Abstract
Under sperm competition, paternity is apportioned by polyandrous females according to the order of matings and the genetic quality of the inseminating males. In order to distinguish between these two effects, we sequentially paired 12 female smooth newts (Lissotriton vulgaris) with each of two males and, where possible, repeated the same procedure in reverse order of the identical males after assumed sperm depletion. For a total of 578 offspring, amplified fragment length polymorphisms genetic markers revealed multiple paternities in all matings, without significant first- or second-male sperm precedence. The paternity share of individual males was transitive across the two trials with male order switch, and successful males had a significantly higher genetic dissimilarity to the female than expected by chance. We argue that patterns of paternity in natural newt populations are determined through a combination of good genes and relatedness.
Keywords: amplified fragment length polymorphisms, genetic compatibility, paternity, sperm competition, Lissotriton vulgaris
1. Introduction
Female mate choice is a key concept in evolutionary biology and has vast consequences for male and female reproductive success. Females can either favour males that signal high quality through condition-dependent traits (good genes hypothesis) or select partners whose genotypes fit best with their own genetic make-up (genetic compatibility hypothesis, Zeh & Zeh 1997). The degree of genetic compatibility depends on specific female–male combinations, and thus represents a relative rather than absolute criterion for mate quality; the genetic basis of compatibility can however be quantified in a rather straightforward way, for example, through measures of genotypic dissimilarity (Mays & Hill 2004).
The aquatic courtship display of European newts (formerly genus Triturus, see Steinfartz et al. 2007) is characterized by a combination of visual and olfactory cues; sperm is transferred via spermatophores, and fertilization is internal. Studies on the mating system of the smooth newt (Lissotriton vulgaris) were among the first to comprehensively reveal the mechanisms of sexual selection and female choice in an amphibian (for a summary see Halliday (1998)). However, as the genetic mating system of L. vulgaris remained unstudied, it is yet unclear to what extent mating strategies are governed by sperm mixing, and/or genetic compatibility mechanisms (as demonstrated for the closely related Mesotriton alpestris, Rafinski & Osikowski (2002) and Garner & Schmidt (2003)).
The reproductive fitness of males with differing genetic backgrounds is often compared by measuring their paternity share after sequential insemination of a single female. However, in such a situation, the males under study compete under differential sperm competition regimes, and a more absolute measure of male quality can only be achieved with methods that disrupt the natural courtship sequence such as artificial fertilization (Birkhead et al. 2004). In this study, we conducted classical sperm competition experiments by sequentially mating one female L. vulgaris with two males. In order to distinguish genetic effects from mating-order effects on the paternity share, we then repeated this experiment for each female after sperm depletion, switching the order of the same two males. Using genetic markers, we document the patterns of sperm precedence in L. vulgaris and demonstrate that the more genetically dissimilar male has a higher paternity share, regardless of the order of access to the female.
2. Material and methods
(a) Mating experiments
To ensure that the study individuals were unmated, they were captured in March 2003 at a breeding pond east of Vienna (Austria) before entering the water. Mating trials were conducted in April–May 2003 and 2004, but no individuals were used successively in both years. For the mating experiments, a single female was placed in an aquarium (50×30×30 cm) and an arbitrarily chosen male was added. After successful spermatophore transfer, the male was removed and a second male was placed into the tank for a second insemination on the following day (completing ‘trial 1’). Thereafter, females (n=12) were housed singly in individual aquaria furnished with anchored plastic strips in which their eggs were wrapped. They immediately started to lay eggs, ceasing after a maximum of approximately 30 days. At least one week after the last egg was laid, we attempted the same procedure as described above, with the same males but switching their order of insemination (‘trial 2’, again with a 1-day interval between the two matings; however, only 8 out of 12 females readily re-mated with both males). Eggs of both trials were raised independently in 5 l plastic tubs until larvae hatched. No carry-over of paternity was observed in trials of females mated successively with two different pairs of males (n=5 females, set-up as described above, data not given), supporting our assumption that they had become sperm depleted.
