A remarkable feature of univalent cation-selective channels is their ability to discriminate between ions of the same charge and similar size. The mechanism of K+ selectivity has been studied extensively. KcsA, the first K+ channel to be characterized by x-ray crystallography, has a narrow 12-Å-long pore, lined symmetrically with main chain carbonyl oxygens from the P loops, and this pore motif is found in all other K+-selective channels that have been crystallized. This region only permits passage of mostly dehydrated K+ ions by close interactions between the cations and the carbonyl oxygens, which compensate energetically for the extensive dehydration of K+ ions (Doyle et al., 1998). Voltage-gated Na+ channels have a similar structure to K+ channels and are thought to have evolved from K+ channels by gene duplications (Hille, 2001). However, the mammalian voltage-gated Na+ channel achieves its selectivity for Na+ over K+ by a very different mechanism. In comparison to the K+ channel P loops, the four Na+ channel P loops are splayed out, leaving only a short selectivity region (Hille, 1975; Lipkind and Fozzard, 1994; Yamagishi et al., 1997). Cation interaction in the Na+ channel selectivity filter is with flexible side chains of negatively and positively charged residues derived from its four nonidentical domains, instead of with the more constrained main chain carbonyls, and the cation remains mostly hydrated (Hille, 1971).
Cloning of the family of mammalian Na+ channel genes has identified four key residues of the selectivity region as an aspartate-glutamate-lysine-alanine (DEKA) motif, consisting of one amino acid from each of the four P loop regions from domains I–IV, respectively (Asp-400, Glu-755, Lys-1237, and Ala-1529, using numbers from rNav1.4, the rat skeletal muscle Na+ channel). The region represents a “signature” pattern (Guy and Seetharamulu, 1986) for all mammalian Na+ channels that have been expressed; pore-binding tetrodotoxin and saxitoxin require these and adjacent residues for high affinity (Terlau et al., 1991; Penzotti et al., 1998); and mutations of these residues alter permeation and selectivity (Terlau et al., 1991; Chiamvimonvat et al., 1996; Favre et al., 1996; Schlief et al., 1996; Tsushima et al., 1997). L-type Ca2+ channels have four glutamates (EEEE motif) in homologous positions. Substitution of lysine in the EEEE motif of L-type Ca2+ channels favors Na+ selectivity (Kim et al., 1993; Yang et al., 1993). Conversely, Heinemann et al. (1992) transformed the Na+ channel into one that showed some Ca2+ channel–selective properties by mutating the KA of DEKA to EE. This essential role of the side chains of the DEKA amino acid residues is in contrast to K+ channels, requiring that the selectivity mechanism be quite different for Na+ channels.
Systematic mutation of these DEKA residues in the Na+ channel has further identified the specific amino acid composition and configuration required for discrimination between K+ and Na+ and for exclusion of Ca2+ permeation (Favre et al., 1996; Schlief et al., 1996; Sun et al., 1997). The Na+ channel is not permeable to Ca2+, and the ionic permeability ratio for K+ over Na+ in Nav1.4 is only 0.03 (Favre et al., 1996), indicating an energetic balance 30-fold in favor of Na+. Table I summarizes many of the experimental measures of K+/Na+ channel permeability ratios. Presence of a positively charged residue in the ring, either lysine or arginine, prevents Ca2+ (or other divalent ion) permeation completely. Selectivity for Na+ vs. K+ absolutely requires the presence of lysine and at least one carboxylate. Simply including a positive charge is not sufficient, because arginine substitution for lysine (DERA) results in a nonselective channel (PK/PNa = 0.9; Favre et al., 1996). Only one carboxylate is absolutely required, because alanine substitution for either aspartate(I) or glutamate(II) preserves Na+ selectivity. The more important carboxylate in the selectivity ring was shown to be the domain II glutamate, which in combination with the domain III lysine (AEKA) maintains the WT selectivity for Na+ vs. K+ (PK/PNa = 0.03; Favre et al., 1996). The domain I aspartate in combination with domain III lysine (DAKA) shows a small loss of Na+ selectivity (PK/PNa = 0.09; Favre et al., 1996). Configuration of lysine and the carboxylate is important; shift of lysine to the domain II position and glutamate to the domain III position (DKEA) reduces selectivity fourfold (Schlief et al., 1996).
