well understood (Lawrence, 2004). This enzyme is relatively unfaithful and appears to be able to extend past a mismatched base pair with higher efficiency than most DNA polymerases (Johnson, et al., 2000). This ability has been invoked to propose a role for Pol ζ in facilitating lesion bypass reactions (Prakash and Prakash, 2002).
C. Family X
Family X polymerases are small, monomeric polymerases that appear to participate in filling in short gaps during DNA repair (Ramadan, et al., 2004). A characteristic feature of most family members is the presence of an N-terminal 8 kDa DNA binding domain, which facilitates binding to gapped substrates (Beard and Wilson, 2006). Family X members are present in different organisms, from certain bacteria and viruses to yeast and mammals (Garcia-Diaz, et al., 2000; Garcia-Diaz, et al., 2005a). This conservation would appear to be related to their ability to conduct gap-filling. The most studied of these enzymes is Pol β, that participates in repair of base damage through the BER process (Wilson, et al., 2000). Other family X polymerases include three enzymes that participate in the V(D)J recombination process: Pol λ, Pol μ (Dominguez, et al., 2000) and the template-independent terminal deoxynucleotidyl transferase (TdT; Nick McElhinny and Ramsden, 2004; Bertocci, et al., 2006). V(D)J recombination is a specialized end-joining reaction that occurs in the cells of the immune system at the antigen receptor gene loci and is responsible for diversification of the antigen recognition site. Interestingly, family X members are also present in organisms devoid of an immune system, such as yeast, viruses or bacteria. The role of these enzymes is thought to be related to DNA repair. In fact, in addition to V(D)J recombination, Pols λ and μ are believed to participate in repair of double strand breaks through the Non-Homologous End-Joining process (Lee, et al., 2004; Nick McElhinny and Ramsden, 2004).
Eukaryotic cells contain yet another family X enzyme. Pol σ, product of the TRF4 gene, was recently described in S. cerevisiae as a DNA polymerase involved in sister chromatid cohesion (Wang, et al., 2000). However, controversy exists as to the nature of its enzymatic activity. It has been suggested that this protein might instead contain a poly-A RNA polymerase activity (Haracska, et al., 2005) involved in processing of certain RNA molecules (Egecioglu, et al., 2006).
D. Family Y
The discovery of family Y polymerases resulted from the almost simultaneous realization that the E.coli proteins UmuC/UmuD’ (Tang, et al., 1999) and DinB (Wagner, et al., 1999) encode DNA polymerases and that eukaryotes contained a related protein, Pol η. In humans, alterations in the Pol η gene result in XP-V (Johnson, et al., 1999; Masutani, et al., 1999), a variant of Xeroderma pigmentosum, an inherited genetic disorder that is associated with photosensitivity and high incidence of skin cancer (Masutani, et al., 2000).
Family Y polymerases have several common characteristics. None of them contains an exonuclease activity, and they have a domain, called the PAD, wrist or little-fingers domain (Ling, et al., 2001; Silvian, et al., 2001; Trincao, et al., 2001), that seems to modulate their substrate specificity (Boudsocq, et al., 2004). Family Y enzymes have low fidelity of synthesis on undamaged DNA (Kunkel, 2004). Unlike polymerases in other families, family Y members have a loose DNA binding pocket for the nascent base pair (Ling, et al., 2001). Thus, these enzymes can accommodate distorted DNA structures in their active site, resulting in the ability of these enzymes to polymerize on damaged DNA. In fact, the main role of family Y polymerases seems to be in DNA lesion tolerance pathways: if the cell fails to repair DNA lesions that can interfere with the replication process and these lesions are encountered by the replication fork, family Y polymerases can bypass those lesions by polymerizing across the damaged site, in a process that has been termed translesion synthesis (Prakash, et al., 2005). For instance, Pol η appears to be specialized in bypass of cyclobutane pyrimidine dimers, a UV- induced lesion.
Additional members of the family were identified subsequently, and these include Pols κ (Gerlach, et al., 1999; Ohashi, et al., 2000b), Ι (McDonald, et al., 1999) and Rev1 (Nelson, et al., 1996). The role of these polymerases is far from clear, but it is generally believed that they participate in translesion synthesis of some specific lesions. Family Y polymerases have also been implicated in the somatic hypermutation process, a phenomenon of targeted mutagenesis at the immunoglobulin gene loci that contributes to the generation of high affinity antibodies (Seki, et al., 2005).
II. Structural analysis of DNA polymerases
The structure of the Klenow fragment of E. coli DNA polymerase I was solved in 1985 (Ollis, et al., 1985). This was the first DNA polymerase structure to be solved, and it provided significant insight into the polymerization mechanism. Since then, a large number of additional structures of DNA polymerases in complex with their substrates have been solved (Kiefer, et al., 1997; Sawaya, et al., 1997; Wang, et al., 1997; Doublié, et al., 1998; Ling, et al., 2001; Trincao, et al., 2001; Garcia-Diaz, et al., 2005b; Nair, et al., 2005), so that we presently know the structure of at least one representative member of each DNA polymerase family (see Figure 1), including family C (Bailey, et al., 2006; Lamers, et al., 2006), represented by E. coli DNA Polymerase III. Interestingly, despite the lack of extensive sequence similarity, the general structure of most of these enzymes shares common features (Steitz, 1999). In all cases their catalytic domain can be likened to a hand, with fingers, palm and thumb subdomains, which contribute to template-primer and dNTP binding. And in all cases the palm subdomain harbors the three catalytic residues that are essential for polymerization. Beyond revealing the general protein fold, structural studies of DNA polymerases are furthering our understanding, at the atomic level, of the nature of the interaction of DNA polymerases with normal (Batra, et al., 2006) and damaged (Krahn, et al., 2003; Ling, et al., 2003; Brieba, et al., 2004; Hsu, et al., 2004; Hsu, et al., 2005), DNA substrates, the specific properties of each DNA polymerase or the molecular mechanisms of mutagenesis (Johnson and Beese, 2004; Garcia-Diaz, et al., 2006). In fact, it has become evident that, in order to precisely understand the functions of the polymerases in a family, it no longer suffices to study a representative model, because DNA polymerases in the same family often present very different properties (such as processivity, fidelity or substrate specificity) that are the result of subtle structural differences (Bebenek and Kunkel, 2004; Kamtekar, et al., 2004; Garcia-Diaz, et al., 2005a; Rodriguez, et al., 2005).
Figure 1. Structural similarity of DNA polymerases.
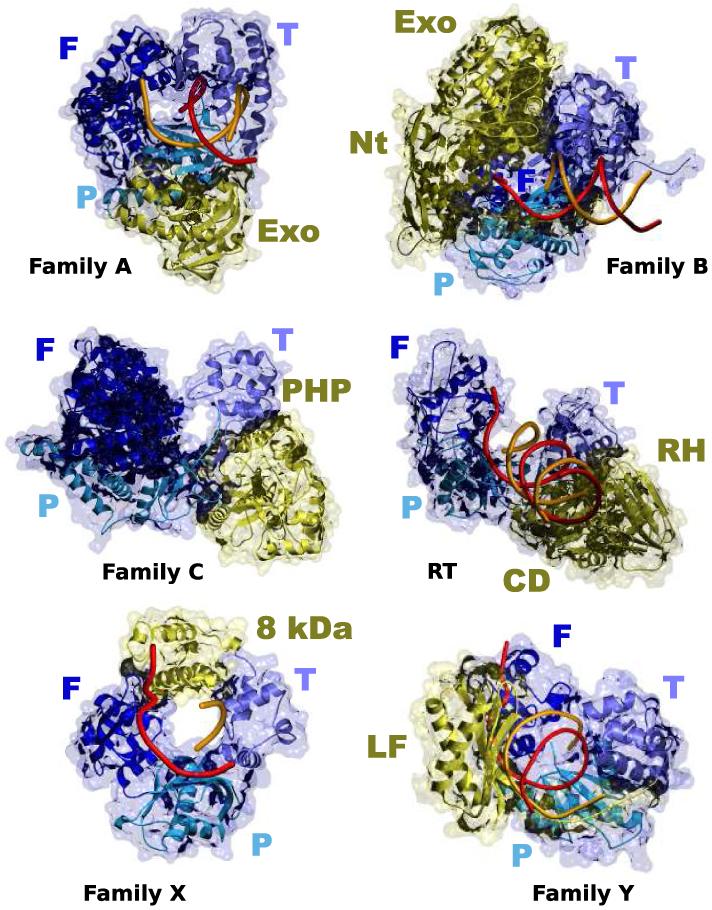
Crystal structures of a representative member of each of the five DNA polymerase families (A, T. aquaticus Pol I; B, RB69 Pol; C, E. coli Pol III; X, Pol λ and Y, S. solfataricus Dpo4) and retrotranscriptases (RT). All DNA polymerases contain a general fold that can be likened to a right hand, with fingers, palm and thumb subdomains, colored in different shades of blue. Additional subdomains, colored yellow, are specific for each family or individual enzyme. Exo, 3′-5′ exonuclease domain; Nt, N-terminal domain; PHP, Polymerase and Histidinol Phosphatase domain; 8 kDa, 8 kDa domain; LF, Little Fingers domain; RH, RNAse H domain; CD, Connecting domain.
