Abstract
The generality of operational species definitions is limited by problematic definitions of between-species divergence. A recent phylogenetic species concept based on a simple objective measure of statistically significant genetic differentiation uses between-species application of statistical parsimony networks that are typically used for population genetic analysis within species. Here we review recent phylogeographic studies and reanalyse several mtDNA barcoding studies using this method. We found that (i) alignments of DNA sequences typically fall apart into a separate subnetwork for each Linnean species (but with a higher rate of true positives for mtDNA data) and (ii) DNA sequences from single species typically stick together in a single haplotype network. Departures from these patterns are usually consistent with hybridization or cryptic species diversity.
Keywords: phylogenetic species, phylogeography, Astraptes, Cypraeidae
1. Introduction
The interruption of gene flow by speciation events is widely expected to produce large observed genetic discontinuities that can be used as operational definitions of species or evolutionarily significant units (ESUs; Mallet 1995; Sites & Marshall 2003, 2004; Vogler & Monaghan 2007). For example, the Barcodes of Life Initiative depends on the measurement of ‘significant’ genetic discontinuities between species for the documentation of biodiversity (Hebert et al. 2003; Moritz & Cicero 2004; Meyer & Paulay 2005). A persistent question is how large such discontinuities are expected to be (Blaxter et al. 2005). The answer has often been based on subjective and taxon-specific experience or patterns. Several recent studies (Wiens & Penkrot 2002; Morando et al. 2003; Cardoso & Vogler 2005; Hart et al. 2006; Monaghan et al. 2006; Pons et al. 2006) have shown in specific contexts that there can be a simple correspondence between the identity of traditional species or ESUs and an objective standard of genetic differentiation: the 95% connection limit in statistical parsimony networks (Posada & Crandall 2001; Templeton 2001). Here we show in a literature survey and a reanalysis of two barcoding studies that this operational species definition appears to be general and quantitatively reliable across a broad range of taxa and genetic markers.
2. Material and methods
We identified from the Science Citation Index Expanded (up to 4 October 2006) a total of 727 publications that cited Clement et al. (2000) and used the statistical parsimony method as implemented in the software application TCS (see electronic supplementary material). We reviewed the results of 517 of these studies (table S1) that were available through our home institution library, included an unambiguous number of parsimony networks and taxa, and used the 95% probability of parsimony. For each network analysis (n=663), we noted the number of taxa and 95% subnetworks. In cases where the parsimony network results led the authors to reinterpret species and ESU designations, we used this revised number of taxa. We also reanalysed data from recent studies of butterflies (Hebert et al. 2004) and marine snails (Cypraeidae or cowries; Meyer & Paulay 2005) related to the mtDNA barcoding approach to documenting biodiversity. We produced 95% parsimony networks from these mtDNA sequence alignments in TCS v. 1.21. We noted the number of subnetworks and taxa, and the 95% connection limit (L95). We used receiver-operating characteristic (ROC) analysis (Metz 2006) to assess the diagnostic accuracy of TCS networks to differentiate species (see definitions in the electronic supplementary material).
3. Results
(a) Review
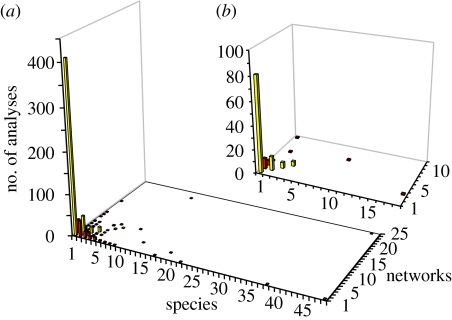
Most of the 663 published network analyses (78%) found the same number of subnetworks as taxa and correctly identified all taxa (figure 1a). The mean (±s.e.) rate of false positives among all analyses was 0.006±0.002 (few errors were made in joining all the members of a sampled species into one subnetwork).
Figure 1.
Frequency distribution of results from (a) all published statistical parsimony network analyses (n=663) or (b) animal mitochondrial COI data only (n=120) as a function of the number of taxa in the analysis and the number of 95% parsimony subnetworks in the result. Yellow bars show the frequency of studies that reported the same number of subnetworks as taxa; red bars show false positives (more subnetworks than taxa) or false negatives (more taxa than subnetworks).
The rate of true positives was only measurable for studies in which more than one species was used in a network analysis. Among 192 such analyses (34%), the mean rate of true positives was 0.521±0.034 (only about half of expected species boundaries were identified).