(b) Genotyping
A random subset of offspring was collected immediately after hatching, and toe-clips were taken from adults and stored in absolute ethanol for subsequent DNA extraction. For paternity determination and relatedness estimates, we applied amplified fragment length polymorphisms (AFLPs), using the fluorescently labelled primer combinations T105, T204, T205 and procedures as outlined in Whitlock et al. (2006). PCRs were performed in Bio-Rad thermal cyclers, and AFLP fragments were separated and visualized using an ABI 3730 capillary sequencer, followed by analysis using the software GeneMapper v. 3.5.
(c) Data analysis
For paternity determination, we identified diagnostic loci where an allele was present in only one father candidate and absent in the mother; at least one diagnostic band was present in every candidate male, as required for paternity assignment using dominant markers (see Whitlock et al. 2006). Pairwise relatedness coefficients between the female and each of the candidate males were obtained using the SM estimator implemented in the software MER (ranging from −1 (least similar) to 1 (identical); Wang 2004); population allele frequencies were estimated on the basis of all genotyped adults, including seven females which were not used for mating experiments due to an insufficient number of males (data not given).
Focusing on relative success of inseminating males rather than the absolute number of offspring produced by each female–male combination, we analysed the difference in SM between the two males (SMdiff). SMdiff was defined to be negative when SM of the more successful male was lower than SM of the less successful male, and positive for the reversed case. In the case of random fertilization (null hypothesis), negative and positive SMdiffs are expected to be randomly distributed around zero. If females chose the genetically more distant or the genetically closer male, however, negative or positive SMdiff values would be substantially more frequent, statistically translating into a significant deviation from the intercept in a logistic regression model. For statistical analysis, we thus followed the logic of a Bernoulli trial, and dichotomized SMdiff values as negative versus positive. To account for the pseudoreplication of our set-up (each female–male trio was used twice), we used a generalized linear mixed effect model (with logit link function, binomial error distribution with estimated dispersion parameter, parameter estimates based on penalized quasi-likelihood). Data points were weighed by total number of offspring genotyped from each trial. We compared a model including male order to the null model without order using AICc (Burnham & Anderson 1998), and the intercept estimate of the superior model was used to formally infer female choice. The analysis was conducted in R (R Development Core Team 2006), in which lmer and glmmPQL procedures gave comparable results.
3. Results
We scored an AFLP size range of 70–280 bp, in which 53 out of 249 (21.3%) loci across three primer combinations were polymorphic across the adult individuals; pairwise relatedness coefficients SM ranged from −0.59 to 0.50, with standard deviations not exceeding 0.16 (table 1). Overall, we assigned the paternity of 578 offspring to their parents. Multiple paternities were observed in all 20 trials conducted with 12 females, with the relative paternity share of the first male averaging at 0.50 (range: 0.14–0.81). The null model focusing on male identity had considerably more support than a model which incorporated male order (ΔAICc=2.35). Thus, the paternity share of specific males was not influenced by whether they were first or second in a specific trial.
Table 1.
Relatedness and paternity in experimental matings of L. vulgaris. (SM, relatedness coefficient (Wang 2004) to the female, with standard deviations in parenthesis. In trials 1 and 2, two identical males are mated with one female in both possible orders. The absolute number of genotyped embryos assigned to the respective males is given.)
| female | relatedness coefficient (SM) | offspring, trial 1 | offspring, trial 2 | |||
|---|---|---|---|---|---|---|
| male 1 | male 2 | male 1 | male 2 | male 2 | male 1 | |
| 1 | −0.28 (0.15) | 0a | 39 | 18 | 1 | 3 |
| 2 | −0.36 (0.16) | 0.50 (0.13) | 30 | 14 | 19 | 12 |
| 3 | 0a | 0.03 (0.15) | 15 | 4 | 7 | 17 |
| 4 | −0.55 (0.15) | −0.01 (0.16) | 22 | 8 | 23 | 36 |
| 5 | −0.40 (0.16) | 0.38 (0.14) | 26 | 6 | 3 | 19 |
| 6 | −0.59 (0.16) | −0.36 (0.15) | 11 | 10 | 15 | 17 |
| 7 | −0.59 (0.13) | 0.22 (0.15) | 20 | 8 | 14 | 15 |
| 8 | 0.26 (0.13) | 0.07 (0.15) | 13 | 19 | 12 | 9 |
| 9 | 0a | −0.20 (0.13) | 8 | 12 | — | — |
| 10 | 0.24 (0.15) | 0.02 (0.14) | 16 | 12 | — | — |
| 11 | −0.13 (0.14) | 0.06 (0.15) | 6 | 21 | — | — |
| 12 | −0.32 (0.15) | −0.16 (0.15) | 5 | 13 | — | — |
For these males, the AFLP profiles were not sufficiently scorable across the whole size range, and SM was set to the population average (zero). Paternity determination was possible based on the partial AFLP screens.
