Abstract
Interspecific hybridization occurs regularly in wild Heliconius butterflies, although hybrid individuals are usually very rare. However, hybridization generally occurs only between the most closely related species. We report a rare naturally occurring hybrid between non-sister species and carry out the first genetic analysis of such distant hybridization. Mitochondrial and nuclear genes indicate that the specimen is an F1 hybrid between a female Heliconius ethilla and a male Heliconius melpomene, originating from a group of 13 species estimated to have diverged over 2.5 Myr ago. The presence of such distant natural hybrids, together with evidence for backcrossing, suggests that gene flow across species boundaries can take place long after speciation. Adaptive genes such as those involved in wing coloration could thus be widely shared among members of this highly mimetic genus.
Keywords: Heliconius ethilla, Heliconius melpomene, hybridization
1. Introduction
Evolutionary biologists generally accept the biological species concept, and this has led to hybridization between species being regarded as uncommon and unimportant (Mayr 1963). Although hybrids are, almost by definition, very rare on a per individual basis, recent surveys have shown that many species do in fact hybridize (Coyne & Orr 2004; Mallet 2005). On average, 10% of animal species and 25% of plant species are known to hybridize in the wild, and in some particularly charismatic groups, such as ducks, birds of paradise and North American Papilio butterflies, 40–75% of species are known to hybridize (Mallet 2005).
Many wild-caught interspecific hybrid specimens between Heliconiina butterflies are known. Hybrids occur in 35% of the 46 species in the genus Heliconius and 27% of the 73 species in the larger sub-tribe Heliconiinae (Mallet et al. 2007). These butterflies have been well studied owing to their bright wing coloration and extensive variation in wing pattern morphology both between species and among geographical races within species. The bright wing colours act as a warning of their unpalatability to potential predators. Many species share similar patterns with unrelated Heliconiinae and Ithomiinae, leading to impressive Müllerian mimicry rings (Bates 1862; Turner 1981; Beccaloni 1997; Joron & Mallet 1998). Although hybrids are rare within most Heliconius populations, the existence of naturally occurring hybrids is intriguing because it suggests that gene flow will be possible between species. Hence, this could lead to the transfer of adaptive genes between species (Gilbert 2003).
Hybridization in Heliconius has hitherto been examined genetically only in a few cases that involve closely related species: Heliconius erato and Heliconius himera, or Heliconius melpomene and Heliconius cydno (Jiggins et al. 1997; Bull et al. 2006; Kronforst et al. 2006). In all other cases, inferences of hybridization have been made on the basis of phenotypes of wild-caught specimens. In contrast, the extremely rare hybrids between H. melpomene and the more distant species in the ‘silvaniform’ Heliconius (Mallet et al. 2007) have never been examined genetically. In this paper, we describe a new rare hybrid formed between two distantly related Heliconius species in the H. melpomene group, recently caught in Peru, and use molecular markers to determine the parental types.
2. Material and methods
We are engaged in an extensive study of heliconiine and ithomiine butterflies in the Departamento de San Martín, Peru (Mallet & Barton 1989; Joron et al. 2001; Whinnett et al. 2005). In addition to the pure species, subspecies and many interracial hybrids sampled, we recently caught a peculiar specimen, a putative interspecific hybrid from the melpomene–silvaniform group (specimen ID 06-921; table 1) near Moyobamba in Northern Peru. The hybrid specimen will be deposited in the collection at the Museo de Historia Natural, Universidad Nacional Mayor de San Marcos (UNMSM), Lima, Peru. The remaining specimens analysed (table 1) are of common species, held at University College London and vouchered at UNMSM.
Table 1.
Details of samples and collection localities.
