Abstract
The mechanisms by which a promiscuous protein can strongly interact with several different proteins using the same binding interface are not completely understood. An example is protein kinase A (PKA), which uses a single face on its docking/dimerization domain to interact with multiple A-kinase anchoring proteins (AKAP) that localize it to different parts of the cell. In the current study, the configurational entropy contributions to the binding between the AKAP protein HT31 with the D/D domain of RII α-regulatory subunit of PKA were examined. The results show that the majority of configurational entropy loss for the interaction was due to decreased fluctuations within rotamer states of the side chains. The result is in contrast to the widely held approximation that the decrease in the number of rotamer states available to the side chains forms the major component. Further analysis showed that there was a direct linear relationship between total configurational entropy and the number of favorable, alternative contacts available within hydrophobic environments. The hydrophobic binding pocket of the D/D domain provides alternative contact points for the side chains of AKAP peptides that allow them to adopt different binding conformations. The increase in binding conformations provides an increase in binding entropy and hence binding affinity. We infer that a general strategy for a promiscuous protein is to provide alternative contact points at its interface to increase binding affinity while the plasticity required for binding to multiple partners is retained. Implications are discussed for understanding and treating diseases in which promiscuous protein interactions are used.
Keywords: energy well, molecular recognition, peptide–protein binding, quasiharmonic approximation, vibrational entropy
Protein–protein interactions control a wide array of cellular functions such as intracellular signaling, transcription, and replication (1). Some proteins are promiscuous in that they can interact with several other proteins, examples of which are prevalent in signaling pathways (2). One example is the regulatory subunit of protein kinase A (PKA), which can interact with multiple partners, collectively referred to as A-kinase-anchoring proteins (AKAPs) that localize PKA to different parts of the cell (3). How one protein can interact with different partners using the same binding interface is not completely understood (4).
Knowledge of the recognition mechanisms of promiscuous protein–protein association is particularly important because the interactions are implicated in many diseases, e.g., Alzhiemer's, heart disease, and some types of cancer (5–7). Accurate descriptions of the interaction mechanisms are currently needed to improve the design of drugs that interfere with aberrant interactions and to improve methods to predict protein interactions to elucidate disease pathways (8, 9). Promiscuous protein interactions use common mechanisms in some diseases, for example, the so-called conformational diseases that owe their etiology to protein aggregation (5). An interesting prospect is that there may be common underlying mechanisms.
At a fundamental level, protein–protein interactions are driven by favorable changes in free energy: an increase in entropy and/or decrease in enthalpy (10). A current challenge is to fully detail the components to the free energy of protein–protein association, and, in particular, the contribution of configurational entropy requires a better description and estimation (11–13). The three-dimensional structure of the protein–protein complex provides an opportunity to resolve the different energetic components, e.g., the bond lengths, the bond angles, and van der Waals interactions. The binding energy can be estimated by taking the difference in the totals of all of the components calculated for the proteins separately compared with the complex (14). To identify the different bound and unbound energetic states of the proteins, molecular dynamics (MD) simulations can be used. The ensemble of structures observed by MD simulations provides a view of protein dynamics and a means to calculate the average energy of interaction. The sampling of the ensemble provides a means to calculate the probability distributions of the energetic states, and these probability distributions can be used to estimate the configurational entropy. An increase in the number of structural/energetic states upon association corresponds to an increase in the configurational entropy and thus a favorable contribution for the free energy of binding.
In the present study, the change in configurational entropy associated with the interaction between the D/D domain of the RIIα subunit of PKA and the binding peptide from the Ht31 AKAP protein was examined. The configurational entropy was decomposed into conformational and vibrational contributions, Sconfig = Sconf + Svib (15–17). According to previous studies, the loss of conformational entropy upon association was estimated to be the more dominant component and was the only term considered (18, 19). The results of the current study, in which both the vibrational and conformational terms were measured, indicate that the approximation is not necessarily valid because the vibrational term was found to be the more dominant factor. Similar results have been reported by recent studies that suggested a narrowing of the energy landscape or the limitation of the flexibility of side-chain conformations contributes significantly to decrease in entropy upon association (17, 20).
The PKA/AKAP interaction interface is primarily composed of hydrophobic interactions (3, 21). To characterize the interaction, we first show that configurational entropy changes in a predictable manner within hydrophobic environments. A direct relationship was found between the number of alternative hydrophobic contacts available to an atom and the entropy associated with that atom. The relationship was found to hold throughout the PKA/AKAP complex. At the interaction interface, the relationship indicated a mechanism to promote an increase in entropy upon protein–protein association: When there is a gain in the number of available alternative contacts upon complex formation, there will be gain in entropy and, hence, an increase of the affinity of the interaction. The mechanism can be used to partially explain how PKA can interact with high affinity with multiple protein-binding partners or AKAPs. Although each AKAP protein has a different set of hydrophobic contacts for the interaction, each of these sets still uses a large number of available alternative hydrophobic contact points provided on the large hydrophobic interaction surface of PKA. Sampling of these alternative contacts increases the entropy of binding and the affinity of the interactions for each of the different AKAP proteins. The mechanism can operate in the context of diseases where promiscuous interactions are implicated.