In 36 analyses, authors attributed apparent discordance between networks and taxa (6 false positives and 80 false negatives) to hybridization (e.g. Kronforst et al. 2006). Other sources of false positives could be inadequate sampling (e.g. figure S1), historical vicariance with rapid lineage sorting or cryptic speciation with undocumented phenotypic divergence (Knowlton 1993).
Failures to detect true positives (i.e. incidents of false negatives) may be caused by limited genetic divergence and incomplete lineage sorting between sister taxa (e.g. figure S2), or by selective constraints on lineage divergence, such as studies that used conserved, expressed nuclear genes.
Among 24 analyses of mtDNA COI sequences (the standard genetic marker in animal barcoding studies; Hebert et al. 2003; figure 1b) in two or more species, the mean false-positive rate was 0.008±0.006. The mean true-positive rate was considerably higher (0.916±0.046) than the rate for all studies (0.521).
(b) Barcoding: butterflies
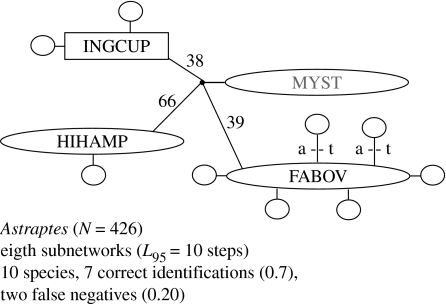
An early and influential barcoding study (Hebert et al. 2004) suggested at least 10 cryptic species in the Astraptes fulgerator complex to which the authors gave provisional acronym identifiers based on larval morphology and food plant use. Statistical parsimony analysis of the mtDNA sequence alignment produced eight subnetworks. Seven of these subnetworks correctly identified a cryptic species: BYTTNER, CELT, LOHAMP, LONCHO, SENNOV, TRIGO and YESSEN (true-positive rate of 0.818). There were no false positives. The eighth subnetwork (figure 2) included three cryptic taxa (FABOV, INGCUP and HIHAMP) that might represent two false negatives. However, these three taxa formed a clade of weakly differentiated lineages separated by small genetic distances with low bootstrap support (figure 2; Hebert et al. 2004). One individual from a host plant characteristically used by INGCUP caterpillars and two from a FABOV host shared the same haplotype (labelled MYST; figure 2). The FABOV, INGCUP and HIHAMP sequences remained in a single subnetwork even at an extremely high probability of parsimony (L99=4 steps). False negatives are generally rare among these butterfly mtDNAs (Hajibabaei et al. 2006). Altogether, the statistical parsimony method correctly identified all of the well-corroborated cryptic species in the A. fulgerator complex, and failed to differentiate the FABOV, INGCUP and HIHAMP phenotypes, probably because they are either conspecific or very recently speciated.
Figure 2.
False negatives in an Astraptes butterfly 95% parsimony subnetwork. Open symbols are sampled haplotypes (square=root haplotype); small filled symbols are unsampled intermediate haplotypes; lines indicate single sequence differences (mutations) joining haplotypes; ‘a–t’ indicates two transversions among FABOV haplotypes. Three numbers beside internal branches indicate bootstrap support for the corresponding clades in a parsimony phylogram. Adapted from Hebert et al. (2004).
(c) Barcoding: cowries
Cypraeid ESUs are well characterized morphologically, ecologically and genetically (Meyer & Paulay 2005). Statistical parsimony analysis of 262 cowrie ESUs produced 227 subnetworks, of which 192 corresponded to single ESUs. The mean true-positive rate (0.857±0.005) among 19 analyses of single genera (table 1) was high and similar to the mean for all COI studies that we reviewed (0.916, see above). The mean false-positive rate (0.005±0.003) was low as in our analyses above.
Table 1.
Sensitivity analyses of parsimony network results for two cowrie genera in which errors can (Palmadusta) or cannot (Cribrarula) be reduced to zero by choosing a probability of parsimony different from the default value of 0.95 (bold); and summed across 15 well-sampled genera (Bistolida, Cribrarula, Cypraeovula, Erosaria, Erronea, Leporicypraea, Luria, Lyncina, Mauritia, Monetaria, Notadusta, Ovatipsa, Palmadusta, Pustularia and Zoila) that included one or more false-negative ESUs or false-positive subnetworks. (No errors were found in four additional genera (Cypraea, Notocypraea, Talostolida and Purpuradusta). Adapted from Meyer & Paulay (2005).)