TABLE I.
Permeability Ratios of Na+ Channel Selectivity Filter Mutants
| Mutant | PK/PNa | ||
|---|---|---|---|
| Favrea | Schliefb | Sunc | |
| DEKA | 0.03 ± 0.01 | 0.10 ± 0.03 | 0.08 ± 0.01 |
| AEKA | 0.03 ± 0.03 | – | – |
| DAKA | 0.09 ± 0.03 | – | – |
| DEKD | – | 0.23 ± 0.05 | – |
| DKEA | – | 0.37 ± 0.03 | – |
| DEAA | 1.1 ± 0.06 | – | 0.87 ± 0.04 |
| DEQA | – | 0.76 ± 0.06 | – |
| DERA | 0.9 ± 0.04 | – | – |
| DAAA | 0.9 ± 0.03 | – | 0.81 ± 0.05 |
| AEAA | 1.1 ± 0.05 | – | 0.91 ± 0.03 |
| AAEA | 1.0 ± 0.06 | – | – |
| DEEE | – | 0.95 ± 0.05 | – |
The mechanism of selectivity for Na+ over K+ remains unresolved, in spite of these mutational studies. Favre et al. (1996) suggested that the butylammonium group of lysine might act as a “tethered cation” within the electric field of the pore. Based on our homology model of the Na+ channel outer vestibule (Lipkind and Fozzard, 2000), we proposed that in the absence of a Na+ ion in the selectivity filter there is a hydrogen bond between the glutamate(II) carboxylate and the lysine(III) amino group. This interaction would block cation interaction with the glutamate oxygens, but Na+ would displace the amino group from the glutamate more easily than K+. These ideas require that selectivity be at least a three-party energetic interaction, including a carboxylate, an amino group, and the cation. Even though exact structural details of the selectivity region for the Na+ channel are not available, it may be possible to explore the energy of interactions expected between univalent cations and amino acid residues in the selectivity region, because the side chains of the critical amino acids are flexible, allowing them to adjust their locations and interactions in the presence of cations.
Results of Molecular Dynamics Simulations
To explore this idea, we have used the structure of the outer vestibule-selectivity filter of the Na+ channel from our model (Lipkind and Fozzard, 2000) to examine the selectivity filter residue interactions with three alkali univalent cations, Na+, K+, and Rb+. The approximate size of the opening at the DEKA level is known from sieving studies (Hille, 1971; Sun et al., 1997) to be ∼3 × 5 Å, and is correctly reproduced in our model. The side chains of the critical amino acids are flexible, allowing them to adjust their location and interactions in the presence of cations. The structure was fixed in all molecular dynamics (MD) calculations except for the side chains of the residues forming the selectivity filter, aspartate(I), glutamate(II), and lysine(III). Molecules of water were introduced inside the pore using the Soak Assembly module of Insight II. Ionic radii for Na+, K+, and Rb+ were 0.95, 1.33, and 1.48 Å, respectively. The MD calculations were performed with the Dynamics module of Insight II at a constant temperature of 300°K with time steps of 1 fs, until the system achieved a steady state (∼0.1 ns simulations). This allowed determination of new configurations produced by introducing individually each of the three univalent cations, with initial positions for the side chains, those in the steady state without a cation in the selectivity filter. To evaluate the short-range electrostatic interactions between the univalent cation and the charged residues in the pore's water environment we have used a typical strategy with an effective dielectric constant Deff of 10 (Simonson and Perahia, 1995; Sansom et al., 1997). The carboxylates are assumed to be fully dissociated, based on the experimental results of Khan et al. (2006).