Results and Discussion
Ht31pep–D/D as a Model System and Overall Dynamics.
The energies, dynamics, and configurational entropy associated with the binding reaction of the D/D domain of the RII α-regulatory subunit of PKA with a binding peptide from the Ht31 AKAP (Ht31pep) were examined. The system provides an example of a promiscuous protein association because the D/D domain is known to bind multiple AKAP partners using the same binding interface. A general characteristic of the interaction is that the D/D domain binds to an amphipathic helix on the AKAP protein, i.e., an α-helix in which one side is predominantly hydrophobic, whereas the other side is polar or charged (22). The interactions typically have high affinity, and the Ht31pep–D/D interaction yields a binding affinity of approximately −11 kcal/mol (Kd = 2–16 nM) (23, 24)
The Ht31 peptide forms a hydrophobic ridge that complements the surface presented by the D/D domain (25). Calculations from the simulations show that the binding of Ht31pep to the RIIα D/D domain is largely driven by van der Waals interactions with a contribution of −41 kcal/mol. Coulombic interactions were favorable at approximately −94 kcal/mol, but most of the Coulombic attraction was cancelled by a electrostatic desolvation penalty of ≈75 kcal/mol [see detailed calculated average energy terms in supporting information (SI) Table S1]. The simulations also showed that some surface areas of both the D/D and Ht31pep become solvent-inaccessible upon binding. The averaged solvent-accessible surface areas (SASA) of Ht31pep, the D/D domain, and the complex are 2,350, 6,116, and 7,000 Å2, respectively; and the average loss of SASA upon binding is ≈1,466 Å2.
Although different AKAP peptides bind to same groove on the RIIα D/D domain (21), a comparison of a crystal structure of DAKAP2–D/D and NMR structures of Ht31pep–D/D revealed a change in helical register between the two AKAPs (Fig. S1). We have analyzed the internal motions of free Ht31pep and the D/D–Ht31pep complex by molecular dynamics simulations and found that the structures obtained from >4-ns MD simulations show a better alignment with the crystal structure, compared with the initial structure solved by NMR (see Figs. S1 and S2). The simulations demonstrate that the C-terminal tail of Ht31pep and N terminus of the D/D domain are highly flexible.
Conformational Changes in D/D–Ht31 Binding.
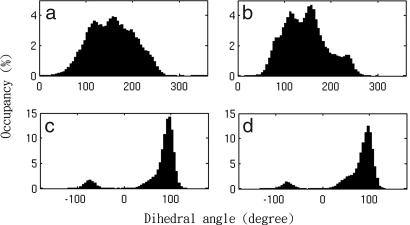
Distinct protein conformations have been observed during the MD simulations. As detailed in Methods, each energy well and protein conformation is defined based on the sampled dihedrals. Our analysis showed that all peptide ω bonds stay in a single energy well, and the mean value of the standard deviation of the ω dihedral angles is only ≈8°. No cis–trans isomerizations were observed. Similarly, most backbone ψ and φ angles have one energy well, and their dihedral distributions show a shape of a one-dimension Gaussian function except a few dihedrals near the N and C termini and in the loop between helices I and II in the D/D. Fig. 1 shows that dihedrals Asp-1ψ of Ht31pep and Pro-7ψ of the protomer II display an irregular peak shape and multiwells in the histograms. The rotameric states and dihedral distributions of backbones usually remain similar in the free proteins and the complex, as illustrated in Fig. 1.
Fig. 1.
Occupancy (%) of backbone dihedral angles. (a) Asp-1ψ of free Ht31pep. (b) Asp-1ψ of bound Ht31pep. (c) Pro-7ψ of the free D/D protomer II. (d) Pro-7ψ of the bound D/D protomer II.
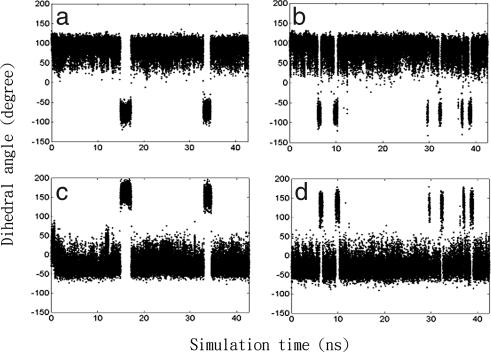
There were a few concerted transitions involving ψ,φ dihedral pairs to flip an amide group, although other backbone rotations near terminal tails do not show strong correlation. These pairs are all near pairs of adjacent prolines: Pro-6,7 and Pro-25,26 from each protomer, which are also in close proximity to each other. In these transitions, the anticorrelated dihedral pairs rotate in opposite direction by ≈180°(Fig. 2) (26), so that the amide group flips over without significantly altering the D/D domain conformations (see Fig. S3). The driving torques can be affected by binding and thus change local kinetics. For example, the amide group between Pro-7ψ and Gly-8φ flips more frequently in the bound protomer II, as illustrated in Fig. 2. In contrast, the amide group near Pro-25 of the bound protomer I does not flip during our simulation, although it flips in its free state. The dynamics changes of these loops show that the D/D domain is asymmetric in the D/D–Ht31pep complex, which may aid in AKAP docking (24).