| specimen ID | species | collection locality | latitude | longitude |
|---|---|---|---|---|
| 06-921 | Heliconius hybrid | Rumiyacu, near Moyobamba | 06°05′23″ S | 076°58′09″ W |
| 04-286 | Heliconius melpomene | Bosque von Humboldt | 08°49′48″ S | 075°03′28″ W |
| 04-288 | Heliconius melpomene | Bosque von Humboldt | 08°49′48″ S | 075°03′28″ W |
| 02-366 | Heliconius melpomene | Davidcillo | 06°14′47″ S | 076°15′58″ W |
| 02-944 | Heliconius melpomene | Puente Serranayacu | 05°40′48″ S | 077°40′50″ W |
| 02-1839 | Heliconius melpomene | Boca Toma Río Shilcayo | 06°27′20″ S | 076°20’40″ W |
| 02-1850 | Heliconius melpomene | Shapaja | 06°34’29″ S | 079°16’48″ W |
| 02-1882 | Heliconius melpomene | Chumia | 06°36’57″ S | 076°11’07″ W |
| 02-1894 | Heliconius melpomene | km 30, Tarapoto-Yurimaguas | 06°24’33″ S | 076°18’24″ W |
| 02-2060 | Heliconius melpomene | km 26, Yurimaguas-Tarapoto | 05°58’30″ S | 076°14’15″ W |
| 02-3 | Heliconius ethilla | Boca Toma Río Shilcayo | 06°27’20″ S | 076°20’40″ W |
| 02-975 | Heliconius ethilla | km 10, Tarapoto-Yurimaguas | 06°27’18″ S | 076°17’46″ W |
| 02-1483 | Heliconius ethilla | La Antena, km 16, Tarapoto-Yurimaguas | 06°27’19″ S | 076°17’54″ W |
| 02-2037 | Heliconius numata | km 26, Yurimaguas-Tarapoto | 05°58’30″ S | 076°14’15″ W |
| 02-364 | Heliconius elevatus | Davidcillo | 06°14’47″ S | 076°15’58″ W |
| 05-1196 | Heliconius pardalinus | Urahuasha | 06°27’43″ S | 076°19’36″ W |
| 02-1330 | Heliconius hecale | km 7.2, Pongo-Barranquita | 06°17’41″ S | 076°13’53″ W |
DNA was extracted from a single leg of the hybrid specimen using a QIAamp DNA Micro Kit (QIAGEN), and from one-third of the thorax of all other specimens using the DNeasy Blood and Tissue Kit (QIAGEN). Approximately 2200 bp of mtDNA comprising CoI and 5′ end of CoII genes were amplified by PCR in three sections for the hybrid individual, representatives of Heliconius melpomene amaryllis, Heliconius elevatus and each of the four sympatric silvaniform species: Heliconius numata; Heliconius ethilla; Heliconius pardalinus; and Heliconius hecale. In addition, five nuclear loci (Tektin, Rpl5, Tpi, Mpi and inv) were amplified in the hybrid, representatives of H. melpomene and H. ethilla. PCR products were cleaned and cycle sequenced with the PCR primers using the Big Dye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems). Table S1 of the electronic supplementary material contains details of the primers used, PCR conditions and sequence accession numbers.
Apart from mtDNA and Tektin, all other loci spanned one or more intron regions. Indels in the intronic regions often resulted in the amplification of alleles with different sizes from a single individual. Unless sequence quality was low, the indel could be readily identified and the two alleles deconvoluted using information from the double-peak signals following the indel (Flot et al. 2006). See the electronic supplementary material for details of this procedure.
3. Results
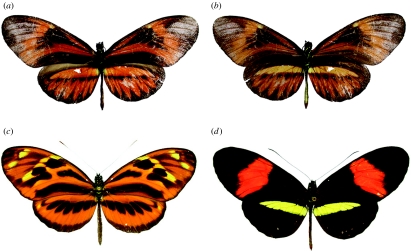
The putative hybrid (figure 1) is a male, and its wing pattern is unlike that of other local melpomene–silvaniform group species. Instead, the markings appear to combine those of H. m. amaryllis (the local ‘postman’-patterned melpomene race) and of a local silvaniform species: H. numata; ethilla; hecale; or pardalinus. Compared with H. m. amaryllis, the forewing crimson colours have become burnt orange and these orange markings extend over both fore- and hindwings, rather than being restricted to a forewing bar. The yellow hindwing bar of the latter species has also become orange on the upperside, although the yellow is almost fully expressed on the underside. Compared with Heliconius ethilla aerotome (identified as the other parent, see below), the orange markings are much reduced, and on the hindwings narrowed into ‘rays’ reminiscent of those found in races of H. melpomene such as Heliconius melpomene aglaope, which occurs over the mountain range in the Amazonian lowlands to the northeast of the capture site. The spotty melanic markings of H. ethilla are also broadened in the putative hybrid to form black smears, particularly in the central part of the forewing. Apart from the underside-expressed yellow hindwing bar, the underside and upperside patterns are similar. This hybrid is similar to a specimen in the Natural History Museum, London, originally named as a separate species ‘Heliconius hippola’ Hewitson (Mallet et al. 2007).
Figure 1.
(a) Dorsal and (b) ventral wing colour patterns of the hybrid specimen 06-921. Dorsal wing colour patterns of the putative parent species (c) H. ethilla aerotome and (d) H. melpomene amaryllis.