The optimal location of the Na+ channel selectivity filter side chains in vacuum in the absence of cation in the model has been shown in Fig. 10 of our previous report (Lipkind and Fozzard, 2000). The MD simulations in a water environment yield an arrangement almost identical to those side chain locations in vacuum, since the configuration of the selectivity filter residues is determined mainly by strong electrostatic interactions between lysine's amino group and the carboxylates in the DEKA motif. Importantly, the amino group of lysine(III) makes close contact with the carboxyl oxygens of glutamate(II) to form a hydrogen bond, while the amino group of lysine is on average separated from aspartate(I). Alanine(IV) is at some distance, with its backbone carbonyl exposed to the pore. Water molecules are largely oriented by the charged residues, as previously reported by Singh et al. (1996). Because the mutational studies have shown that it is not necessary to have two negatively charged amino acids, aspartate (I) and glutamate (II), simultaneously in the selectivity filter in order to achieve high selectivity for Na+ over K+ (Favre et al., 1996), we consider a simplified model problem of interaction of added cations Na+, K+, and Rb+ with only two side chains, one negative and one positive, including AEKA + Na+, AEKA + K+, AEKA + Rb+, AERA + K+, DAKA + Na+, DAKA + K+, DAKA + Rb+, and DARA + K+.
AEKA + Na+.
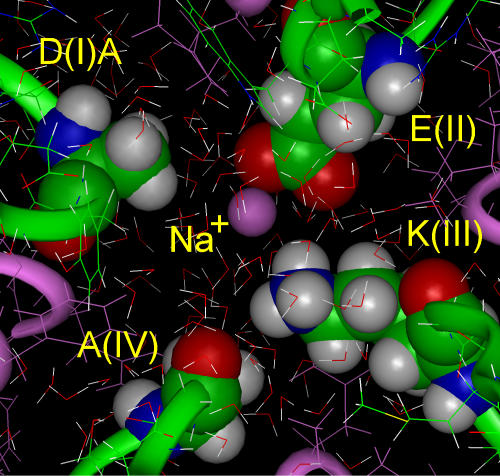
Under physiological conditions the Na+ channel is occupied by only one Na+ ion (Ravindran et al., 1991). The beginning position for the test Na+ ion inside the AEKA motif was adjacent to the left oxygen of the carboxylate group of glutamate(II) in the selectivity filter opening, while both oxygens of glutamate interacted with the amino group of lysine(III). During MD simulation the Na+ ion (r = 0.95 Å) first oriented water molecules in its vicinity, with typically five waters interacting (Zhu and Robinson, 1992; Varma and Rempe, 2007). It then moved to interact with both oxygens of glutamate(II), easily breaking the hydrogen bond with the larger amino group of lysine. The lysine moved away from glutamate(II) in the direction of alanine(IV), where it was in van der Waals contact (Fig. 1). In this arrangement, the permeating Na+ ion generated a new configuration of the selectivity filter by breaking the hydrogen bond and displacing the side chain of lysine(III) from the carboxylate group of glutamate(II), which then formed the binding site for Na+. The carboxylate group substituted for one molecule of water around Na+, so that the cation remained almost completely hydrated (approximately four molecules of water remaining).
Figure 1.
A molecular model of the AEKA motif of the selectivity filter of the Na+ channel in a steady-state configuration formed by MD simulation with the Na+ ion inside. I-IV P loops are shown by green ribbons, residues of AEKA by space-filled images, Na+ by a pink ball. During MD simulation the Na+ ion interacts with oxygens of glutamate(II), losing one water of hydration and easily breaking its initial hydrogen bond with lysine(III).
An approximation of the electrostatic energy of interaction of Na+ with glutamate(II) is −12 kcal/mol and of Na+ with lysine (III) is +7.5 kcal/mol, so that the net binding energy for Na+ was −4.5 kcal/mol. Although the presence of lysine substantially reduced the net energy for Na+ binding in the selectivity filter, it did not prevent direct interaction of Na+ with the oxygens of the glutamate(II), because the electrostatic attraction of Na+ to glutamate is stronger than its repulsion by the side chain of lysine. The energetics of this new stable configuration can be compared with those of other univalent cations.