Fig. 2.
Flipping of an amide group. (a) Pro-7ψ of the free D/D protomer II. (b) Pro-7ψ of the bound D/D protomer II. (c) Gly-8 φ of the free D/D protomer II. (d) Gly-8 φ of the bound D/D protomer II.
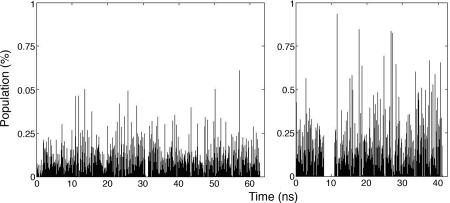
The combination of different rotameric states of side-chain dihedrals results in a large number of distinct conformations. As suggested previously, the backbone dihedral is less rotatable compared with side-chain dihedrals, so we focused on the side-chain rotations. Fig. 3 shows conformations of Ht31pep found during three MD simulations. Only the conformations of Ht31pep are displayed, because there are too many different conformations of the D/D domain (data not shown). Each peak indicates a distinct conformation and the time that it was first sampled during the simulations. The higher peak means the more populated conformation, which presumably has lower energy and is an important conformation. Even after a total 64 or 42.5 ns, new conformations of the free and bound Ht31pep were observed. This suggested that sampling of the accessible phase space for side-chains of Ht31pep was not fully converged. In addition, the total number of distinct conformations of bound Ht31pep is not largely reduced. We sampled 2,116 free-Ht31pep conformations versus 1,337 bound-Ht31pep conformations, resulting in a conformational entropy loss of only −0.33 kcal/mol. The small difference may due to incomplete sampling, but it also suggests that Ht31pep in the complex can still maintain flexibility to some degree.
Fig. 3.
Distinct side-chain conformations found during the MD simulations. (Left) Free Ht31pep. (Right) Bound Ht31pep. x axis, sample saved during the simulations at time x; y axis, population for each distinct conformation (%). The x axis combines three independent simulations. These samples were taken every 1 ps based on the time evolution during the MD simulation. If a sample has 0 population (y value = 0) at sample x, then it means that the snapshot saved at a certain time is a repeat.
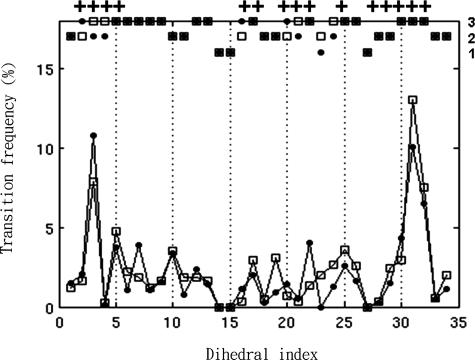
The rotameric states and the dynamics of the side-chain dihedrals were examined. Most side-chain dihedrals of linear residues had more than one rotamer state, but residues with aromatic rings usually had only one rotameric state in our simulations (Fig. 4). A cluster of Leu, Thr, and Val residues from each protomer of the D/D domain creates the preformed surface where the Ht31pep helix docks. The hydrophobic groove of the domain and the Ht31pep do not have any Trp, Phe, or Met residues, which are commonly found in protein–protein-binding sites (27). Binding of Ht31pep to the D/D domain does not notably decrease rotamers per dihedral, even within the hydrophobic binding groove. The absence of larger aromatic rings may efficiently prevent such clashes in this system.
Fig. 4.
Number of rotameric states and their transition frequency (%) observed during the simulations. Open squares, side-chain dihedrals of free Ht31pep; filled circles, side-chain dihedrals of bound Ht31pep; x axis, dihedral index; left y axis, transition frequency; right y axis, number of rotameric states. Dihedrals of residues that interact with the D/D domain are indicated by a cross aligned at the top of the plot. Note that the hydrophobic environment may allow a side-chain dihedral to have more rotameric states in its bound state (see dihedrals 2, 16, and 20) or to have more frequent transitions between different states, e.g., dihedral 3.
A few Ht31pep side-chain dihedrals exhibit new rotamer states when they dock into the D/D domain, e.g., Leu-2χ1, Ile-10χ1, and Val-14χ1 (see Fig. 4 and Fig. S4). To test whether these states were present only in the complex and not in the free peptide, the new Ht31pep conformation from its bound state was used to initiate another MD simulation in explicit water. After 1 ns, those dihedrals all rotated to rotamers seen in the free state, which suggests that the rotameric states are stable only in the D/D–Ht31pep complex. We infer that the hydrophobic environment provided by the D/D domain allowed a side-chain dihedral to sample energy wells that were not favorable in aqueous solvent. For instance, Val-14χ1 can rotate more outwards in the hydrophobic groove provided by the D/D domain, where no waters were found in our simulations, which is consistent with another AKAP-D/D crystal structure, DAKAP–D/D (28). The proposed mechanism for the increase in rotameric states will be further described.