Protein-protein interactions are an important component of DNA polymerase function and regulation. Structural studies of DNA polymerases in complex with protein partners are beginning to shed light on how the polymerase catalytic subunit interacts with its accessory subunits and other protein factors that are crucial to the different DNA transaction. For example, a significant understanding has been gained on the nature of the interactions between the different subunits of Pol ε (Asturias, et al., 2006), or the interaction of a family B enzyme with PCNA, its processivity factor (Shamoo and Steitz, 1999). In addition, structural studies have provided insight into the process of protein-primed initiation of DNA replication (Kamtekar, et al., 2006), a mechanism employed by certain phages that appears to also be used for the replication of mitochondrial plasmids, including some present in plants (Bernad, et al., 1987).
III. Nuclear replication in plants
Perhaps the most important function of DNA polymerases is to synthesize an exact replica of the genome during the replication process. Although plants, as other eukaryotes, contain semi-autonomous organelles, the vast majority of the genetic material is contained in the nucleus. Eukaryotic nuclear replication is a bidirectional process initiated at one of several origins of replication, where a carefully regulated complex of proteins unwinds the DNA and facilitates the assembly of a replication fork (Garg and Burgers, 2005). Unwinding is followed by binding of the single-strand binding protein (RPA) and loading of the Polα/primase complex. The primase subunit contains a unique activity that is capable of synthesizing de novo a short RNA primer. This primer is then extended by the Pol α polymerase activity to generate a DNA primer for subsequent elongation by one of the two processive polymerases, Pols δ and ε. These polymerases, with assistance of the sliding clamp PCNA, carry out most DNA synthesis (Garg and Burgers, 2005). The process of Polα/primase binding and switching to a different DNA polymerase is repeated at each origin and for every Okazaki fragment in the lagging strand. How Pol δ and ε share the task of replicating the eukaryotic genome is a largely open question, but both polymerases are critical for the process.
The molecular details of the replication process in plants have not been clearly elucidated. It appears, however, that the basic principles that apply to other eukaryotes also apply to plants. Plants seem to have several origins and possess at least some of the proteins responsible for recognizing and unwinding the origins (Bryant, et al., 2001; Dambrauskas, et al., 2003; Li, et al., 2005). RPA is also conserved in plants, and genetic evidence suggests that it participates in replication. Interestingly, at least in some cases, other RPA types exist and seemingly participate in DNA repair and in plastid replication (Ishibashi, et al., 2006). Moreover, the sequence of a PCNA homolog has also been identified in several plant species (Hata, et al., 1992; Lopez, et al., 1997; Yamamoto, et al., 2005). Thus, in addition to DNA polymerases, ancillary proteins contributing to replication are also present in plant organisms.
For several decades, significant attention has been placed in understanding the plant enzymes that ultimately conduct replication. The isolation of DNA polymerase activities in plants precedes the availability of extensive DNA sequence information. Early biochemical work resulted in the isolation of several polypeptides with polymerase activity and properties that were similar to that of their animal counterparts (Castroviejo, et al., 1975; Tarrago-Litvak, et al., 1975; Coello and Vazquez-Ramos, 1995; Seto, et al., 1998). Among these, several polypeptides with primase activity were identified in a wide variety of organisms (Litvak, et al., 1984; Nielsen, et al., 1991; Laquel, et al., 1994; Garcia-Maya and Buck, 1998). The analysis of different sequenced plant genomes suggests that, like in other eukaryotes, this activity corresponds to a Polα/primase protein complex, and that initiation of replication shares the same components in plants as in other eukaryotes.
As already discussed, the elongation phase of replication mainly involves family B polymerases δ and ε. Both polymerases are present in plants. Genetic evidence in Arabidopsis suggests that Pol ε is essential for replication (Tzafrir, et al., 2004; Ronceret, et al., 2005). Similarly, the expression patterns of rice Pol δ are compatible with a role in the replication process (Uchiyama, et al., 2002). It thus seems that the elongation machinery is conserved in plants. However, the fine details of polymerase dynamics at the replication fork remain to be understood even in other eukaryotes, and those details might very well differ for plant organisms because of the particularities of plant physiology (like, for instance, the high exposure to UV radiation; see below).
Polymerization is not the only role of DNA polymerases. DNA replication is a complicated process that depends on a myriad of other factors that control almost every aspect of cellular metabolism. As such, it is natural that DNA polymerases are implicated in controlling some of these processes. For instance, a link is emerging between the replicative polymerases and chromatin remodeling (Bolognese, et al., 2006). In addition, there is a more clearly established interplay between replicative DNA polymerase and cell-cycle control (D’Urso, et al., 1995; Navas, et al., 1995; Navas, et al., 1996; Marini, et al., 1997; Datta, et al., 2000). It is critical for cell viability to ensure the proper timing of the replication process, because the replication process can and should only take place at a specific stage of the cell cycle. But, at the same time, cell cycle progression needs to be sensitive to the outcome of the replication process. In fact, several checkpoints exist that respond to abnormal events during replication. Perhaps the better understood is the damage checkpoint, which is triggered when the replication fork encounters DNA damage that slows progression of the replication fork (Harrison and Haber, 2006; Ishikawa, et al., 2006). It is therefore natural that the core replication proteins would be involved in checkpoint control. It is known that the C-terminal domain of Pol ε has a not fully clarified role in checkpoint control (Navas, et al., 1995; Navas, et al., 1996). Interestingly, some genetic evidence suggests that a similar role of Pol ε might be conserved as well in Arabidopsis (Jenik, et al., 2005).
IV. Translesion synthesis
In addition to Pols α, δ and ε, it seems clear that other polymerases can have access to the replication fork. This is the case of family Y DNA polymerases during TLS (Prakash, et al., 2005). As stated earlier, the discovery of family Y polymerases resulted partly from the realization that two E. coli proteins that are part of the DNA damage-induced SOS response encode two previously unidentified DNA polymerases. E. coli DinB and UmuC/UmuD’ (now known as Pol IV and Pol V, respectively) have low processivity and DNA synthesis fidelity (Kobayashi, et al., 2002; Wagner, et al., 2002), but in return they have the ability to synthesize across templates containing different DNA-lesions that block other polymerases (Shcherbakova and Fijalkowska, 2006). This ability seems to be at the core of the physiological role of family Y polymerases. A large number of DNA lesions block DNA synthesis when present in the template strand. Different repair mechanisms exist to repair those lesions (see below), but if the cell is engaged in the replication process and the amount of damage exceeds its repair capacity, the replication fork will be blocked at the site of the lesion. Under such circumstances, it appears that family Y polymerases can temporarily take the place of the replicative polymerases to polymerize across (bypass) the lesion, after which normal replication can resume. Two ubiquitin-binding domains that are conserved in family Y enzymes appear to be crucial to regulate access of these polymerases to the replication fork (Kannouche and Lehmann, 2004). However, it is not clear whether the role of these domains is dependent on their capacity to be ubiquitinated (Lehmann, 2006) or to their ability to bind monoubiquitinated proteins, such as PCNA (Hoege, et al., 2002). As an alternative model, some evidence suggests that, at least in some cases, normal replication could restart past the lesion, after which the remaining gaps could be filled by a family Y polymerase, temporally dissociating TLS from replication fork progression (Heller and Marians, 2006; Lopes, et al., 2006).