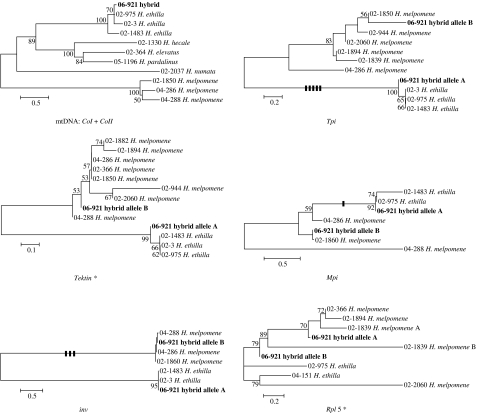
A Blast search of the mtDNA sequence revealed 99.5% similarity to H. ethilla. Subsequent comparison with mtDNA sequences obtained from locally caught specimens of the four potential silvaniform species showed unambiguously that the hybrid possessed a H. ethilla mitochondrial sequence (figure 2; table S2, electronic supplementary material). Diagnostic sequence differences between H. melpomene and H. ethilla were not found at Rpl5, but were present at the other four nuclear loci. The hybrid individual was found to possess both a melpomene- and an ethilla-type allele at each of these four diagnostic nuclear loci (figure 2).
Figure 2.
Neighbour-joining trees for six loci showing the relationship between alleles found in the hybrid and other melpomene–cydno–silvaniform group species. Nodes with 50% or greater bootstrap support are labelled. Bold vertical lines indicate diagnostic indels between H. melpomene and H. ethilla. Scale bars represent percentage sequence divergence. Asterisks, additional information in the electronic supplementary material.
An F1 hybrid should be heterozygous at all nuclear genes, bearing alleles of each parental type. In contrast, a backcross hybrid should only be heterozygous at half its nuclear genes and homozygous for one parental type over its remaining nuclear genes. As the hybrid is heterozygous at four diagnostic nuclear loci, this indicates that the individual is unlikely to be a backcross or F2 hybrid (binomial p=0.54=0.0625); an F1 hybrid between H. ethilla and H. m. amaryllis is 16 times more likely than any second-generation hybrid.
4. Discussion
Although interspecific hybridization is a common phenomenon among heliconiine butterflies, at the individual level, hybridization is rare, usually comprising less than 1 in 1000 wild individuals (Mallet et al. 2007). Most wild hybrids are either intraspecific (between different wing pattern races) or between closely related, usually sister taxa (Mallet et al. 2007). Only 10 putative hybrid specimens have been documented between melpomene and silvaniform species, and only four are putatively between H. melpomene and H. ethilla (Mallet et al. 2007). However, no molecular verification of these rare and distant hybrids has hitherto been carried out, so the identity of the parents and whether they are in fact hybrids or aberrations is in doubt (Mallet et al. 2007). Here, genetic evidence for natural hybridization between such distant non-sister Heliconius species (H. melpomene and H. ethilla) has been obtained for the first time.
Heliconius ethilla and H. melpomene are approximately 5% different at the mtDNA studied. Assuming that this gene evolves in a clock-like manner at 2% Myr−1 (Brower 1994), this suggests hybridization events occurring ca 2.5 Myr after speciation has occurred. The occurrence of a wild adult F1 hybrid between these species indicates that such hybrids can develop normally and survive in the wild. Some putative wild hybrids between melpomene and silvaniform group butterflies are considered to be backcrosses (Mallet et al. 2007), providing evidence that some such hybrids may be fertile and capable of reproduction. Two of the four putative hybrids between H. melpomene and H. ethilla known from Colombia are clearly not F1 hybrids, and are presumably backcrosses to H. ethilla (Mallet et al. 2007). Although those putative hybrids have not been analysed genetically, the more ethilla-like patterns, the strong expression of yellow coloration in the forewing (which is recessive in hybrids) and strongly ethilla-like hindwing pattern all provide clear evidence of backcrossing to ethilla in nature. The other two hybrids are H. hippola-like and are presumably F1 hybrids (Mallet et al. 2007). Backcrosses via male F1 hybrids in a complex cross involving H. melpomene and the silvaniforms H. hecale and H. atthis (the latter close to H. ethilla) have been obtained in captivity, although female hybrids were reported to be sterile (Mallet et al. 2007), indicating that such backcrossing is possible. This potential for backcrossing may result in transfer of genes between species.
As hybridization is regular, species boundaries in Heliconius have the potential to be porous. Within the melpomene–cydno group, hybridization and backcrossing has led to interspecific introgression at some, but not all, genomic regions (Bull et al. 2006; Kronforst et al. 2006), and has apparently produced at least one hybrid species (Mavárez et al. 2006). If extensive hybridization among these two closely related species can cause adaptive genes to introgress as a result of hybridization, rarer hybridization between more distant species, such as between the silvaniform and the melpomene–cydno group species, as here, may also play a role. Closely related Heliconius species are often members of different colour pattern mimicry rings, and very similar and apparently homoplasious mimetic patterns are often found between related non-sister species; for instance, H. elevatus (a silvaniform) shares almost identical ray patterns with some races of H. melpomene and Heliconius timareta. Heliconius besckei (also a silvaniform), in contrast, shares a melpomene amaryllis-like postman pattern with races of H. melpomene and H. timareta. Our data therefore contribute evidence for the intriguing possibility that mimetic wing patterns may be shared via introgression among distant as well as closely related species (Gilbert 2003; Mallet et al. 2007).