Changes in Configurational Entropy upon Binding.
Because bond stretching and angle bending contribute little to the entropy change of D/D–Ht31pep complex formation (see Fig. S5), we focused on the dihedral degrees of freedom. Entropies were calculated first by using the Gibbs formula and the QH approximation for dihedrals shown in Figs. 1–3. The computed Gibbs entropies (TSG) for Asp-1ψ dihedral of free Ht31pep and the complex were 0.72 and 0.68 kcal/mol, respectively. The QH approximation (TSQH) also yielded similar results, 0.74 and 0.72 kcal/mol for the free and bound peptide. For example, the entropy of Pro-7ψ dihedral was computed, and the difference between the free D/D and the bound state was found to be small too. Although Pro-7ψ sampled between rotamers more frequently in the bound state, both states result in the same probability distribution (Figs. 1 and 2), which yielded the same entropy. The distribution was not close to Gaussian, but had two peaks. As a result, for Pro-7ψ of the bound D/D domain, the QH approximation merges the two peaks into a single Gaussian, thus TSQH yields higher entropy, 0.7 kcal/mol, compared with TSG, 0.3 kcal/mol.
Similarly, the entropy computed by both methods, TSG, TSQH, is nearly zero for Val-14χ1 of free Ht31pep. The entropy increases in the bound state because the dihedral samples one more energy well, yielding 0.16 and 0.84 kcal/mol for TSG and TSQH, respectively. The errors generated from the QH approximation cannot always be cancelled, and TΔSQH is overestimated by ≈0.5 kcal/mol here, compared with the more accurate TΔSG. Although the Gibbs formula (Eq. 4) can be directly applied to a multidimensional probability distribution and solved numerically, adequate convergence is not possible in its present form (29). Thus, the approximation to the total TSG becomes the sum of the entropy computed from each dihedral angle, and the coupling among dihedrals is neglected if one considers more than one dihedral. As a result, TSQH is reported because it considers coupling between the given dihedral angles. The diagonal elements of the covariance matrix (TSQH_diag) were examined. The difference between TSQH and TSQH_diag indicates how significant the coupling is; if TSQH equals TSQH_diag, then there is no coupling among different dihedrals. Table ST2 presents the configurational entropy change upon binding. Although we observed entropy increases for some dihedrals, the entire system loses 16.3 kcal/mol of Gibbs entropy upon binding Ht31pep to the D/D domain. Because correlated motions between Ht31pep and the D/D domain are neglected, TΔSG presumably yields a smaller value. Not surprisingly, then, TΔSQH yields a larger entropy loss of 26.1 kcal/mol. However, neglecting off-diagonal terms causes the computed entropy loss to fall by only 0.4 kcal/mol to 25.7 kcal/mol, suggesting that most correlations may come from intramolecular interactions and may be partially cancelled when computing the entropy change.
It has been approximated that binding entropy loss upon protein–protein interaction and protein folding results chiefly from a drop in the number of side-chain conformations (18, 19). The present results indicate that this approximation can be incorrect because only ≈3 kcal/mol of the side-chain and backbone entropy losses are found to result from a drop in the number of conformations sampled (Eq. 3). We found that the width of energy wells or the vibrational range of rotatable bonds can be slightly narrower in the complex compared with that in their free states. Although the change of vibrational range of one dihedral is small, the total effect can be significant, because there are many rotatable bonds in the protein system. The findings indicated that the loss of vibrational entropy cannot be neglected. Note that the loss of external translational and rotational entropy is not computed here, and it is likely to cost from a few to ≈15 kcal/mol in aqueous solution at standard concentration.
Both the backbone ψ and φ angles and side-chain dihedrals decrease flexibility when forming the complex. This shows that considering either backbone or side-chain dynamics alone can lead to a different description of the dynamics behavior of a protein system. Nevertheless, we observe entropy increases in a few regions, in particular, the hydrophobic side-chains of Ht31pep, backbone ω, ψ and φ angles of the residues near N terminus that form the hydrophobic binding groove (Ile-5, Pro-6, Pro-7, and Gly-8 of both protomers and Leu-9 of protomer I), and C-terminal backbone dihedrals of the D/D domain. The simulations are in agreement with NMR studies, which found increases in the D/D backbone flexibility, especially near the hydrophobic binding groove (24).
Structural Mechanisms for the Observed Changes in Configurational Entropy.
Studies of the backbone and/or side-chain dynamics of molecular complexes have shown a decreased flexibility upon complex formation (30–32), and the increase in flexibility upon binding at and near the hydrophobic binding interface contrasts with these studies. It has been demonstrated, however, that the hydrophobic interactions can be highly dynamic, for example those inside the protein hydrophobic core (33–35). Furthermore, there can be correlated changes in dynamics in the backbone or distal regions in the backbone because of hydrophobic interactions (24). To account for the increased flexibility, it has been proposed that there is a highly ordered water cluster in the hydrophobic cavity that restricts protein motions, and the release of these structured waters upon ligand binding can increase protein flexibility (24, 36). We propose an additional mechanism for how the increased flexibility can be achieved.