There are four family Y polymerases in vertebrates: Pol η, Pol Ι, Pol κ and Rev1 (Ohmori, et al., 2001; Shcherbakova and Fijalkowska, 2006). All these enzymes have been shown to possess some type of lesion bypass capacity in vitro that is usually specific for a particular type of lesion. For example, while Pol η performs highly efficient bypass of a T-T dimer (McCulloch, et al., 2004b), Pol Ι has a much lower efficiency when bypassing this lesion (Johnson, et al., 2000; Tissier, et al., 2000). However, Pol Ι seems to efficiently bypass certain minor groove DNA adducts (Perrino, et al., 2005; Wolfle, et al., 2005). In addition, prokaryotic family Y polymerases have clearly been implicated in spontaneous and/or damage-induced mutagenesis (Tang, et al., 2000), while some evidence indicates that the same might be true in eukaryotes (Bavoux, et al., 2005; Abdulovic and Jinks-Robertson, 2006; Dumstorf, et al., 2006), suggesting that they participate in an error-prone process. All this suggests a role of family Y polymerases in TLS. However, strong genetic evidence supporting this hypothesis only exists in the case of Pol η. In eukaryotes, Pol η appears to be responsible for bypass of cyclobutane pyrimidine dimers, and, as stated above, mutations in its gene cause Xeroderma pigmentosum (Masutani, et al., 1999). It is widely accepted that the disorder is a consequence of the defect in the ability of Pol η to bypass UV-induced DNA lesions, as cells lacking Pol η are deficient in their ability to conduct replication after UV damage (Lehman, et al., 1975). This corresponds very well with the high efficiency of bypass by Pol η of a cis-syn thymine dimer in vitro (Johnson, et al., 1999; Masutani, et al., 1999; McCulloch, et al., 2004b).
Plant cells are subject to a constant exposure to UV radiation. Yet, like mammalian cells, plant cells are sensitive to high energy UV radiation, and this sensitivity varies widely among species and among cultivars (Hidema and Kumagai, 2006). In order to cope with this exposure, plants have developed a number of mechanisms to limit the effect of UV radiation. A first mechanism of defense is in the form of surface barriers to the radiation, such as reflective structures (resins), hairs or the cell wall (Julkunen-Tiitto, et al., 2005). In addition, plants accumulate several phenolic compounds in the epidermal cell layers that are capable of absorbing UV radiation (Kootstra, 1994). Finally, plants perform DNA repair, either through NER (Kimura and Sakaguchi, 2006) or in a light-dependent pathway through photolyase enzymes that are absent in mammals but can directly remove the major UV-induced lesions (Jiang, et al., 1997). Despite these repair systems, plant cells presumably cannot keep their genomes free of UV-induced lesions and thus, as other eukaryotes, would have a requirement for TLS. In fact, plant genomes encode different family Y polymerases (see Table 2). Pol η, in particular, seems to be present in several plant organisms. Interestingly, the A. thaliana Pol η ortholog can complement the UV-sensitivity caused by a Pol η deficiency in S. cerevisiae, suggesting that this enzyme could participate in TLS in plants (Santiago, et al., 2006). Besides Pol η, other family Y polymerases seem to participate in TLS of UV lesions in plants. In A. thaliana, a defect in rev1 results in moderate UV sensitivity (Takahashi, et al., 2005) and, in addition, a Pol κ homolog has been biochemically characterized, although its role is not yet clear (Garcia-Ortiz, et al., 2004).
Table II.
DNA Polymerases in Arabidopsis thaliana
| Family | Polymerase | Homo Sapiens | Saccharomyces cerevisiae | Arabidopsis thaliana |
|---|---|---|---|---|
| A | γ | POLG | MIP1 | - |
| θ | POLQ | - | AtPolθ (Kimura, et al., 2002) | |
| ν | POLN | - | - | |
|
Pol I-like A Pol I-like B |
- - |
- - |
AtPolI-like A (Mori, et al., 2005) AtPolI-like B (Mori, et al., 2005) |
|
| B | α | POLA | POL1 (CDC17) | AtPolα (Kimura, et al., 2002) |
| δ | POLD1 | POL3 (CDC3) | AtPolδ GenBank: NM_125792 | |
| ε | POLE | POL2 | AtPolε (Kimura, et al., 2002) | |
| ζ | POLZ (REV3) | REV3 | AtREV3 (Sakamoto, et al., 2003) | |
| X | β | POLB | - | - |
| λ | POLL | POLIV (POLX) | AtPolλ (Garcia-Diaz, et al., 2000) | |
| μ | POLM | - | - | |
| TdT | TdT | - | - | |
| Y | η | POLH | RAD30 | AtRAD30 (Kunz, et al., 2005) |
| κ | POLK (DINB) | - | AtPolκ (Kunz, et al., 2005) | |
| Ι | POLI (RAD30B) | - | - | |
| Rev1 | REV1 | REV1 | AtREV1 (Takahashi, et al., 2005) |
In addition to family Y polymerases, one family B polymerase, Pol ζ, appears to be critical for TLS. Pol ζ seems to be responsible for a large amount of all spontaneous (Harfe and Jinks-Robertson, 2000) and, together with rev1 (Lawrence and Hinkle, 1996; Gibbs, et al., 2000; Lawrence and Maher, 2001), UV-induced mutagenesis in eukaryotes (Lawrence, 2004). In mamalian cells, its defect leads to chromosomal instability (Wittschieben et al., 2006). In vitro, Pol ζ is relatively accurate compared to other family Y polymerases, although its fidelity is lower than that of other family B enzymes (Johnson, et al., 2000; Zhong, et al., 2006). In addition, it is promiscuous for mismatch extension, leading to the suggestion that its role in TLS could be in extending the intermediates generated by family Y polymerases (Johnson, et al., 2000). In A. thaliana, deletion of one of the Pol ζ subunits (rev3) causes severe UV sensitivity (Sakamoto, et al., 2003), suggesting that Pol ζ also plays a role in plant TLS.
V. Polymerases in organelles
Plastids and mitochondria are self-proliferating organelles considered to be descendents of endosymbiotic prokaryotes.
A. Polymerases in Chloroplasts
In contrast to mitochondria, plastids are not present in animal cells. Among different types of plastids chloroplasts, with the capacity to perform photosynthesis, have been the most studied. Chloroplasts contain multiple copies of a ∼150kb chromosome. The number of copies of the chloroplast chromosome changes during development and in a tissue-specific manner. The highest copy numbers are observed in actively photosynthesizing leaves and the lowest in roots. Proteins responsible for the replication of the chloroplast DNA, including the polymerase(s) are encoded by the cell nucleus. DNA polymerase activity has been purified from the chloroplasts of several species of higher plants including: spinach (Sala, et al., 1980; Keim and Mosbaugh, 1991), soybean (Heinhorst, et al., 1990; Bailey, et al., 1995) and pea (McKown and Tewari, 1984; Gaikwad, et al., 2002). The size of these polymerases was reported to be 70-90 kDa, they co-purified with a 3′→5′ exonuclease activity and based on the initial biochemical characterization they were classified as pol γ-like, thus family A members. A 43 kDa DNA binding protein that interacts with and stimulates the activity of the pea chloroplast polymerase was also reported (Chen, et al., 1996).
Recent sequence analyses of the genome from rice (Oryza sativa) and A. thaliana lead to the identification and cloning of the nuclear genes encoding the chloroplast polymerases (Kimura, et al., 2002; Mori, et al., 2005). Two highly homologous polymerases, designated PolI-like A and PolI-like B have been identified in both organisms. The AtPolI-like A and AtPolI-like B open reading frames localized to Arabidopsis chromosomes 1 and 3, and encoded predicted products of 1049 amino acids (117 kDa) and 1034 amino acids (115 kDa), respectively. AtPolI-like A and AtPolI-like B share over 70% amino acid sequence identity. Though they are most closely related to Pol I from Cyanobacteria, AtPolI -like A and AtPolI-like B are 35- and 33% identical with E. coli PolI and are less closely related to mammalian pol γ (and the Arabidopsis Pol θ). Homology-based modeling of the structure of AtPolI-like A and AtPolI-like B predicted a fold of the C-terminal 3′→5′ exonuclease and polymerase domains similar to that of E. coli Klenow fragment. The PolI-like polymerases were found to localize to plastids of proliferating cells, although they where not detected in mature leaves. In addition increased level of AtPolI-like B expression was observed after treatment with H2O2. These findings suggest that the PolI-like polymerases are responsible for the replication and repair of the chloroplast DNA (Kimura and Sakaguchi, 2006).
B. Polymerases in Mitochondria
DNA polymerase γ is a member of family A. As the only polymerase in mammalian and yeast mitochondria, Pol γ is responsible for both replication and repair of the mitochondrial genome (Graziewicz, et al., 2006). Pol γ, like the family prototype, E. coli PolI, is endowed with a 3′→5′ exonuclease proofreading activity. As expected of a replicative polymerase, Pol γ has high fidelity and high processivity (Longley, et al., 2001). It forms a complex with the p55 accessory subunit, which increases the binding affinity to DNA, increases processivity and stimulates the polymerase and exonuclease activitites of the catalytic subunit. In addition, pol γ has been shown to have a dRP lyase activity, which suggests that it is involved in mitochondrial BER (Longley, et al., 1998).