| group | probability of parsimony | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.90 | 0.91 | 0.92 | 0.93 | 0.94 | 0.95 | 0.96 | 0.97 | 0.98 | 0.99 | |
| Palmadusta (16 ESUs) | ||||||||||
| false negatives | 5 | 5 | 5 | 4 | 3 | 2 | 1 | 0 | 0 | 0 |
| false positives | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| total errors | 5 | 5 | 5 | 4 | 3 | 2 | 1 | 0 | 0 | 8 |
| Cribrarula (18 ESUs) | ||||||||||
| false negatives | 9 | 7 | 7 | 6 | 6 | 5 | 5 | 5 | 3 | 3 |
| false positives | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| total errors | 9 | 7 | 7 | 6 | 6 | 5 | 5 | 5 | 3 | 6 |
| 15 genera (178 ESUs) | ||||||||||
| false negatives | 55 | 50 | 50 | 46 | 42 | 35 | 31 | 29 | 25 | 20 |
| false positives | 2 | 2 | 2 | 2 | 3 | 3 | 4 | 6 | 14 | 54 |
| total errors | 57 | 52 | 52 | 48 | 45 | 38 | 35 | 35 | 39 | 74 |
In some cowrie genera (table 1), the total number of errors could be reduced to zero by adjusting the probability of parsimony to 0.97–0.98. Across 15 well-sampled genera in which false positives or false negatives occurred, the fewest errors were found at a probability of parsimony of 0.96–0.97, slightly better (35 errors total) than at 0.95 (38 errors; table 1). These results suggest that network identifications of cowrie ESUs are conservative and weakly sensitive to the probability of parsimony (except at very high probabilities).
4. Discussion
Two recent steps towards a statistically objective phylogenetic species concept have used bifurcating trees in a maximum likelihood or Bayesian framework. Matz & Nielsen (2005) and Nielsen & Matz (2006) developed a likelihood ratio test for inclusion of a test sequence in a single species sample. Pons et al. (2006) identified the boundary between two species as a shift from short branches within taxa (caused by coalescence processes in populations) to long branches between taxa (caused by speciation and extinction events), analogous to the difference between within- and among-species lineage sorting evident in statistical parsimony networks.
Recent analyses of specific taxa (cited above) show that the 95% parsimony connection limit can provide an additional and simple quantitative standard for phylogenetic species (Monaghan et al. 2006). Our review and reanalyses suggest that this standard has a low rate of false-positive errors across a broad range of taxa, speciation problems and genetic markers, and that it may be generally useful for assigning unknown specimens to known, well-sampled taxa (DNA barcoding as defined by Vogler & Monaghan 2007). This approach might be particularly useful for barcoding studies in which morphological or ecological species markers are labile (such as caterpillar host plant use by INGCUP and FABOV forms of A. fulgerator; Hebert et al. 2004). The parsimony connection limit appears to have a higher true-positive rate for successful identification of known species boundaries, and by extension for discovering new cryptic species from sequence data (DNA taxonomy; Vogler & Monaghan 2007), when applied to non-recombining loci with rapid lineage sorting (mtDNA; Moore 1995). In contrast, frequent recombination between nuclear alleles (and the associated intraspecific homoplasy) may limit the rate at which ancestral polymorphisms shared between recently diverged species are lost from one (or both) of them by lineage sorting and thus reduce the rate at which haplotype differences between sister species approach the parsimony connection limit. Within these constraints, our observations and those of other recent studies suggest that the 95% parsimony connection limit might provide a useful general tool in species assignment (conventional DNA barcoding), species discovery (more controversial DNA taxonomy) and other applications in evolutionary ecology and conservation.
Acknowledgments
Thanks to the members of FAB*lab (the organic evolution group at Simon Fraser University), Alfred Vogler and an anonymous reviewer for constructive criticism. Suzanne Edmands, Mike Hickerson, Haris Lessios, Peter Marko, Misha Matz, Chris Meyer, Rasmus Nielsen, David Posada, Kate Sanders and Eric Sotka provided data or discussed analyses with us. This work was supported by the Natural Sciences and Engineering Research Council of Canada and by Simon Fraser University.
Supplementary Material
Methodological notes on statistical parsimony network analyses
Background on statistical parsimony methods, details of network analyses of several barcoding studies, definitions of true positive and false positive rates using ROC methods and examples of false positive and false negative errors.
