Abstract
Many species inhabiting the Peninsular Desert of Baja California demonstrate a phylogeographic break at the mid-peninsula, and previous researchers have attributed this shared pattern to a single vicariant event, a mid-peninsular seaway. However, previous studies have not explicitly considered the inherent stochasticity associated with the gene-tree coalescence for species preceding the time of the putative mid-peninsular divergence. We use a Bayesian analysis of a hierarchical model to test for simultaneous vicariance across co-distributed sister lineages sharing a genealogical break at the mid-peninsula. This Bayesian method is advantageous over traditional phylogenetic interpretations of biogeography because it considers the genetic variance associated with the coalescent and mutational processes, as well as the among-lineage demographic differences that affect gene-tree coalescent patterns. Mitochondrial DNA data from six small mammals and six squamate reptiles do not support the perception of a shared vicariant history among lineages exhibiting a north–south divergence at the mid-peninsula, and instead support two events differentially structuring genetic diversity in this region.
Keywords: approximate Bayesian computation, comparative biogeography, statistical phylogeography
1. Introduction
Comparative phylogeography is a powerful approach for elucidating the historical processes responsible for shaping genetic diversity across co-distributed species (Avise 2000; Arbogast & Kenagy 2001). Baja California, a peninsula 1300 km long with a complex tectonic history, is inhabited by many co-distributed species that share the common phylogeographic pattern of genetic divergence across the middle of the peninsula (Riddle et al. 2000a). A seaway spanning the mid-peninsula is most often invoked as the vicariant event responsible for producing this shared genealogical pattern (Lindell et al. 2006), but this hypothesis is controversial owing to a lack of any geological evidence (Crews & Hedin 2006). Despite the effort expended on debating the existence of a putative seaway, previous studies did not consider that the appreciable differences in genetic divergence observed across different lineage pairs separated at the mid-peninsula are an indication that diversification may have occurred in multiple waves. Thus, whether the shared genealogical divisions at the mid-peninsula occurred simultaneously or non-synchronously due to multiple events remains untested.
Inferring the number of events responsible for structuring genetic diversity is imperative for understanding evolutionary processes, especially when the outcome of such analyses fuels conclusions concerning historical biogeography. Multiple temporally and spatially distinct historical events can produce similar phylogeographic patterns among co-distributed taxa and thus appear as a single event (Riddle & Hafner 2006). These alternatives are indistinguishable without an explicit examination of the timing of population divergence. The objective of this study is to statistically test the simultaneous vicariance hypothesis for the small mammals and squamate reptiles sharing concordant mitochondrial DNA (mtDNA) divergence at the mid-peninsula of Baja California.
We use a hierarchical Bayesian model to test multi-taxa hypotheses while explicitly accounting for uncertainty in taxon-specific demographic parameters that will differentially affect gene-tree coalescence. Instead of calculating the likelihood function in this cumbersome parameter-rich hierarchical model, we use approximate Bayesian computation (ABC) to replace the data with a summary statistic vector (Beaumont et al. 2002). The core advantage gained by this replacement is the ability to evaluate more complex models, such as those that apply to multi-species datasets (Hickerson et al. 2006a).
2. Material and methods
(a) Samples
We analysed 12 mtDNA datasets, representing six small mammals and six squamate reptiles that share concordant phylogeographic breaks at the mid-peninsula. Properties of the sampling intensity and nucleotide composition of these datasets and their sources are listed in table 1.
Table 1.
Mammals and reptiles with phylogeographic breaks at the mid-peninsula of Baja California used to test the simultaneous vicariance hypothesis. (Transition/transversion estimates were calculated in PAUP v. 4.0b10 under the HKY model of nucleotide substitution.)