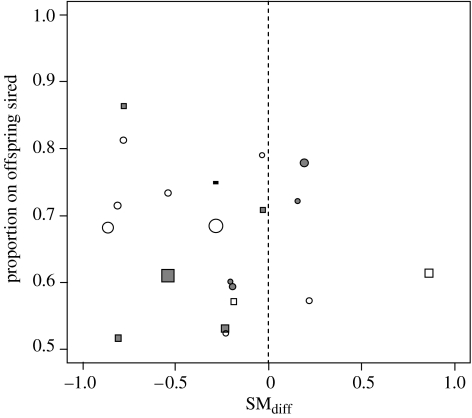
In 16 out of 20 trials, the male that was genetically more dissimilar to the female fathered the majority of the offspring (figure 1). As a consequence, the model intercept had a significantly positive deviation from zero (glmmPQL routine: t12=−2.54, intercept estimate ±95% CI: 0.05–0.44, p=0.026), demonstrating that genetic dissimilarity had a significant influence on the paternity share.
Figure 1.
Difference in relatedness coefficient (SMdiff) for those male smooth newts (L. vulgaris), which sired more offspring than the competitor when inseminating one female. The dotted line indicates the expected average when relatedness plays no role in determining paternity. Circles represent trial 1 and squares represent trial 2 (see text). Colour indicates mating order (white, first male; grey, second male). Symbol size scales linearly with total number of offspring.
4. Discussion
We demonstrate a prevalent effect of genetic dissimilarity on paternity in sperm competition experiments with L. vulgaris. Moreover, although last-male sperm precedence was suggested based on morphological evidence (Sever et al. 1999), we show that the paternity share between two competing males is independent of their order of insemination.
Given our modest sample size and accounting for the fact that several factors are potentially confounding any genetic effects (for example, we had no information about the number of sperm transferred by each male), the influence of genetic dissimilarity seems surprisingly strong. Out of 20 cases, only one male with higher paternity share was strikingly more closely related to the female than the competitor; in three other cases, SMdiff of the more successful male was only marginally positive (figure 1). This suggests that paternity is more likely to be determined by a threshold effect rather than in a gradual way. Female sensitivity to differences in genetic dissimilarity of their mating partners is unexpectedly high. As typical effective population sizes in European newts are approximately 10–20 individuals (Jehle et al. 2005), it is possible that we created experimental matings between closely related individuals. Indeed, SM values between 12 known mother–offspring relationships (one offspring for each female used) were between 0.21 and 0.55 (mean 0.36). It is possible, albeit unlikely, that setting SM of three males to zero biased our results: in two cases (involving females 1 and 9), known SM values of the competing males are several standard errors below the population average (−0.20 and −0.28 at s.e.=0.068), rendering it probable that the unknown SM values are above these values; excluding the case where the known male has an SM value close to the average (involving female 3), for example, results in an overall p value of 0.034.
Mate choice decisions are generally assumed to be shaped by both good and compatible genes in a complementary way (Mays & Hill 2004; Neff & Pitcher 2004). In natural situations, however, the temporal gaps between matings are more variable, and females have the opportunity to mate with more than two males; we are therefore uncertain whether the strong dissimilarity effects we obtained in our experiments are directly translatable to field populations. As our experiments excluded the possibility of overt mate choice, we also cannot draw conclusions on the efficiency of trading up for good genes in L. vulgaris (see Gabor & Halliday 1997). Future research should look into a potential influence of condition-dependant traits on the outcome of paternity under sperm competition, whether the importance of dissimilar genes is related to population size and inbreeding regimes, and whether the differential paternity share is due to the differential mortality of offspring, and/or cryptic female choice.
Acknowledgments
The study was conducted under the ethics guidelines of the Austrian Institutional authorities, using licence 68. 210/29-Pr/4/2002 for tissue sampling and licence RU5-SB-105/000 for animal experimentation.