AEKA + K+.
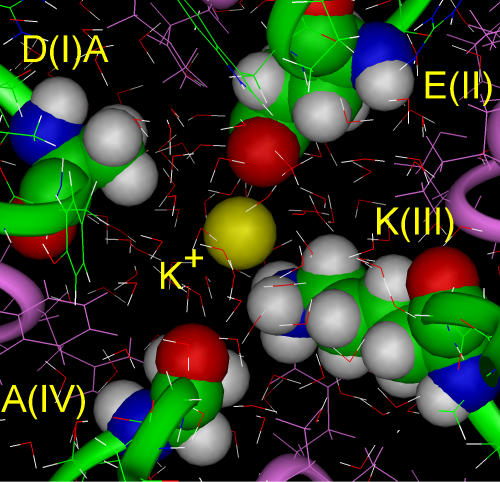
Initiation of the MD simulation with a K+ ion in the AEKA model also produced a new configuration of the selectivity filter, with K+ interacting with the carboxylate group of glutamate(II) and repelling lysine(III) in the direction of alanine(IV) (Fig. 2). However, the K+ ion (r = 1.33 A) is larger than Na+ and its electrostatic attraction to glutamate(II) is consequently weaker. The K+ ion is located midway between the carboxylate and the amino group. In this situation the energies of attraction to glutamate(II) and repulsion for lysine(III) are approximately equal (∼8 kcal/mol), interfering with the interaction of K+ with glutamate(II). In contrast to Na+, the electrostatic interaction of K+ with glutamate(II) does not overcome electrostatic repulsion from the side chain of lysine(III). K+ binding is therefore energetically less favorable than Na+ binding, but not completely precluded inside the selectivity filter, presumably reducing K+ current.
Figure 2.
A steady-state configuration for AEKA + K+ (yellow ball). During MD simulation the K+ ion breaks the hydrogen bond between glutamate(II) and lysine(III), but remains midway between glutamate(II) and lysine(III), making K+ binding less favorable.
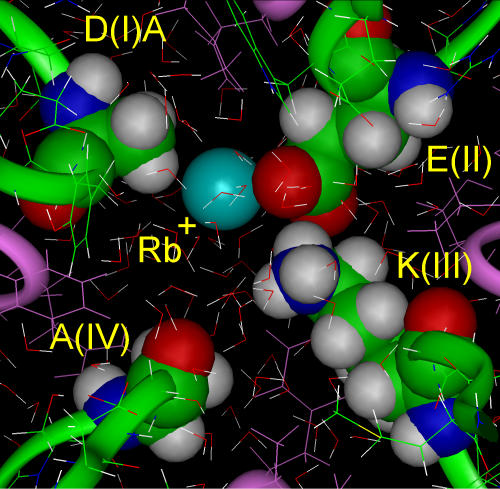
AEKA + Rb+.
In the row of univalent alkali metal ions the next larger ion is Rb+ (r = 1.48 Å), which is not permeable in the Na+ channel (Sun et al., 1997). MD simulation with Rb+ showed that it interacted partially with glutamate(II) inside AEKA (Fig. 3), but this electrostatic interaction is weaker than that of lysine(III). Consequently, the side chains of glutamate(II) and lysine(III) maintained the same optimal configuration found for the AEKA motif in the absence of cations. Rb+ did not compete successfully with the glutamate–lysine interaction and lysine(III) is able to exclude Rb+ from binding inside the selectivity filter.
Figure 3.
A steady-state configuration for AEKA + Rb+ (blue ball). The relationship between glutamate(II) and lysine(III) is unchanged by presence of Rb+.