| mtDNA gene | length (bp) | sample size (north/south) | percentage of mtDNA divergence | source | |
|---|---|---|---|---|---|
| mammals | |||||
| Ammospermophilus leucurus | Cytb | 548 | 52/25 | 4.7 | Whorley et al. (2004) |
| Chaetodipus arenarius | COIII | 699 | 22/12 | 10.1 | Riddle et al. (2000a) |
| Chaetodipus baileyi | COIII | 699 | 34/5 | 2.1 | Riddle et al. (2000c) |
| Dipodomys merriami | COIII | 699 | 6/6 | 1.1 | Riddle et al. (2000a) |
| Peromyscus eremicus | COIII | 699 | 10/8 | 3.4 | Riddle et al. (2000b) |
| Thomomys bottae | Cytb | 500 | 13/123 | 2.0 | Trujano-Alvarez (2007) |
| squamates | |||||
| Callisaurus draconoides | Cytb, ATPase 6 | 1953 | 14/23 | 4.8 | Lindell et al. (2005) |
| Phrynosoma coronatum | ND1, ND2 | 2002 | 22/10 | 4.0 | Leaché et al. (submitted) |
| Uta stansburiana | ATPase 6 | 890 | 8/7 | 3.9 | Upton & Murphy (1997) |
| Sceloporus zosteromus | ND4 | 825 | 8/4 | 7.0 | Leaché & Mulcahy (in press) |
| Pituophis catenifer | ND4 | 893 | 9/4 | 5.1 | Rodriguez-Robles & Jesús-Escobar (2000) |
| Trimorphodon biscutatus | ND4 | 817 | 15/3 | 1.6 | Devitt (2006) |
(b) Hierarchical Bayesian model of multi-species divergence
The hierarchical Bayesian model tests for simultaneous vicariance under the ABC framework, which is implemented in the MSBayes software pipeline (Hickerson et al. 2007). For full details of the model, see Hickerson et al. (2006b) and the electronic supplementary material. We use the hierarchical ABC model to estimate the hyper-parameters that quantify variability in the Y divergence times. This includes the number of possible divergence times per Y taxon-pairs (Ψ), the mean divergence time across the Y taxon-pairs (E(τ)) and the ratio of the variance of τ to the mean of τ(Ω=var(τ)/E(τ)). Although Ψ is a discrete hyper-parameter, it is estimated as a continuous hyper-posterior due to our implementation of ABC employing the local regression method of Beaumont et al. (2002). Each set of hyper-posteriors was constructed from 1000 accepted draws from K=500 000 simulated draws from the hyper-prior using the acceptance/rejection with local regression algorithm (Beaumont et al. 2002). We express divergence times in units of millions of years by assuming a generation time of 2 years and use a range of calibration rates (1, 2.0 and 3.0% Myr−1) that reflect our uncertainty in the actual rate of molecular evolution.
(c) Hypothesis testing
We use the Bayes factor (Kass & Raftery 1995) to compare posterior support for competing models (simultaneous H1 and variable divergence times H2) while accounting for prior support for either model. To calculate Bayes factors, we represent these alternative models as two arbitrary partitions of hyper-parameter space, such that H1=(Ω<0.05) and H2=(Ω≥0.05).
3. Results
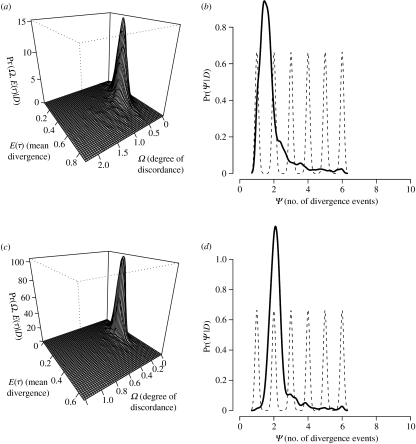
The ABC analysis of six mammals and six reptiles indicates that the data are most consistent with two temporally displaced divergence events. For the mammals, the estimate of Ψ(1.39) indicates that one or two divergence times are nearly equally probable. However, the values obtained for Ω(0.30), and its corresponding Bayes factor (0.07), provide strong evidence supporting multiple divergence times in mammals (figure 1). For reptiles, the estimate of Ψ(2.11) and Ω(0.15) (figure 1) indicate multiple divergence times, and this is moderately supported by a Bayes factor of 0.25. Although these Bayes factors are based on the hyper-parametric threshold of Ω=0.05, using Ω thresholds of 0.1 and 0.01 yielded similar Bayes factor support ranging from 0.02 to 0.52 in mammals and 0.18 to 0.40 in reptiles. Although these results are likely to be sensitive to the prior of τ used (τmax=10.0), estimates based on simulations have shown that overall conclusions based on hyper-parameter estimates rarely change when τmax varies from 5.0 to 10.0.
Figure 1.