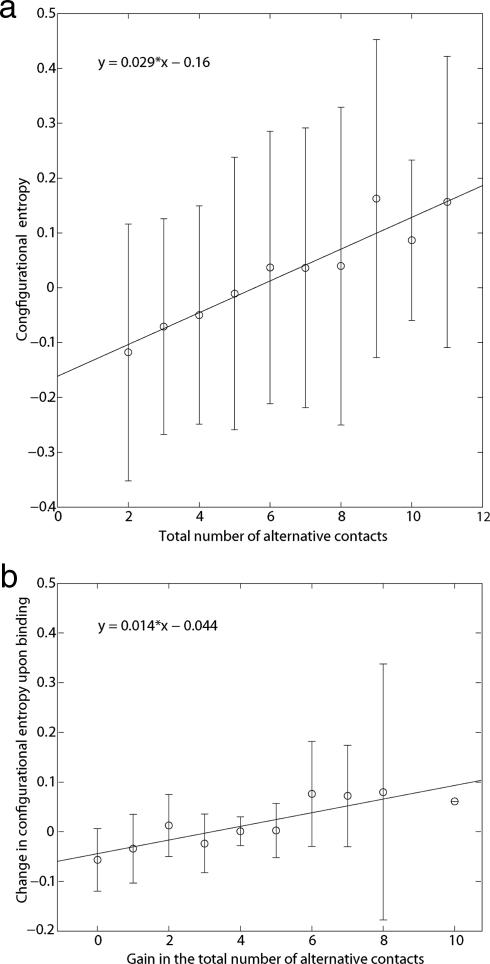
In the free Ht31 peptide, the hydrophobic side chains tend to form intramolecular contacts with other hydrophobic side chains nearby, and that limits their mobility. Upon complex formation, the hydrophobic surface of the D/D domain provides the hydrophobic side chains of Ht31pep various ways to satisfy their hydrophobic contact. Sampling of these different contacts increases the number of bound configurations. Note that bulky hydrophobic residues may be less preferable in such a system, because they are usually more rigid and may require more specific interactions. As an initial validation of the mechanism, a general relationship between configurational entropy and the number of mutually exclusive alternative hydrophobic contacts or simply alternative contacts was found. A plot of the configurational entropy associated with atoms in the complex versus the number of alternative hydrophobic contacts that can be made with those atoms is shown in Fig. 5a. The linear fit of the plot had a correlation coefficient of 0.95 and negligible P value, which indicated that there is dependence of configurational entropy on the number of available alternative contacts within hydrophobic environments. Environments that provide a larger number of available alternative contacts have higher associated entropy.** When the protein–protein interaction interface was examined, a similar relationship was found. Fig. 5b shows a plot of the gain of the number of alternative contacts for the atoms within the HT31 peptide upon complex formation versus the configurational entropy associated with those atoms. The relationship indicates that the number of bound configurations can be increased when the number of available alternative contacts is increased.
Fig. 5.
Relationship between alternative contacts and configurational entropy. (a) Plot of the total number of mutually exclusive alternative contacts for atoms that participate in hydrophobic contacts versus their associated configurational entropy. The correlation coefficient for the linear fit of the averages was 0.95, with a P value of 0. The correlation was a property of the average entropy at each alternative contact. The result indicates that the configurational entropy of a hydrophobic atom is, on average, directly proportional to the number of alternative hydrophobic contacts available to that atom. (b) Plot of the total number of alternative hydrophobic contacts of hydrophobic atoms in the Ht31 peptide that were gained upon complex formation versus the average configurational entropy change associated with these atoms upon binding. The correlation coefficient for the linear fit of the averages was 0.88, with a P value of 0.0008. The result demonstrates that when there is a gain in the number of available alternative contacts available to an atom upon complex formation, there will be higher configurational entropy associated with that atom.
The relationship between the number of alternative contacts and configuration entropy provides a means to rationalize the thermodynamic consequences of the increase in the number of rotameric states for some of the hydrophobic residues of the peptide because these residues interact with the hydrophobic groove on the domain. For example, as described above, Val-14 and Ile-10 have an increase in their number of rotameric states upon complex formation. This is due to an increase in the number of available alternative contacts that the atoms of these residues can participate as illustrated in Fig. S6 for one of the atoms in the Val-14 residue. The general interpretation is that flexibility induced by binding can be due to a hydrophobic environment that provides a fluid-like interface and makes new rotameric states accessible that are unstable in the aqueous environment. Given the nature of the interaction interface of the PKA/AKAP system, i.e., composed in part by branched chain hydrocarbons of side chains such as valine and isoleucine, it can be expected from studies of simple organic compounds that there will be higher entropy of association when a higher number of structures are exhibited by the interactions (37). For the specific interaction interface of PKA/Ht31pep studied here, the larger number of alternative contacts for the side chains corresponds to a larger number of structures or configurations, and higher configurational entropy was observed when more alternative contacts were available. In addition, the concept of minimal frustration in an energy landscape can be invoked to further understand the structural mechanism of the entropy change upon binding (38). The hydrophic interface of PKA/Ht31pep provides a favorable environment for a set of structures, each of which is minimally frustrated and thus energetically stable.