Mutations in the gene for the catalytic subunit of pol γ have been associated with progressive external ophtalmoplegia (PEO) and other heritable mitochondrial diseases (Longley, et al., 2005). PEO is a rare disorder resulting in muscle dysfunction caused by loss of mitochondrial activity due to accumulation of mutations and depletion of mitochondrial DNA.
Though polymerase activity was purified from mitochondria of several higher plants (Heinhorst, et al., 1990; Daniell, et al., 1995) the gene(s) encoding the polymerase(s) is not yet unequivocally determined. The isolated polymerse activities had similar properties to those purified from plastids, and they were originally classified as pol γ-like. A property of pol γ from animal cells is high sensitivity to inhibition by ddNTP. However, this trait is not shared by the plant mitochondrial or plastid polymerases. Consistently, sequence analysis of the genome revealed that a homolog of animal POLG is not present in O. sativa and A. thaliana (Kimura and Sakaguchi, 2006). In a recent study Mori et al. (Mori, et al., 2005) show that one of the plastid-associated PolI-like polymerases, PolI-like B, also localizes to mitochondria. Thus it appears that the same polymerase may be responsible for replication and/or repair in mitochondria and plastids.
VI. DNA Polymerases in Repair
The genome is constantly exposed to endogenous and environmental factors that damage DNA. In consequence DNA bases may be modified, cross-linked or lost, backbone modifications can occur, and single or double strand breaks may be introduced. Some of these lesions may result in mutations or block the replication fork posing a threat to genome integrity, and some may impede transcription affecting protein expression and disrupting cellular processes. To remove these lesions multiple repair pathways operate in cells (Bray and West, 2005). Which repair pathway(s) is used depends on the type of lesion and the stage of the cell cycle. Most repair processes require a synthesis step to restore the integrity of the DNA strand(s). Because DNA substrates generated in different repair pathways vary, so do the requirements for the polymerase(s).
A. NER
NER recognizes and removes helix-distorting lesions like those caused by ultraviolet (UV) light radiation or bulky chemical adducts (Reardon and Sancar, 2005). Two major subpathways operate in NER, Transcription Coupled Repair and Global Genome Repair. Regardless of the subpathway, removal of damage follows the same steps: recognition of the damage, unwinding of the DNA duplex, incision on both sides of the lesion and displacement of the lesion-containing oligonucleotide followed by filling of the single strand gap and ligation. The excision tracts range in size from 12 to 13 nucleotides in E. coli and from 23 to 30 nucleotides in eukaryotes. In E. coli the gap-filling is performed by PolI. It is generally accepted that in eukaryotes the gap is filled by a B family polymerase: pol δ and/or ε. However, a recent study provided evidence that also Pol κ, a family Y member functions in NER-associated gap filling in mammalian cells (Ogi and Lehmann, 2006). This finding is surprising, given the fact that NER is considered an error free process, while Pol κ is a low fidelity enzyme implicated in bypass of bulky adducts. Nonetheless, it suggests the existence of a distinct NER subpathway and provides yet another example of a Y family polymerase with a function outside TLS.
Genetic studies have indicated that photoreactivation is not the only pathway for repair of UV-damage in plants (Nakajima, et al., 1998). Consistently, sequence analysis of the Arabidopsis and rice genomes has revealed that most of the yeast and mammalian genes whose products have been implicated in NER, including DNA polymerases δ, ε and κ are conserved in plants, suggesting a conservation of function.
B. Polymerases in BER
DNA bases modified by oxidation, alkylation or deamination, inappropriate bases (e.g. dU), as well as sites of base loss (AP sites) are repaired by base excision repair (BER), which involves several subpathways (Fromme and Verdine, 2004). Repair is initiated by removal of the damaged base by a DNA glycosylase, which cleaves the N-glycosylic bond generating an AP site. Depending on whether the AP site is generated by a mono- or bifunctional (containing also an intrinsic AP-lyase activity) DNA glycosylase, subsequent cleaveage of the sugar-phosphate backbone by an AP endonuclease will create a nick with a 3′ -OH and a 5′-dRP termini or a single nucleotide gap with a 3′ -OH and a 5′-phosphate termini, respectively. In both cases completion of repair requires DNA polymerase activity followed by ligation. Though several polymerases have been implicated in mammalian BER, the major subpathway relies on DNA pol β. Pol β incorporates a nucleotide onto the 3′ -OH terminus and removes the 5′-dRP group with its dRP-lyase activity. An alternative to “single-nucleotide” BER is the “long-patch” repair pathway. If the nature of the 5′-moiety is different (the dRP group is modified) or if the dRP group is not removed by pol β’s dRP-lyase activity, strand displacement synthesis will generate a single-stranded DNA flap (∼2-13 nucleotides) that is cleaved by FEN1 flap endonuclease. The polymerases implicated in “long-patch” BER are pol β, pol δ and pol ε. A new study by Yoshimura et al. (Yoshimura, et al., 2006) indicates the involvement of DNA pol θ, a recently identified member of family A, in BER. As demonstrated, chicken cells deficient in pol θ are hypersensitive to oxidative base damage induced by H2O2 and extracts of these cells are defective in long-patch and to a lesser extend in short-patch BER. This suggests that pol θ may have a function redundant with that of pol β in repair of oxidative base damage.
In contrast to mammalian cells, plants and yeast lack DNA polymerase β. Thus, the majority of BER in yeast appears to proceed through the long-patch pathway, with the involvement of pols ε, δ and α. The same may be true in plants. A POLQ homolog has been identified in O. sativa and A. thaliana (Kimura and Sakaguchi, 2006). Thus, like in animal cells, pol θ may contribute to BER in plants.
In addition to pol β and pol γ, two other human DNA polymerases, pol Ι and λ, have dRP lyase activity (Bebenek, et al., 2001b; Garcia-Diaz, et al., 2001), indicating that they may be involved in repair processes that require removal of the dRP group.
Pol Ι, present only in animal cells, has an unusual nucleotide incorporation specificity it incorporates dTMP opposite template A much more efficiently than it forms the three remaining Watson-Crick base pairs, while it misinserts dGMP opposite a template T at a rate that exceeds that of correct dAMP incorporation. Given this specificity it was hypothesized that polΙ could participate in BER of UTP incorporated during replication and/or that it may function in a BER reaction replacing dGs that are inadvertently removed by a DNA glycosylase from G-T or G-U mismatches (Bebenek, et al., 2001b).
Mammalian polλ is closely related to pol β, sharing the same structural organization (Garcia-Diaz, et al., 2004) and a number of enzymatic properties (Garcia-Diaz, et al., 2002). It can substitute for pol β in repair of uracil-containing DNA in a reconstituted reaction in vitro. Furthermore, pol λ can carry backup repair in Pol β -deficient cells and polλ-deficient mouse fibroblasts are hypersensitive to oxidative DNA damaging agents (Braithwaite, et al., 2005a; Braithwaite, et al., 2005b). Together these results suggest that pol λ is involved in some form of BER in mammalian cells.
Pol λ is conserved in plants, and, like Pol IV in yeast, it is the only family X enzyme. The POLL ortholog has been identified in rice and Arabidopsis (Garcia-Diaz, et al., 2000) and the recombinant rice protein has been purified (Uchiyama, et al., 2004). Like the mammalian and yeast enzymes, plant pol λ has dRP lyase activity suggesting that it may function in BER. However, so far there is no clear evidence that pol IV, the yeast homolog of pol λ, is involved in BER.
C. Polymerases in interstrand crosslink repairs
Covalent DNA interstrand crosslinks (ICLs) are highly cytotoxic lesions produced by agents such as nitrogen mustard, cisplatin, psoralens, and mitomycin C (Scharer, 2005). Because ICLs cause the loss of genetic information on both DNA strands, repair of these lesions presents a difficult task for the cells, requiring a coordinated action of multiple repair pathways. Models for repair of ICLs are based on studies in bacteria. In E. coli a well characterized repair pathway involves NER, homologous recombination (HR) and gap-filling synthesis with pol I (McHugh, et al., 2001). A distinct pathway involving NER and the activity of pol II, a family B enzyme, has also been described (Berardini, et al., 1999). So far ICL repair in eukaryotes is less well understood. Studies showing that mutant alleles of some genes confer sensitivity to cross-link-inducing agents implicated yeast pol ζ (Lawrence, 2004) and Drosophila melanogaster mus308 gene (Boyd, et al., 1990) product in repair of ICLs. Mus308 is an ortholog of the recently identified human pol θ (Seki, et al., 2003). Pol θ contains an N-terminal ATPase-helicase domain and a polymerase domain with homology to E. coli pol I in its C-terminal region. Unlike what was reported with mus308 in Drosophila (Boyd, et al., 1990), chicken DT40 cells deficient in pol θ did not show hypersensitivity to cross-linking agents, suggesting that in vertebrates pol θ does not participate in repair of ICLs (or that another polymerase has a redundant function).