References
- Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. Defining operational taxonomic units using DNA barcode data. Phil. Trans. R. Soc. B. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. doi:10.1098/rstb.2005.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso A, Vogler A.P. DNA taxonomy phylogeny and Pleistocene diversification of the Cicindela hybrida species group (Coleoptera: Cicindelidae) Mol. Ecol. 2005;14:3531–3546. doi: 10.1111/j.1365-294X.2005.02679.x. doi:10.1111/j.1365-294X.2005.02679.x [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen D.H, Burns J.M, Hallwachs W, Hebert P.D.N. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA. 2006;103:968–971. doi: 10.1073/pnas.0510466103. doi:10.1073/pnas.0510466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M.W, Keever C.K, Dartnall A.J, Byrne M. Morphological and genetic variation indicate cryptic species within Lamarck's little sea star, Parvulastra (=Patiriella) exigua. Biol. Bull. 2006;210:158–167. doi: 10.2307/4134604. [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. doi:10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;101:14 812–14 817. doi: 10.1073/pnas.0406166101. doi:10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton N. Sibling species in the sea. Annu. Rev. Ecol. Syst. 1993;24:189–216. doi:10.1146/annurev.es.24.110193.001201 [Google Scholar]
- Kronforst M.R, Young L.G, Blume L.M, Gilbert L.E. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Mallet J. A species definition for the modern synthesis. Trends Ecol. Evol. 1995;10:294–298. doi: 10.1016/0169-5347(95)90031-4. doi:10.1016/0169-5347(95)90031-4 [DOI] [PubMed] [Google Scholar]
- Matz M.V, Nielsen R. A likelihood ratio test for species membership based on DNA sequence data. Phil. Trans. R. Soc. B. 2005;360:1969–1974. doi: 10.1098/rstb.2005.1728. doi:10.1098/rstb.2005.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C.E. Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems. J. Am. Coll. Radiol. 2006;3:413–422. doi: 10.1016/j.jacr.2006.02.021. doi:10.1016/j.jacr.2006.02.021 [DOI] [PubMed] [Google Scholar]
- Meyer C.P, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan M.T, Balke M, Pons J, Vogler A.P. Beyond barcodes: complex DNA taxonomy of a south Pacific island radiation. Proc. R. Soc. B. 2006;273:887–893. doi: 10.1098/rspb.2005.3391. doi:10.1098/rspb.2005.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W.S. Inferring phylogenies from mtDNA variation: mitochondrial-gene versus nuclear-gene trees. Evolution. 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. doi:10.2307/2410325 [DOI] [PubMed] [Google Scholar]
- Morando M, Avila L.J, Sites J.W. Sampling strategies for delimiting species: genes, individuals, and populations in the Liolaemus elongates–kriegi complex (Squamata: Liolaemidae) in the Andean–Patagonian South America. Syst. Biol. 2003;52:159–185. doi: 10.1080/10635150390192717. doi:10.1080/10635150390192717 [DOI] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PLoS Biol. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. doi:10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Matz M. Statistical approaches for DNA barcoding. Syst. Biol. 2006;55:162–169. doi: 10.1080/10635150500431239. doi:10.1080/10635150500431239 [DOI] [PubMed] [Google Scholar]
- Pons J, Barraclough T.G, Gomez-Zurita J, Cardoso A, Duran D.P, Hazell S, Kamooun S, Sumlin W.D, Vogler A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. doi:10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. doi:10.1016/S0169-5347(00)02026-7 [DOI] [PubMed] [Google Scholar]
- Sites J.W, Marshall J.C. Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 2003;18:462–470. doi:10.1016/S0169-5347(03)00184-8 [Google Scholar]
- Sites J.W, Marshall J.C. Operational criteria for delimiting species. Annu. Rev. Ecol. Syst. 2004;35:199–227. doi:10.1146/annurev.ecolsys.35.112202.130128 [Google Scholar]
- Templeton A.R. Using phylogeographic analyses of gene trees to test species status and processes. Mol. Ecol. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. doi:10.1046/j.1365-294x.2001.01199.x [DOI] [PubMed] [Google Scholar]
- Vogler A.P, Monaghan M.T. Recent advances in DNA taxonomy. J. Zool. Syst. Evol. Res. 2007;45:1–10. doi:10.1111/j.1439-0469.2006.00384.x [Google Scholar]
- Wiens J.J, Penkrot T.A. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus) Syst. Biol. 2002;51:69–91. doi: 10.1080/106351502753475880. doi:10.1080/106351502753475880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methodological notes on statistical parsimony network analyses
Background on statistical parsimony methods, details of network analyses of several barcoding studies, definitions of true positive and false positive rates using ROC methods and examples of false positive and false negative errors.