Acknowledgments
We are thankful to Gerardo Lamas of the Museo de Historia Natural, UNMSM, Lima, for his lepidopteran expertise, INRENA for permits, and NERC and DEFRA–Darwin Initiative for funding different aspects of this project.
Supplementary Material
Details of PCR conditions
Comparison of nuclear and mtDNA sequences of the Heliconius hybrid with sequences of the putative parent and other sympatric melopomene/cydno/silvaniform species
Comparison of hybrid sequences with putative parental sequences
References
- Bates H.W. Contributions to an insect fauna of the Amazon valley, Lepidoptera: Heliconidae. Trans. Linn. Soc. Lond. 1862;23:495–566. [Google Scholar]
- Beccaloni G.W. Ecology, natural history and behaviour of ithomiine butterflies and their mimics in Ecuador (Lepidoptera: Nymphalidae: Ithomiinae) Trop. Lepid. 1997;8:103–124. [Google Scholar]
- Brower A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. doi:10.1073/pnas.91.14.6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull V, Beltrán M, Jiggins C.D, McMillan W.O, Bermingham E, Mallet J. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biol. 2006;4:11. doi: 10.1186/1741-7007-4-11. doi:10.1186/1741-7007-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Flot J.-F, Tillier A, Samadi S, Tillier S. Phase determination from direct sequencing of length-variable DNA regions. Mol. Ecol. Notes. 2006;6:627–630. doi:10.1111/j.1471-8286.2006.01355.x [Google Scholar]
- Gilbert L.E. Adaptive novelty through introgression in Heliconius wing patterns: evidence for a shared genetic “toolbox” from synthetic hybrid zones and a theory of diversification. In: Boggs C.L, editor. Ecology and evolution taking flight: butterflies as model systems. University of Chicago Press; Chicago, IL: 2003. pp. 281–318. [Google Scholar]
- Jiggins C.D, McMillan W.O, King P, Mallet J. The maintenance of species differences across a Heliconius hybrid zone. Heredity. 1997;79:495–505. doi:10.1038/sj.hdy.6882230 [Google Scholar]
- Joron M, Mallet J.L.B. Diversity in mimicry: paradox or paradigm? Trends Ecol. Evol. 1998;13:461–466. doi: 10.1016/s0169-5347(98)01483-9. doi:10.1016/S0169-5347(98)01483-9 [DOI] [PubMed] [Google Scholar]
- Joron M, Wynne I.R, Lamas G, Mallet J. Variable selection and the coexistence of multiple mimetic forms of the butterfly Heliconius numata. Evol. Ecol. 2001;13:721–754. doi:10.1023/A:1010875213123 [Google Scholar]
- Kronforst M.R, Young L.G, Blume L.M, Gilbert L.E. Multilocus analysis of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. doi:10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Mallet J, Barton N.H. Strong natural selection in a warning color hybrid zone. Evolution. 1989;43:421–431. doi: 10.1111/j.1558-5646.1989.tb04237.x. doi:10.2307/2409217 [DOI] [PubMed] [Google Scholar]
- Mallet J, Beltrán M, Neukirchen W, Linares M. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. doi:10.1186/1471-2148-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavárez J, Salazar C.A, Bermingham E, Salcedo C, Jiggins C.D, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. doi:10.1038/nature04738 [DOI] [PubMed] [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1963. Animal species and evolution. [Google Scholar]
- Turner J.R.G. Adaptation and evolution in Heliconius: a defense of neo-Darwinism. Annu. Rev. Ecol. Syst. 1981;12:99–121. doi:10.1146/annurev.es.12.110181.000531 [Google Scholar]
- Whinnett A, Zimmermann M, Willmott K.R, Herrera N, Mallarino R, Simpson F, Joron M, Lamas G, Mallet J. Strikingly variable divergence times inferred across an Amazonian butterfly ‘suture zone’. Proc. R. Soc. B. 2005;272:2525–2533. doi: 10.1098/rspb.2005.3247. doi:10.1098/rspb.2005.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of PCR conditions
Comparison of nuclear and mtDNA sequences of the Heliconius hybrid with sequences of the putative parent and other sympatric melopomene/cydno/silvaniform species
Comparison of hybrid sequences with putative parental sequences