Implications for Biology and Pharmacology.
As was found in the crystal structure of the complex D/D domain of the RII α-subunit and another AKAP peptide (DAKAP) (28), during the molecular dynamics simulation, the hydrophobic residues within the flexible N terminus of the D/D domain were recruited for interaction with the HT31 peptide. These residues add to the group of hydrophobic residues that form the hydrophobic binding pocket that interfaces with AKAP peptides. The pocket provides a physicochemical environment where energetic barriers among energy wells are lower, and dihedral transitions between energy wells are more frequent. In most cases, the population distributions, and thus solute entropy and free energy, were altered. However, in specific cases, the environment of the free and bound states can provide different viscosity of the system and, therefore, influence the rate of a dihedral transition but not the population distribution. In these cases, the thermodynamic properties such as entropy or free energy do not change. One example, which is just outside of the binding pocket, shows increased backbone transitions of an amide flip that was controlled by Pro-7ψ and Gly-8φ angles. The more frequent amide flip in the bound form did not alter side-chain conformations but results in small, but significant, increase in backbone flexibility near Pro-6 and Pro-7. Based on its structural context, the changes may affect the kinetic control of the recruitment of the hydrophobic N-terminal residues to the binding pocket. These and related changes of the rates of backbone dihedral transitions in and around the binding pocket can affect the kinetic behavior of protein–peptide binding.
We observed that the hydrophobic binding pocket of the RII subunit allows for increased flexibility of hydrophobic side chains of the Ht31 peptide and a decrease in the configurational entropy cost of binding, as was found experimentally (24). An additional structural mechanism was described in which that stabilization can be achieved through the sampling of alternative contacts, and the mechanism aids in understanding how the RII subunit can achieve high affinity with each one of its diverse set of partners. The RII subunit can accommodate various AKAP proteins that each have a different set of hydrophobic side chains used for the interaction (24, 28, 39). Although different in primary sequence, a property needed for a high-affinity interaction, namely a set of hydrophobic residues that can use the alternative contacts on the PKA-binding surface, is present within each of the different interaction partners.
The use of a hydrophobic interaction surface may be a general strategy for the thermodynamic stabilization of promiscuous protein–protein interactions (24), and there are a many diseases that are associated with such interactions (5–7). The use of alternative contacts to stabilize the interactions suggests that the design of drugs to treat diseases caused by the aberrant interactions will be a challenge. The drug may be able to adapt the different conformations that are necessary to participate in the alternative contacts available on the binding surface of the promiscuous target.
Methods
Theory.
The standard chemical potential of a molecule in solution can be written as (40)
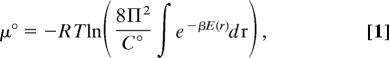 |
where R is the gas constant, T is the absolute temperature, and C° is the standard concentration (typically 1 M). The energy term, E(r) is the sum of the potential energy (U) and solvation free energy (W) of a molecular system as a function of its conformation, where the conformation is specified by given coordinates r. The configurational entropy S°can be obtained by −TS° = μ° − (〈U〉 + 〈W〉), where the angle brackets indicate averaged potential energy and solvation free energy. By introducing the solute probability distribution function p(r), the entropy can be rewritten as (10, 29)
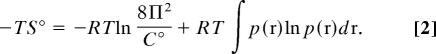 |
S° is defined here as the configurational entropy of the solute, and the solvent entropy is already included in the solvation free energy W(r). The configurational entropy itself consists of a conformational part Sconf, which reflects the number of occupied energy wells, and a vibrational part Svib, which reflects the width of the occupied wells (15, 17). To estimate the conformational entropy for the special case of M equally stable (low) energy wells, Sconf equals simply R lnM. When the energy wells are not equally stable, the probability of occupying energy well j, pj, is first obtained, and Sconf is approximated by
Similarly, the configurational entropy, Sconfig = Sconf + Svib, may be written as the Gibbs entropy formula (10). For example, if only one degree of freedom x is considered where the probability distribution is denoted as p(x), the entropy can be expressed as
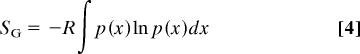 |
The probability distribution may be approximated by using a Gaussian distribution function, which is
where μ is the mean value of x, and σ is the variance or the covariance matrix if x is replaced by a multidimensional set x. Thus, Eq. 3 can be approximated as (41):
where n is the number of degrees of freedom in a given system, and ρ is the determinant of the covariance matrix σ. Eq. 5 is usually called the quasiharmonic (QH) approximation.