Recent studies in yeast have demonstrated the requirement for pol ζ in ICL repair in G1 phase of haploid cells (Sarkar, et al., 2006). According to this model, repair is initiated by NER on one DNA strand, to generate a single strand gap and an oligonucleotide cross-linked to the other strand. Next, the gap is filled by pol ζ, which is recruited by pol δ and monoubiquitinated PCNA. This pathway is analogous to repair in E. coli involving NER and TLS synthesis by Pol II (Berardini, et al., 1999). A few studies have addressed the function of the Arabidopsis pol ζ, AtREV3 (Sakamoto, et al., 2003; Takahashi, et al., 2005), and an ortholog of animal pol θ appears to be present in plants (see Table II). However, there is as yet no information on the possible function of these polymerases in ICL repair in plants.
D. Repair of Double strand breaks
Double strand DNA breaks (DSBs) create a particularly dangerous threat to the stability of the genome leading to different chromosomal abnormalities an even cell death. They may be induced by such external factors as X-rays and ionizing radiation or they may arise as a consequence of a stalled replication fork. Two major mechanisms exist for the repair of DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). Both pathways are conserved in cells from bacteria to humans. Like mammals, higher plants show a bias towards the use of NHEJ rather than HR for repair of DSBs (Britt, 1999). Though several subpathways of NHEJ have been recognized, in general the repair involves alignment and joining of broken DNA ends with minimal (just a few complementary base pairs) homology. Depending on the NHEJ subpathway the structure of the DNA ends vary, so that in some cases the alignment - mediated duplex will have short gaps that need to be filled by a DNA polymerase. In mammalian cells, the family X polymerases λ and μ have been implicated in NHEJ (Nick McElhinny and Ramsden, 2004). Both polymerases contain an N-terminal BRCT domain, which mediates interactions with protein partners. Lee et al. (Lee, et al., 2004) demonstrated that pol λ is required for filling short gaps during XRCC4-LigaseIV-dependent joining of DSBs in extracts of HeLa cells, and that this activity of pol λ depends on its BRCT domain. Consistently, pol λ was shown to perform gap filling in a reconstituted NHEJ reaction in vitro, in the presence of end-joining factors: Ku, XRCC4-LigaseIV (Ma, et al., 2004; Nick McElhinny, et al., 2005). In addition, CHO cells over expressing a catalytically inactive form of pol λ exhibit increased sensitivity and genomic instability in response to ionizing radiation, a phenotype characteristic of cells with a NHEJ defect (Capp, et al., 2006). Like pol λ, its yeast homolog, pol IV has also been implicated in NHEJ (Tseng and Tomkinson, 2002). Preferred substrates for yeast pol IV are small gaps formed by alignment of linear duplex DNA molecules. Furthermore, pol IV interacts physically and functionally with the Dn14/Lif1 complex (homologous to the mammalian XRCC4/LigaseIV complex) and this interaction is dependent on pol IV’s BRCT domain. Different components of the plant NHEJ sub-pathways have been isolated and analyzed, including Arabidopsis AtKu80 protein, DNA ligase IV (LIG4) and AtXRCC4. Arabidopsis mutants defective in Ku80 and LIG4 are hypersensitive to ionizing radiation and arrest in development after treatment with doses of gamma rays that do not affect the wild-type plants (Friesner and Britt, 2003). Considering that the function of the main NHEJ factors appears to be conserved in plants it is likely that so is the role of pol λ. However, there is no experimental evidence so far to support this hypothesis.
Several lines of evidence also indicate pol μ’s role in mammalian NHEJ. Pol μ interacts with Ku and stably associates with DNA in the presence of Ku and XRCC4/LigaseIV. In this context, pol μ can fill in 1-2 nucleotide gaps generated during annealing of partially overlapping DNA ends (Mahajan, et al., 2002). Moreover, in the context of NHEJ, pol μ, in contrast to pol λ, has the remarkable ability to incorporate a nucleotide onto a primer terminus that lacks its complementary template counterpart, using as a template a nucleotide from the other DNA molecule (Nick McElhinny, et al., 2005). In addition, exposure of human cells to ionizing radiation results in increased levels of pol μ and the polymerase localizes in nuclear foci containing double-strand breaks, consistent with a role in NHEJ (Mahajan, et al., 2002).
DSBs resulting from replication-fork collapse during normal replication are usually repaired by homologous recombination. In this case any DNA sequence lost due to the break is restored by synthesis using the intact sister chromatid as a template and the invading broken end as primer. Studies in yeast indicated that pols δ and ε conduct synthesis associated with DSB repair (Holmes and Haber, 1999). Recent studies with human cell extracts (McIlwraith, et al., 2005) provided evidence that HR-dependent replication restart from a D-loop structure formed by strand invasion depends on synthesis by the TLS pol η. Subsequent switch to pol δ would reestablish the replication fork. These transactions likely depend on interactions with PCNA. Furthermore, Kawamoto et al. (Kawamoto, et al., 2005) demonstrated that the frequency of DSB-induced HR in chicken DT40 cells defective in pol η is reduced relative to that in wild type cells, and that this phenotype could be reversed by complementation with the human enzyme. Together, these results indicate that, in addition to a role in TLS of UV induced pyrimidine dimmers, pol η functions in repair of DSBs by HR.
VII. DNA Polymerase Fidelity
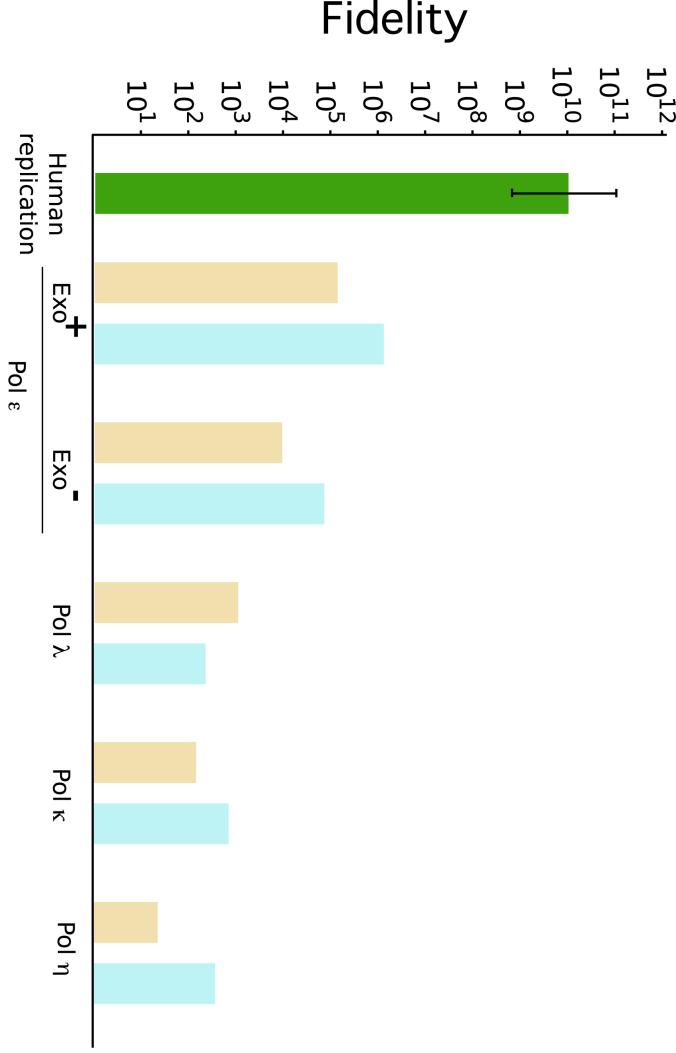
The maintenance of the integrity of the genome through multiple rounds of replication depends largely on the fidelity of DNA polymerases. DNA synthesis errors result in mutations leading to disruption of normal cellular processes, disease and aging. On the other hand, low fidelity DNA synthesis is a prerequisite for such processes as the development of the immune system, or the rapid adaptation of microbes to environmental changes. Maintaining the balance between stability and change makes special demands of DNA polymerases. Polymerases from different families and even from the same family have different properties (see Hübscher, et al., 2002; Bebenek and Kunkel, 2004; Kunkel, 2004; Prakash, et al., 2005). These differences include striking differences in the fidelity of synthesis on undamaged DNA: depending on the polymerase, the rates measured in vitro for a simple replication error such as a base substitution can differ as much as 106 to107 (Kunkel, 2004; see Figure 2). These differences in fidelity reflect the adaptation to fulfill specific functions in the cell. For example, polymerases that only fill short gaps during repair processes, such as the family X members pol β and pol λ, have only modest fidelity, with average error rates between 10-3 to 10-4. However, pol λ has a remarkably high rate for single base deletions, a property that is believed to reflect the enzyme’s ability to use imperfect (misaligned) DNA ends during NHEJ (Bebenek, et al., 2003). Family Y polymerases implicated in TLS are the least accurate (10-1-10-3). This is considered to be a consequence of their adaptation to accommodate damaged DNA in their active sites. These high error rates can be tolerated because family X and Y polymerases are not likely to conduct an extensive amount of DNA synthesis. In contrast, polymerases like pol δ, ε or γ, responsible for the bulk of synthesis during DNA replication are highly accurate, with average rates for single base errors on the order of 10-6 to 10-5. This high fidelity is partially achieved by the presence of an intrinsic 3→5′ exonuclease, or proofreading activity, which increases fidelity 10 to 100- fold (see Figure 2). Proofreading activity is intrinsic to a few polymerases, but several additional exonuclease-containing proteins exist in the cell, bringing up the possibility that a proofreading exonuclease might correct errors introduced by a different protein, a mechanism known as proofreading in trans. In fact, it has been shown that errors made by one polymerase can be corrected by another (Perrino and Loeb, 1989; Pavlov, et al., 2006), a mechanism thought to operate during TLS to reduce the error-proneness of the process (Bebenek, et al., 2001a; McCulloch, et al., 2004a). In addition, it has been suggested that the 3′-5′ exonuclease of Pol δ is essential for Okazaki fragment maturation, suggesting that proofreading replication errors might not be the only physiological role of 3′-5′ exonucleases (Jin, et al., 2005).