Posterior estimates of temporal concordance among six lineage pairs of (a,b) mammals and (c,d) reptiles spanning the mid-peninsula of Baja California. (a,c) Joint posterior probability densities for E(τ). (b,d) Posterior probability densities for Ψ, the number of divergence times per six lineage pairs. (a) Ω=0.30, Bayes factor=0.07; (b) Ψ=1.39; (c) Ω=0.15, Bayes factor=0.25; (d) Ψ=2.11.
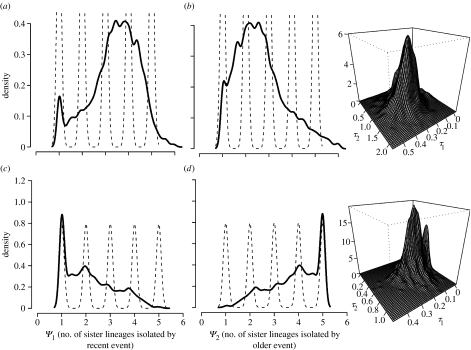
We ran the ABC model conditional on there being two divergence times (Ψ=2) to obtain estimates on τ1, the recent divergence time, τ2, the more ancient divergence time and the number of taxon pairs that diverged at these two respective times (Ψ1 and Ψ2). This suggested that two mammals (Ammospermophilus leucurus and Chaetodipus arenarius) and five reptiles (Uta stansburiana, Phrynosoma coronatum, Callisaurus draconoides, Pituophis catenifer and Sceloporus zosteromus) diverged approximately 2.3–15.3 Myr ago when assuming a broad spectrum of divergence rates (figure 2). A recent divergence at approximately 0.6–3.4 Myr ago is supported for the remaining reptile (Trimorphodon biscutatus) and mammals (Chaetodipus baileyi, Dipodomys merriami, Peromyscus eremicus and Thomomys bottae; figure 2). Although rates are likely to vary between the two taxonomic groups, it is striking that both groups yield two similar divergence time estimates if one naively assumes equal rates. Furthermore, if rate variation within the two groups were causing signatures of multiple divergence events, we would not expect the same pattern of temporal discordance within both groups.
Figure 2.
Posterior densities of Ψ1, Ψ2, τ1 and τ2 for the mammals and reptiles under the model constrained for two divergence times (τ1 and τ2). The parameters Ψ1 and Ψ2 are the number of taxon pairs that split at times τ1 and τ2, respectively. (a) Four mammals (recent event), (b) two mammals (older event), (c) one reptile (recent event) and (d) five reptiles (older event).
4. Discussion
As a consequence of focusing on the population-level genealogical relationships of organisms across landscapes, studies of comparative phylogeography can reveal cryptic patterns of vicariance that are invisible to historical biogeographic approaches based solely on taxonomy (Riddle et al. 2000a). The paradigm under the traditional approach is to accept a shared biogeographic history as the most parsimonious explanation for concordant patterns of geographical subdivision, or conversely, to assume that unequal divergence times implies multiple divergence events. As advocated in the nascent field of statistical phylogeography, comparative phylogeographic inference requires hypothesis testing with statistical models that incorporate variance in demographic parameters that strongly influence gene-tree coalescent times (Rosenberg & Nordborg 2002; Knowles 2004). By modelling uncertainty into the inherent demographic variance across species with a hierarchical Bayesian model, we have quantified multi-species concordance across the mid-peninsula of Baja California.
The traditional view of a single seaway is insufficient to explain the two episodes of diversification at the mid-peninsula of Baja California and provides an overly simplistic view of evolutionary history. Although marine incursions are supported by geological data (Holt et al. 2000), they probably never submerged the Gulf Escarpment, and therefore are unlikely to have acted as a complete barrier to gene flow. Major episodes of volcanism from Middle to Late Miocene to Holocene have produced more than 2500 km2 of igneous rocks stretching from the Sea of Cortéz to the Pacific Coast (Sawlan & Smith 1984; Conly et al. 2005). If a geological explanation is sought for the mid-peninsular divergences, then it is possible that the combined forces of volcanism and marine incursions created the physical barriers necessary for population subdivision. Alternatively, a strong ecological and climatic gradient occurs at the mid-peninsula and limits the distribution of some species (Grismer 1994). Thus, the strength of adaptive divergence appears to affect species differentially at the mid-peninsula, and this might have resulted in the observed differences in mtDNA divergences apart from the coalescent model that we explicitly incorporate (Edwards & Beerli 2000).