Computational Details.
Simulations were performed by using the program NAMD with the CHARMM22 force field (42, 43). Explicit water molecules were included, and standard simulation procedures were followed (SI Text). A snapshot was recorded in Cartesian coordinates every 1 ps during the simulations. The output snapshots were converted into bond–angle–torsion (BAT) coordinates, which use a bond length (bi), bond angle (θi), and torsional angle (φi) to define an atom position, as illustrated in Fig. S7 (44). Based on the BAT coordinates, we selected only side-chain dihedral anglesχ, backbone ψ,φ angles, and peptide bond ω angles for detailed analysis. If more than one atom shared the same dihedral rotation (e.g., atoms 4 and 5 in Fig. S7), we chose one of the torsions as our selected dihedral, because these atoms rotate as a group. The dihedral angles for rotating hydrogen atoms, e.g., dihedrals of methyl (CH3) and hydroxyl groups, were not considered.
To identify different conformations of the protein, the energy wells (rotameric states) and their ranges of a dihedral of interest are defined based on snapshots from the MD simulations (see Fig. S7 for details). For example, a molecule can have two dihedrals, A and B. Dihedral A can have two energy wells Ia and IIa, and dihedral B can have three energy wells Ib, IIb, and IIIb. If dihedrals A and B of a snapshot have Ia and IIb wells, and another snapshot also has Ia and IIb wells for both dihedrals, we cluster them as the same conformation. Otherwise, they are different conformations. A dihedral is labeled as “located near energy barriers” if it does not locate within a defined energy well, and this snapshot is not considered as an energy well but as a transition state. The probability distribution p(x) in Eq. 4 was generated by constructing a histogram from the actual trajectory data. For the current study, 72 bins were used for each dihedral distribution to construct a histogram, and SG was then computed numerically. Note that we found that one may choose 36–100 bins, but the use of 72 bins produced the most stable numerical results.
Contacts were found for each atom that, by definition, moves when a dihedral angle is changed, e.g., atom 4 in Fig. S9. The contacts for each moving atom (MA) were identified by using the Contacts of Structural Units (CSU) program (45) from snapshots that were taken during the last 10 ns of a 20-ns trajectory. Hydrophobic contacts for each MA were extracted from the program's output. Mutually-exclusive alternative hydrophobic contacts, or simply alternative contacts, were then identified for each MA in the following manner. All of the contact pairs in which the MA participates were found. For each contact pair, all of the other contact pairs that cooccurred with that pair within one or more snapshots of the trajectory were identified. The set of alternative contacts was then the contact pairs that did not cooccur with the others throughout the trajectory analyzed. Fig. S9 shows a schematic of the method to find alternative contacts.
Supplementary Material
Acknowledgments.
We thank the San Diego Supercomputer Center and National Center for Supercomputing Applications for computational resources and Drs. Susan Taylor, Choel Kim, and Tushar Jain for helpful discussions. W.A.M. was supported by National Institutes of Health (NIH) Training Grant 5 T32DK07233. Additional support was provided by the National Science Foundation, NIH, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, the Center for Theoretical Biological Physics, and Accelrys, Inc.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800452105/DCSupplemental.
We noted a complication at high numbers of alternative contacts, e.g. between 12 and the maximum value of 16, where additional alternative contacts can be gained through large scale movements within the complex. The entropy changes associated with these movements were not completely reflected by the local entropy measurements, and the corresponding linear relationship was not apparent at very high values of alternative contacts.
References
- 1.Alberts B. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 2.Buck E, Iyengar R. Organization and functions of interacting domains for signaling by protein–protein interactions. Sci STKE. 2003;2003:re14. doi: 10.1126/stke.2092003re14. [DOI] [PubMed] [Google Scholar]
- 3.Colledge M, Scott JD. AKAPs: From structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 4.Kim WK, Ison JC. Survey of the geometric association of domain–domain interfaces. Proteins. 2005;61:1075–1088. doi: 10.1002/prot.20693. [DOI] [PubMed] [Google Scholar]
- 5.Chow MK, Lomas DA, Bottomley SP. Promiscuous beta-strand interactions and the conformational diseases. Curr Med Chem. 2004;11:491–499. doi: 10.2174/0929867043455936. [DOI] [PubMed] [Google Scholar]
- 6.Kammerer S, et al. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: A disease susceptibility polymorphism. Proc Natl Acad Sci USA. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxevanis CN, Sotiropoulou PA, Sotiriadou NN, Papamichail M. Immunobiology of HER-2/neu oncoprotein and its potential application in cancer immunotherapy. Cancer Immunol Immunother. 2004;53:166–175. doi: 10.1007/s00262-003-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns-Hamuro LL, et al. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc Natl Acad Sci USA. 2003;100:4072–4077. doi: 10.1073/pnas.2628038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kann MG. Protein interactions and disease: Computational approaches to uncover the etiology of diseases. Brief Bioinform. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- 10.Dill KA, Bromberg S. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. New York: Garland Science; 2003. [Google Scholar]