Figure 2. DNA synthesis fidelity.
In vitro polymerization fidelity (defined as 1/error rate) of representative DNA polymerases. The fidelity for base substitutions (light brown bars) and insertion/deletion mutation (blue bars) is shown for of exo-deficient polymerases η (Matsuda, et al., 2001), κ (Ohashi, et al., 2000a) and λ (Bebenek, et al., 2003), for exo-proficient Pol ε and a Pol ε mutant deficient in proofreading (Shcherbakova, et al., 2003). An estimate of the error rate of the replication process in human cells (Drake, et al., 1998; Loeb, 2001) is shown in green. The error bar indicates the range of measured rates.
VIII. Mismatch Repair
It is thought that even DNA polymerases endowed with an associated proofreading activity generate errors during DNA synthesis with a frequency that far exceeds the in vivo error rate of an organism (see Figure 2). This difference is explained in part by the large number of repair processes that have been described above, which are sometimes independent of replication and contribute to guarantee that the replication fork will encounter “clean” substrates. However, a major contribution to replication fidelity comes from a postreplicative repair pathway, mismatch repair (MMR; Kunkel and Erie, 2005; Iyer, et al., 2006; Jiricny, 2006). MMR eliminates replication errors that are left behind in the nascent strand by the replication fork. Repair involves excision of a region of the newly synthesized strand (∼300 nucleotides in human cells; see Figure 3) followed by accurate resynthesis. In human cells, DNA polymerase δ has been implicated in the resynthesis step. A distinct repair pathway exists for removal of mismatches such as G-U and G-T resulting from deamination of cytosine and 5-methylcythosine. In this case repair is initiated by a DNA glycosylase and the gap is filled by a BER polymerase such as pol β.
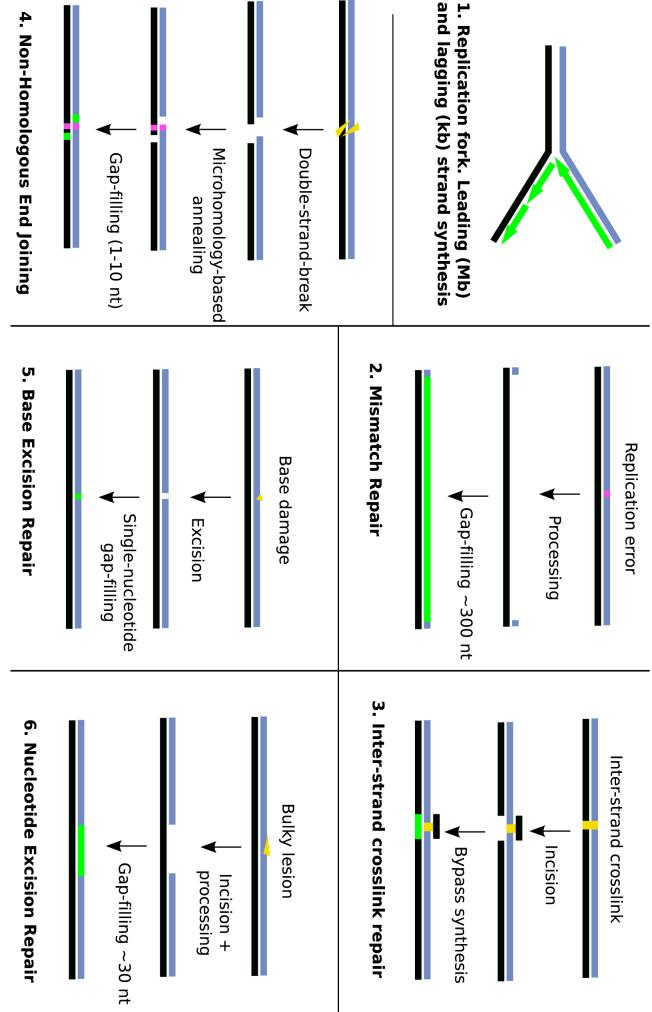
Figure 3. Different modes of synthesis during replication and repair.
DNA polymerases carry out synthesis during many different processes. The requirements for the polymerase are very different in each pathway. Replication requires an extremely long, processive synthesis in the leading strand and shorter, interrupted patches of synthesis in the lagging strand. Different repair processes require patch synthesis anywhere from one to a few hundred nucleotides, and may also require synthesis on non-canonical substrates, such as DNA containing mismatches (shown in magenta) or different types of damage (yellow). The patch of synthesis performed by the polymerase is shown in green.
The postreplicative mismatch repair pathway is conserved in plants. Studies in Arabidopsis have demonstrated that efficient MMR activity is crucial during meristematic growth for maintaining of genome integrity and consequently species stability (Hoffman, et al., 2004). Homologs of the mammalian and yeast MSH and MLH proteins are present in plants. In addition a MSH7 protein, that is unique to plants, and its ortholog Mus2, have been identified in Arabidopsis (Culligan and Hays, 2000) and Zea mays (Horwath, et al., 2002) respectively, indicating functional differences between the plant, yeast and mammalian MMR pathways.
IX. Concluding remarks
Replication and maintenance of the genome depends on a large number of complex DNA transactions that involve different protein partners and very different DNA substrates (see Figure 3). To cope with the many scenarios, these processes utilize multiple DNA polymerases with very different properties. Our growing understanding of the many roles carried out by different DNA polymerases allows us to begin to appreciate how the subtle (and sometimes not so subtle) differences in their properties are intimately linked to their physiological role. The classification of DNA polymerases into families has been often thought to imply that polymerases in a family possess similar characteristics and participate in related processes. This is true to some degree, but evidence accumulates that polymerases in the same family can behave in radically different ways and even have a role in totally different processes. A paradigmatic example is the relatively low fidelity family B Pol ζ, which unlike other members in the B family, has a role in TLS. At the same time, it is becoming clear that most DNA polymerases play more than a single role. This is the case not only for pols δ and ε but also for a number of recently identified polymerases. For example in addition to their function in TLS pol κ has been implicated in NER, Pol ζ in ICL repair, pol η in homologous recombination-dependent rescue of stalled replication forks, and pol θ in BER. Also, a dual role, in BER and in NHEJ, has been suggested for pol λ. It is worth noting that all these polymerases (as well as the complex processes in which they participate) appear to be conserved in plants.
With the exception of a few pathways that are specific to certain cell lineages, such as those related to the development of the vertebrate immune system, plant organisms appear to share the same requirements for DNA polymerase activities as mammalian cells. However, plant cells may very well have additional plant-specific requirements. Plant cells are subject to a constant UV exposure, need to ensure replication of the genome of plastids and are capable of resuming cell division after years of being in a dormant state in seeds. In addition, plant cells sometimes undergo a process of genetic and epigenetic variation called somaclonal variation (Evans, 1989). The molecular details of this mutagenic process are not well understood, but it is possible that it involves one or more DNA polymerase activities. Moreover, there exists a seemingly crucial difference related to the transmission of genetic information. Presumably, animal cells can tolerate a higher mutational load on somatic tissues because they contain a specialized cell lineage that is reserved for the transmission of the genome through generations, the germ line. Plant organisms do not contain such lineage, and instead, gametes are produced from meristematic cells after many rounds of somatic division. This suggests that the replication and/or repair processes in plant cells may have a larger requirement for fidelity. In this context, it will be interesting to analyze how the polymerases of plant cells differ from their animal counterparts, and how their roles are adapted to the specific conditions of plant physiology.