The study was supported by FWF grant P-14799. We thank Klaus Reinhold for constructive comments and Andreas Baierl for statistical advice.
References
- Birkhead T.R, Chaline N, Biggins J.D, Burke T, Pizzari T. Nontransitivity of paternity in a bird. Evolution. 2004;58:416–420. [PubMed] [Google Scholar]
- Burnham K.P, Anderson D.R. Springer; New York, NY: 1998. Model selection and inference: a practical information–theoretic approach. [Google Scholar]
- Gabor C.R, Halliday T.R. Sequential mate choice by multiply mating smooth newts: females become more choosy. Behav. Ecol. 1997;8:162–166. doi:10.1093/beheco/8.2.162 [Google Scholar]
- Garner T.W.J, Schmidt B.R. Relatedness, body size and paternity in the alpine newt Triturus alpestris. Proc. R. Soc. B. 2003;270:619–624. doi: 10.1098/rspb.2002.2284. doi:10.1098/rspb.2002.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday T.R. Sperm competition in amphibians. In: Birkhead T.R, Møller A.P, editors. Sperm competition and sexual selection. Academic Press; London, UK: 1998. pp. 465–502. [Google Scholar]
- Jehle R, Wilson G.A, Arntzen J.W, Burke T. Contemporary gene flow and the spatio-temporal genetic structure of subdivided newt populations (Triturus cristatus, Triturus marmoratus) J. Evol. Biol. 2005;18:619–628. doi: 10.1111/j.1420-9101.2004.00864.x. doi:10.1111/j.1420-9101.2004.00864.x [DOI] [PubMed] [Google Scholar]
- Mays H.L, Hill G.E. Choosing mates: good genes versus genes that are a good fit. Trends Ecol. Evol. 2004;19:554–559. doi: 10.1016/j.tree.2004.07.018. doi:10.1016/j.tree.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Neff B.D, Pitcher T.E. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol. Ecol. 2004;14:19–38. doi: 10.1111/j.1365-294X.2004.02395.x. doi:10.1111/j.1365-294X.2004.02395.x [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2006. R: a language and environment for statistical computing.http://www.r-project.org [Google Scholar]
- Rafinski J, Osikowski A. Sperm mixing in the alpine newt (Triturus alpestris) Can. J. Zool. 2002;80:1293–1298. doi:10.1139/z02-099 [Google Scholar]
- Sever D.M, Halliday T, Waights V, Brown J, Davies H.A, Moriarty E.C. Sperm storage in females of the smooth newt (Triturus v. vulgaris L.): I. Ultrastructure of the spermathecae during the breeding season. J. Exp. Zool. 1999;283:51–70. doi: 10.1002/(sici)1097-010x(19990101)283:1<51::aid-jez7>3.0.co;2-i. doi:10.1002/(SICI)1097-010X(19990101)283:1<51::AID-JEZ7>3.0.CO;2-I [DOI] [PubMed] [Google Scholar]
- Steinfartz S, Vicario S, Arntzen J.W, Caccone A. A Bayesian approach on molecules and behaviour: reconsidering phylogenetic and evolutionary patterns of the Salamandridae with emphasis on Triturus newts. J. Exp. Zool. B (Mol. Dev. Evol.) 2007;308B:139–162. doi: 10.1002/jez.b.21119. doi:10.1002/jez.b.21119 [DOI] [PubMed] [Google Scholar]
- Wang J. Estimating pairwise relatedness from dominant genetic markers. Mol. Ecol. 2004;13:3169–3178. doi: 10.1111/j.1365-294X.2004.02298.x. doi:10.1111/j.1365-294X.2004.02298.x [DOI] [PubMed] [Google Scholar]
- Whitlock A, Sztatecsny M, Jehle R. AFLPs: genetic markers for paternity studies in newts (Triturus vulgaris) Amphibia-Reptilia. 2006;27:126–129. doi:10.1163/156853806776052029 [Google Scholar]
- Zeh J.A, Zeh D.W. The evolution of polyandry 2. Post-copulatory defences against genetic incompatibility. Proc. R. Soc. B. 1997;264:69–75. doi:10.1098/rspb.1997.0010 [Google Scholar]