- 11.Steinberg IZ, Scheraga HA. Entropy changes accompanying association reactions of proteins. J Biol Chem. 1963;238:172–181. [PubMed] [Google Scholar]
- 12.Meirovitch H. Recent developments in methodologies for calculating the entropy and free energy of biological systems by computer simulation. Curr Opin Struct Biol. 2007;17:181–186. doi: 10.1016/j.sbi.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Baron R, van Gunsteren WF, Hünenberger PH. Estimating the configurational entropy from molecular dynamics simulations: Anharmonicity and correlation corrections to the quasi-harmonic approximation. Trends Phys Chem. 2006;11:87–122. [Google Scholar]
- 14.Kollman PA, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 15.Karplus M, Ichiye T, Pettitt BM. Configurational entropy of native proteins. Biophys J. 1987;52:1083–1085. doi: 10.1016/S0006-3495(87)83303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doig AJ, Sternberg MJ. Side-chain conformational entropy in protein folding. Protein Sci. 1995;4:2247–2251. doi: 10.1002/pro.5560041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CE, Chen W, Gilson MK. Ligand configurational entropy and protein binding. Proc Natl Acad Sci USA. 2007;104:1534–1539. doi: 10.1073/pnas.0610494104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson RM, Sternberg MJ. A continuum model for protein–protein interactions: Application to the docking problem. J Mol Biol. 1995;250:258–275. doi: 10.1006/jmbi.1995.0375. [DOI] [PubMed] [Google Scholar]
- 19.Camacho CJ, Gatchell DW, Kimura SR, Vajda S. Scoring docked conformations generated by rigid-body protein–protein docking. Proteins. 2000;40:525–537. doi: 10.1002/1097-0134(20000815)40:3<525::aid-prot190>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 20.Thorpe IF, Brooks CL. Molecular evolution of affinity and flexibility in the immune system. Proc Natl Acad Sci USA. 2007;104:8821–8826. doi: 10.1073/pnas.0610064104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banky P, et al. Related protein–protein interaction modules present drastically different surface topographies despite a conserved helical platform. J Mol Biol. 2003;330:1117–1129. doi: 10.1016/s0022-2836(03)00552-7. [DOI] [PubMed] [Google Scholar]
- 22.Carr DW, et al. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 23.Alto NM, et al. Bioinformatic design of A-kinase anchoring protein in silico: A potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Natl Acad Sci USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fayos R, et al. Induction of flexibility through protein–protein interactions. J Biol Chem. 2003;278:18581–18587. doi: 10.1074/jbc.M300866200. [DOI] [PubMed] [Google Scholar]
- 25.Newlon MG, et al. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mccammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 27.Ma BY, Elkayam T, Wolfson H, Nussinov R. Protein–protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinderman FS, et al. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Killian BJ, Kravitz JY, Gilson MK. Extraction of configurational entropy from molecular simulations via an expansion approximation. J Chem Phys. 2007;127 doi: 10.1063/1.2746329. 024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnan VV, Sukumar M, Gierasch LM, Cosman M. Dynamics of cellular retinoic acid binding protein I on multiple time scales with implications for ligand binding. Biochemistry. 2000;39:9119–9129. doi: 10.1021/bi000296l. [DOI] [PubMed] [Google Scholar]
- 31.Dyson HJ, Wright PE. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 2001;339:258–270. doi: 10.1016/s0076-6879(01)39317-5. [DOI] [PubMed] [Google Scholar]
- 32.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: Role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 33.Wong KB, Daggett V. Barstar has a highly dynamic hydrophobic core: Evidence from molecular dynamics simulations and nuclear magnetic resonance relaxation data. Biochemistry. 1998;37:11182–11192. doi: 10.1021/bi980552i. [DOI] [PubMed] [Google Scholar]
- 34.Cole C, Warwicker J. Side-chain conformational entropy at protein–protein interfaces. Protein Sci. 2002;11:2860–2870. doi: 10.1110/ps.0222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 36.Zidek L, Novotny MV, Stone MJ. Increased protein backbone conformational entropy upon hydrophobic ligand binding. Nat Struct Biol. 1999;6:1118–1121. doi: 10.1038/70057. [DOI] [PubMed] [Google Scholar]
- 37.Hildebrand JH. Liquid structure and entropy of vaporization. J Chem Phys. 1939;7:233–235. [Google Scholar]
- 38.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Hundsrucker C, et al. High-affinity AKAP7delta-protein kinase A interaction yields novel protein kinase A-anchoring disruptor peptides. Biochem J. 2006;396:297–306. doi: 10.1042/BJ20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: A critical review. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karplus M, Kushick JN. Method for estimating the configurational entropy of macromolecules. Macromolecules. 1981;14:325–332. [Google Scholar]
- 42.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitzer KS. Energy Levels and thermodynamic functions for molecules with internal rotation. 2. Unsymmetrical tops attached to a rigid frame. J Chem Phys. 1946;14:239–243. [Google Scholar]
- 45.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.