Acknowledgments
The authors would like to thank Drs. Olga Kozyreva, Dmitry Gordenin and Thomas Kunkel for critical reading of the manuscript and helpful discussions. This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Sciences.
References
- Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol zeta- and Pol eta-dependent frameshift mutagenesis. Genetics. 2006;172:1487–1498. doi: 10.1534/genetics.105.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- Autexier C, Lue NF. The Structure and Function of Telomerase Reverse Transcriptase. Annu. Rev. Biochem. 2006 doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Bailey JC, 2nd, Heinhorst S, Cannon GC. Accuracy of Deoxynucleotide Incorporation by Soybean Chloroplast DNA Polymerases Is Independent of the Presence of a 3[prime] to 5[prime] Exonuclease. Plant Physiol. 1995;107:1277–1284. doi: 10.1104/pp.107.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoux C, Hoffmann JS, Cazaux C. Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase kappa. Biochimie. 2005;87:637–646. doi: 10.1016/j.biochi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein. Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. Proofreading of DNA polymerase eta-dependent replication errors. J. Biol. Chem. 2001a;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001b;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A, Zaballos A, Salas M, Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. Embo J. 1987;6:4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Blanco L, Salas M. Relating structure to function in phi29 DNA polymerase. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- Bolognese F, Forni C, Caretti G, Frontini M, Minuzzo M, Mantovani R. The Pole3 bidirectional unit is regulated by MYC and E2Fs. Gene. 2006;366:109–116. doi: 10.1016/j.gene.2005.07.046. [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- Boyd JB, Sakaguchi K, Harris PV. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, Wilson SH. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 2005a;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005b;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE. DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005;168:511–528. doi: 10.1111/j.1469-8137.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. Embo J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt AB. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 1999;4:20–25. doi: 10.1016/s1360-1385(98)01355-7. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Moore K, Aves SJ. Origins and complexes: the initiation of DNA replication. J. Exp. Bot. 2001;52:193–202. [PubMed] [Google Scholar]
- Cann IK, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castroviejo M, Tarrago-Litvak L, Litvak S. Partial purification and characterization of two cytoplasmic DNA polymerases from ungerminated wheat. Nucleic Acids Res. 1975;2:2077–2090. doi: 10.1093/nar/2.11.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gaikwad A, Mukherjee SK, Choudhary NR, Kumar D, Tewari KK. A 43 kDa DNA binding protein from the pea chloroplast interacts with and stimulates the cognate DNA polymerase. Nucleic Acids Res. 1996;24:3953–3961. doi: 10.1093/nar/24.20.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P, Vazquez-Ramos JM. Studies on the Processivity of Maize DNA Polymerase 2, an [alpha]-Type Enzyme. Plant Physiol. 1995;109:645–650. doi: 10.1104/pp.109.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Hays JB. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell. 2000;12:991–1002. doi: 10.1105/tpc.12.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J. Cell Sci. 1995;108(Pt 9):3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- Dambrauskas G, Aves SJ, Bryant JA, Francis D, Rogers HJ. Genes encoding two essential DNA replication activation proteins, Cdc6 and Mcm3, exhibit very different patterns of expression in the tobacco BY-2 cell cycle. J. Exp. Bot. 2003;54:699–706. doi: 10.1093/jxb/erg079. [DOI] [PubMed] [Google Scholar]
- Daniell H, Zheng D, Nielsen BL. Isolation and characterization of an in vitro DNA replication system from maize mitochondria. Biochem. Biophys. Res. Commun. 1995;208:287–294. doi: 10.1006/bbrc.1995.1336. [DOI] [PubMed] [Google Scholar]
- Datta A, Schmeits JL, Amin NS, Lau PJ, Myung K, Kolodner RD. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol. Cell. 2000;6:593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez M, A Kirchhoff, T., Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. Embo J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. see comments. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase {iota} in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. U S A. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. Rna. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA. Somaclonal variation--genetic basis and breeding applications. Trends Genet. 1989;5:46–50. doi: 10.1016/0168-9525(89)90021-8. [DOI] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Base excision repair. Adv. Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- Gaikwad A, Hop DV, Mukherjee SK. A 70-kDa chloroplast DNA polymerase from pea (Pisum sativum) that shows high processivity and displays moderate fidelity. Mol. Genet. Genomics. 2002;267:45–56. doi: 10.1007/s00438-001-0631-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Gao G, Pedersen LC, London RE, Kunkel TA. Structure-function studies of DNA polymerase lambda. DNA Repair (Amst) 2005a;4:1358–1367. doi: 10.1016/j.dnarep.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat. Struct. Mol. Biol. 2005b;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J. Biol. Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- Garcia-Maya MM, Buck KW. Purification and properties of a DNA primase from Nicotiana tabacum. Planta. 1998;204:93–99. doi: 10.1007/s004250050234. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortiz MV, Ariza RR, Hoffman PD, Hays JB, Roldan-Arjona T. Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 2004;39:84–97. doi: 10.1111/j.1365-313X.2004.02112.x. [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl. Acad. Sci. U S A. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl. Acad. Sci. U S A. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Tippin B. The expanding polymerase universe. Nat. Rev. Mol. Cell. Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Prakash L, Prakash S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol. 2005;25:10183–10189. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. Surviving the Breakup: The DNA Damage Checkpoint. Annu. Rev. Genet. 2006 doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Hata S, Kouchi H, Tanaka Y, Minami E, Matsumoto T, Suzuka I, Hashimoto J. Identification of carrot cDNA clones encoding a second putative proliferating cell-nuclear antigen, DNA polymerase delta auxiliary protein. Eur. J. Biochem. 1992;203:367–371. doi: 10.1111/j.1432-1033.1992.tb16559.x. [DOI] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC, Weissbach A. Chloroplast and Mitochondrial DNA Polymerases from Cultured Soybean Cells. Plant. Physiol. 1990;92:939–945. doi: 10.1104/pp.92.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Hidema J, Kumagai T. Sensitivity of rice to ultraviolet-B radiation. Ann. Bot. (Lond) 2006;97:933–942. doi: 10.1093/aob/mcl044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hoffman PD, Leonard JM, Lindberg GE, Bollmann SR, Hays JB. Rapid accumulation of mutations during seed-to-seed propagation of mismatch-repair-defective Arabidopsis. Genes Dev. 2004;18:2676–2685. doi: 10.1101/gad.1217204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Horwath M, Kramer W, Kunze R. Structure and expression of the Zea mays mutS-homologs Mus1 and Mus2. Theor. Appl. Genet. 2002;105:423–430. doi: 10.1007/s00122-002-0955-8. [DOI] [PubMed] [Google Scholar]
- Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Kimura S, Sakaguchi K. A Higher Plant Has Three Different Types of RPA Heterotrimeric Complex. J. Biochem. (Tokyo) 2006;139:99–104. doi: 10.1093/jb/mvj014. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Ishii H, Saito T. DNA damage-dependent cell cycle checkpoints and genomic stability. DNA Cell. Biol. 2006;25:406–411. doi: 10.1089/dna.2006.25.406. [DOI] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Jurkuta RE, Barton MK. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell. 2005;17:3362–3377. doi: 10.1105/tpc.105.036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CZ, Yee J, Mitchell DL, Britt AB. Photorepair mutants of Arabidopsis. Proc. Natl. Acad. Sci. U S A. 1997;94:7441–7445. doi: 10.1073/pnas.94.14.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Garg P, Stith CM, Al-Refai H, Sterling JF, Murray LJ, Kunkel TA, Resnick MA, Burgers PM, Gordenin DA. The multiple biological roles of the 3′-->5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. 2005;25:461–471. doi: 10.1128/MCB.25.1.461-471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell. Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Julkunen-Tiitto R, Haggman H, Aphalo PJ, Lavola A, Tegelberg R, Veteli T. Growth and defense in deciduous trees and shrubs under UV-B. Environ. Pollut. 2005;137:404–414. doi: 10.1016/j.envpol.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. Embo J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Keim CA, Mosbaugh DW. Identification and characterization of a 3′ to 5′ exonuclease associated with spinach chloroplast DNA polymerase. Biochemistry. 1991;30:11109–11118. doi: 10.1021/bi00110a013. [DOI] [PubMed] [Google Scholar]
- Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- Kimura S, Sakaguchi K. DNA repair in plants. Chem. Rev. 2006;106:753–766. doi: 10.1021/cr040482n. [DOI] [PubMed] [Google Scholar]
- Kimura S, et al. A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.) Nucleic Acids Res. 2002;30:1585–1592. doi: 10.1093/nar/30.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Valentine MR, Pham P, O’Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- Kootstra A. Protection from UV-B-induced DNA damage by flavonoids. Plant Mol. Biol. 1994;26:771–774. doi: 10.1007/BF00013762. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker T. DNA replication. Freeman; New York: 1992. [Google Scholar]
- Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Anderson HJ, Osmond MJ, Vonarx EJ. Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ. Mol. Mutagen. 2005;45:115–127. doi: 10.1002/em.20094. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Laquel P, Litvak S, Castroviejo M. Wheat DNA Primase (RNA Primer Synthesis in Vitro, Structural Studies by Photochemical Cross-Linking, and Modulation of Primase Activity by DNA Polymerases) Plant Physiol. 1994;105:69–79. doi: 10.1104/pp.105.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Hinkle DC. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:41–46. doi: 10.1098/rstb.2000.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- Lehman AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl. Acad. Sci. U S A. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J. Biol. Chem. 1958;233:163–170. [PubMed] [Google Scholar]
- Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell. Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Li KG, Yang JS, Attia K, Su W, He GM, Qian XY. Cloning and characterization of OsORC2, a new member of rice origin recognition complex. Biotechnol. Lett. 2005;27:1355–1359. doi: 10.1007/s10529-005-0937-4. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Litvak S, Graveline J, Zourgui L, Carvallo P, Solari A, Aoyama H, Castroviejo M, Tarrago-Litvak L. Studies on the initiation of DNA synthesis in plant and animal cells. Adv. Exp. Med. Biol. 1984;179:249–262. doi: 10.1007/978-1-4684-8730-5_26. [DOI] [PubMed] [Google Scholar]
- Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Lopez I, Khan S, Vazquez J, Hussey PJ. The proliferating cell nuclear antigen (PCNA) gene family in Zea mays is composed of two members that have similar expression programmes. Biochim. Biophys. Acta. 1997;1353:1–6. doi: 10.1016/s0167-4781(97)00072-9. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern DF, Foiani M. A role for DNA primase in coupling DNA replication to DNA damage response. Embo J. 1997;16:639–650. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, J OW. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006 doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Yuasa M, Araki M, Nogimori T, Yokoi M, Eki T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant: from a human genetic disorder to a novel DNA polymerase. Cold Spring Harb. Symp. Quant. Biol. 2000;65:71–80. doi: 10.1101/sqb.2000.65.71. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004a;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004b;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- McKown RL, Tewari KK. Purification and properties of a pea chloroplast DNA polymerase. Proc. Natl. Acad. Sci. U S A. 1984;81:2354–2358. doi: 10.1073/pnas.81.8.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Kimura S, Saotome A, Kasai N, Sakaguchi N, Uchiyama Y, Ishibashi T, Yamamoto T, Chiku H, Sakaguchi K. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005;334:43–50. doi: 10.1016/j.bbrc.2005.06.052. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nakajima S, Sugiyama M, Iwai S, Hitomi K, Otoshi E, Kim ST, Jiang CZ, Todo T, Britt AB, Yamamoto K. Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998;26:638–644. doi: 10.1093/nar/26.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas TA, Sanchez Y, Elledge SJ. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- Nielsen BL, Rajasekhar VK, Tewari KK. Pea chloroplast DNA primase: characterization and role in initiation of replication. Plant Mol. Biol. 1991;16:1019–1034. doi: 10.1007/BF00016074. [DOI] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell. Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000a;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000b;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Harvey S, Blans P, Gelhaus S, Lacourse WR, Fishbein JC. Polymerization past the N2-isopropylguanine and the N6-isopropyladenine DNA lesions with the translesion synthesis DNA polymerases eta and iota and the replicative DNA polymerase alpha. Chem. Res. Toxicol. 2005;18:1451–1461. doi: 10.1021/tx050114u. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Proofreading by the epsilon subunit of Escherichia coli DNA polymerase III increases the fidelity of calf thymus DNA polymerase alpha. Proc. Natl. Acad. Sci. U S A. 1989;86:3085–3088. doi: 10.1073/pnas.86.9.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher RS, Wilson TE, Doherty AJ. New insights into NHEJ repair processes in prokaryotes. Cell Cycle. 2005;4:675–678. doi: 10.4161/cc.4.5.1676. [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Ramadan K, Shevelev I, Hubscher U. The DNA-polymerase-X family: controllers of DNA quality? Nat. Rev. Mol. Cell. Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic. Acid. Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Lazaro JM, Blanco L, Kamtekar S, Berman AJ, Wang J, Steitz TA, Salas M, de Vega M. A specific subdomain in phi29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. U S A. 2005;102:6407–6412. doi: 10.1073/pnas.0500597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A, Guilleminot J, Lincker F, Gadea-Vacas J, Delorme V, Bechtold N, Pelletier G, Delseny M, Chaboute ME, Devic M. Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 2005;44:223–236. doi: 10.1111/j.1365-313X.2005.02521.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Lan VT, Hase Y, Shikazono N, Matsunaga T, Tanaka A. Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell. 2003;15:2042–2057. doi: 10.1105/tpc.012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala F, Amileni AR, Parisi B, Spadari S. A gamma-like DNA polymerase in spinach chloroplasts. Eur. J. Biochem. 1980;112:211–217. doi: 10.1111/j.1432-1033.1980.tb07196.x. [DOI] [PubMed] [Google Scholar]
- Santiago MJ, Alejandre-Duran E, Ruiz-Rubio M. Analysis of UV-induced mutation spectra in Escherichia coli by DNA polymerase eta from Arabidopsis thaliana. Mutat. Res. 2006;601:51–60. doi: 10.1016/j.mrfmmm.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Prasad P, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase ß complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase. Q Embo J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto H, Hatanaka M, Kimura S, Oshige M, Tsuya Y, Mizushina Y, Sawado T, Aoyagi N, Matsumoto T, Hashimoto J, Sakaguchi K. Purification and characterization of a 100 kDa DNA polymerase from cauliflower inflorescence. Biochem. J. 1998;332(Pt 2):557–563. doi: 10.1042/bj3320557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 2006;11:2496–2517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- Steitz TA. DNA polymerases: Structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Sakamoto A, Sato S, Kato T, Tabata S, Tanaka A. Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol. 2005;138:870–881. doi: 10.1104/pp.105.060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrago-Litvak L, Castroviejo M, Litvak S. Studies on a DNA polymerse gamma-like enzyme from wheat embryos. FEBS Lett. 1975;59:125–130. doi: 10.1016/0014-5793(75)80356-5. [DOI] [PubMed] [Google Scholar]
- Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota. Embo J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Hatanaka M, Kimura S, Ishibashi T, Ueda T, Sakakibara Y, Matsumoto T, Furukawa T, Hashimoto J, Sakaguchi K. Characterization of DNA polymerase delta from a higher plant, rice (Oryza sativa L.) Gene. 2002;295:19–26. doi: 10.1016/s0378-1119(02)00822-3. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Kimura S, Yamamoto T, Ishibashi T, Sakaguchi K. Plant DNA polymerase lambda, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 2004;271:2799–2807. doi: 10.1111/j.1432-1033.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- Wagner J, Etienne H, Janel-Bintz R, Fuchs RP. Genetics of mutagenesis in E. coli: various combinations of translesion polymerases (Pol II, IV and V) deal with lesion/sequence context diversity. DNA Repair (Amst) 2002;1:159–167. doi: 10.1016/s1568-7864(01)00012-x. [DOI] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- Wang Z, Castano IB, De Las Penas A, Adams C, Christman MF. Pol kappa: A DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA polymerase beta and mammalian base excision repair. Cold. Spring Harb. Symp. Quant. Biol. 2000;65:143–155. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Human DNA polymerase iota promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA. Mol. Cell. Biol. 2005;25:8748–8754. doi: 10.1128/MCB.25.19.8748-8754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Mori Y, Ishibashi T, Uchiyama Y, Ueda T, Ando T, Hashimoto J, Kimura S, Sakaguchi K. Interaction between proliferating cell nuclear antigen (PCNA) and a DnaJ induced by DNA damage. J. Plant Res. 2005;118:91–97. doi: 10.1007/s10265-005-0197-3. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, et al. Vertebrate POLQ and POLbeta Cooperate in Base Excision Repair of Oxidative DNA Damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM, McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]