Acknowledgments
We thank B. Riddle, L. Alexander, T. Devitt, J. Rodriguez-Robles, J. Whorley and A. Lilia Trujano-Alvarez for supplying their datasets; R. Murphy and B. Hollingsworth for tissue samples; B. Hausback and J. Crews for discussion of Baja geology; and J. McGuire, R. Murphy, J. Patton, D. Wake and three anonymous reviewers for their useful comments. This research was funded by a NSFDDIG to A.D.L. (DEB-0508929) and an NSF postdoctoral fellowship in interdisciplinary informatics to M.J.H. (DEB-0305966).
Supplementary Material
S1. Hierarchical approximate Bayesian computation; S2. Hierarchical population divergence model; S3. Summary statistic vector; S4. References; S5. Multiple population-pair divergence model; S6. Parameters and their prior distributions
References
- Arbogast B.S, Kenagy G.J. Comparative phylogeography as an integrative approach to historical biogeography. J. Biogeogr. 2001;28:819–825. doi:10.1046/j.1365-2699.2001.00594.x [Google Scholar]
- Avise J.C. The history and formation of species. Harvard University Press; Cambridge, MA: 2000. Phylogeography. [Google Scholar]
- Beaumont M.A, Zhang W, Balding D.J. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conly A.G, Brenan J.M, Bellon H, Scott S. Arc to rift transitional volcanism in the Santa Rosalía Region Baja California Sur, Mexico. J. Volcanol. Geotherm. Res. 2005;142:303–341. doi:10.1016/j.jvolgeores.2004.11.013 [Google Scholar]
- Crews S.C, Hedin M. Studies of morphological and molecular phylogenetic divergence in spiders (Araneae: Homalonychus) from the American southwest, including divergence along the Baja California Peninsula. Mol. Phylogenet. Evol. 2006;38:470–487. doi: 10.1016/j.ympev.2005.11.010. doi:10.1016/j.ympev.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Devitt T.J. Phylogeography of the western lyresnake (Trimorphodon biscutatus): testing aridland biogeographical hypotheses across the Nearctic–Neotropical transition. Mol. Ecol. 2006;15:4387–4407. doi: 10.1111/j.1365-294X.2006.03015.x. doi:10.1111/j.1365-294X.2006.03015.x [DOI] [PubMed] [Google Scholar]
- Edwards S.V, Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescent time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Grismer L.L. The origin and evolution of the peninsular herpetofauna of Baja California, Mexico. Herpetol. Nat. Hist. 1994;2:51–106. [Google Scholar]
- Hickerson M.J, Dolman G, Moritz C. Comparative phylogeographic summary statistics for testing simultaneous vicariance across taxon-pairs. Mol. Ecol. 2006a;15:209–224. doi: 10.1111/j.1365-294X.2005.02718.x. doi:10.1111/j.1365-294X.2005.02718.x [DOI] [PubMed] [Google Scholar]
- Hickerson M.J, Stahl E.A, Lessios H.A. Test for simultaneous divergence using approximate Bayesian computation. Evolution. 2006b;60:2435–2453. [PubMed] [Google Scholar]
- Hickerson M.J, Stahl E, Takebayashi N. MSBayes: pipeline for testing comparative phylogeographic histories using hierarchical approximate Bayesian computation. BMC Bioinform. 2007;8:268. doi: 10.1186/1471-2105-8-268. doi:10.1186/1471-2105-8-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J.W, Holt E.W, Stock J.M. An age constraint on Gulf of California rifting from the Santa Rosalia Basin Baja California Sur, Mexico. GSA Bull. 2000;112:540–549. [Google Scholar]
- Kass R.E, Raftery A. Bayesian factors. J. Am. Stat. Assoc. 1995;90:773–795. doi:10.2307/2291091 [Google Scholar]
- Knowles L.L. The burgeoning field of statistical phylogeography. J. Evol. Biol. 2004;17:1–10. doi: 10.1046/j.1420-9101.2003.00644.x. doi:10.1046/j.1420-9101.2003.00644.x [DOI] [PubMed] [Google Scholar]
- Leaché A.D, Mulcahy D.G. Phylogeny, divergence times, and species limits of spiny lizards (Sceloporus magister species group) in Western North American deserts and Baja California. Mol. Ecol. In press. doi: 10.1111/j.1365-294X.2007.03556.x. [DOI] [PubMed] [Google Scholar]
- Leaché, A. D., Koo, M. S., Papenfuss, T. J., Fisher, R. N. & McGuire, J. A. Submitted. Quantifying phylogenetic and ecological divergence in the coast horned lizard (Phrynosoma coronatum).
- Lindell J, Mendez-de la Cruz F.R, Murphy R.W. Deep genealogical history without population differentiation: discordance between mtDNA and allozyme divergence in the zebra-tailed lizard (Callisaurus draconoides) Mol. Phylogenet. Evol. 2005;2005:682–694. doi: 10.1016/j.ympev.2005.04.031. doi:10.1016/j.ympev.2005.04.031 [DOI] [PubMed] [Google Scholar]
- Lindell J, Ngo A, Murphy R.W. Deep genealogies and the mid-peninsular seaway of Baja California. J. Biogeogr. 2006;33:1327–1331. doi:10.1111/j.1365-2699.2006.01532.x [Google Scholar]
- Riddle B.R, Hafner D.J. A step-wise approach to integrating phylogeographic and phylogenetic biogeographic perspectives on the history of a core North American warm deserts biota. J. Arid Environ. 2006;66:435–461. doi:10.1016/j.jaridenv.2006.01.014 [Google Scholar]
- Riddle B.R, Hafner D.J, Alexander L.F, Jaeger J.R. Cryptic vicariance in the historical assembly of a Baja California Peninsular Desert biota. Proc. Natl Acad. Sci. USA. 2000a;97:14 438–14 443. doi: 10.1073/pnas.250413397. doi:10.1073/pnas.250413397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle B.R, Hafner J, Alexander L.F. Phylogeography and systematics of the Peromyscus eremicus species group and the historical biogeography of North American warm regional deserts. Mol. Phylogenet. Evol. 2000b;17:145–160. doi: 10.1006/mpev.2000.0841. doi:10.1006/mpev.2000.0841 [DOI] [PubMed] [Google Scholar]
- Riddle B.R, Hafner D.J, Alexander L.F. Comparative phylogeography of Baileys' pocket mouse (Chaetodipus baileyi) and the Peromyscus eremicus species group: historical vicariance of the Baja California Peninsular Desert. Mol. Phylogenet. Evol. 2000c;17:161–172. doi: 10.1006/mpev.2000.0842. doi:10.1006/mpev.2000.0842 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Robles J.A, De Jesus-Escobar J.M. Molecular systematics of new world gopher, bull, and pinesnakes (Pituophis: colubridae), a transcontinental species complex. Mol. Phylogenet. Evol. 2000;14:35–50. doi: 10.1006/mpev.1999.0698. doi:10.1006/mpev.1999.0698 [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A, Nordborg M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 2002;3:380–390. doi: 10.1038/nrg795. doi:10.1038/nrg795 [DOI] [PubMed] [Google Scholar]
- Sawlan M.G, Smith J.G. Geology of the Baja California Peninsula. The Pacific Section of the Society of Economic Paleontologists and Mineralogists; Los Angeles, CA: 1984. Petrologic characteristics, age and tectonic setting of Neogene volcanic rocks in northern Baja California Sur, Mexico. [Google Scholar]
- Trujano-Alvarez A.L, Álvarez-Castañeda S.T. Taxonomic revision of Thomomys bottae in the Baja California Sur lowlands. J. Mamm. 2007;88:343–350. doi:10.1644/05-MAMM-A-291R3.1 [Google Scholar]
- Upton D.E, Murphy R.W. Phylogeny of the side-blotched lizards (Phrynosomatidae: Uta) based on mtDNA sequences: support for a midpeninsular seaway in Baja California. Mol. Phylogenet. Evol. 1997;8:104–113. doi: 10.1006/mpev.1996.0392. doi:10.1006/mpev.1996.0392 [DOI] [PubMed] [Google Scholar]
- Whorley J.R, Alvarez-Castaneda S.T, Kenagy G.J. Genetic structure of desert ground squirrels over a 20-degree-latitude transect from Oregon through the Baja California Peninsula. Mol. Ecol. 2004;13:2709–2720. doi: 10.1111/j.1365-294X.2004.02257.x. doi:10.1111/j.1365-294X.2004.02257.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Hierarchical approximate Bayesian computation; S2. Hierarchical population divergence model; S3. Summary statistic vector; S4. References; S5. Multiple population-pair divergence model; S6. Parameters and their prior distributions