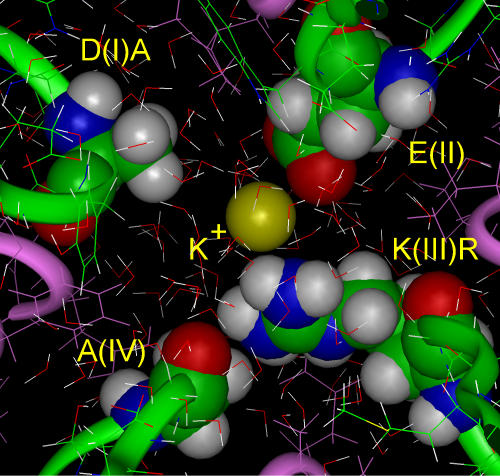
AERA + K+.
Because substitution of lysine(III) by arginine with the same positive charge leads to the loss of selectivity for Na+ over K+ (PK/PNa = 0.9; Favre et al., 1996), we tested the model AERA. MD simulation of K+ in the AERA selectivity filter shows that K+ interacted with glutamate(II) and successfully repelled arginine(III), displacing it toward alanine(IV) (Fig. 4). The reason for this success is that the arginine-positive charge is distributed on five hydrogens of the wide and flat guanidinium group, so that some of the partial charges are at a greater distance from K+ than with the amino group of lysine(III). Consequently, arginine(III) is less able to interfere with K+ interaction with glutamate(II). The approximation of the energy of glutamate(II) interaction with K+ is −8 kcal/mol and arginine(III) interaction is +6.5 kcal/mol. In contrast to lysine(III), arginine(III) does not prevent interaction of K+ with its binding site on glutamate(II) within the selectivity filter, presumably thereby permitting K+ permeation.
Figure 4.
A steady-state configuration for AERA + K+. During MD simulation K+ successfully repels arginine(III) from its interaction with glutamate(II).
DAKA Interactions
The DAKA channel has a PK /PNa ratio of 0.09, threefold less than the ratio for WT of 0.03 (Favre et al., 1996), but the channel is still quite Na+ selective. As with AEKA we have one carboxylate and a lysine, although they are located opposite to each other in the modeled DAKA configuration. Electrostatic forces result in their side chains extending toward each other. In the MD simulations they do not form a hydrogen bond, but are separated by a molecule of water (Lipkind and Fozzard, 2000). Consequently, Na+ could interact directly with the aspartate(I), with less interference by lysine(III). When Na+ was introduced in the MD simulation, it interacted with aspartate(I) with more attractive energy than the repulsive energy with lysine(III) (−12.5 and +7.5 kcal/mol), favoring binding of Na+. In contrast, the larger K+ ion is located farther away from aspartate(I) and the electrostatic energy calculation showed that the interactions were about equal with aspartate(I) and lysine(III) (−9 and +8.5 kcal/mol). This suggests that in the DAKA selectivity filter Na+ is able to overcome the interaction between aspartate(I) and lysine(III) and bind to aspartate, but K+ is less effective. Even though relatively high selectivity remains in the absence of glutamate(II), Schlief et al. (1996) reported that the mutant DQKA has a low single channel conductance, and Chiamvimonvat et al. (1996) reported that DCKA had a fourfold reduction in conductance. As before, replacement of lysine(III) by arginine in MD simulations of DARA decreased the repulsion of K+, allowing K+ to bind to the aspartate(I). The DARA mutant has not been studied experimentally, but we would predict that it also would be freely K+ permeant.
Cation Interaction in the WT DEKA Model
Where is the Na+ binding site in the WT Na channel that contains both carboxylates? Either aspartate(I) or glutamate(II), in conjunction with lysine, results in a channel that is selective for Na+ over K+, so the combination is also selective for Na+. From mutational data it seems most likely that the carboxylate of glutamate(II) is the binding site for Na+, while aspartate(I) may only produce a negative potential that increases permeation rate by offsetting the negative field of lysine(III). Replacement of aspartate(I) by alanine did not alter the PK/PNa ratio (Favre et al., 1996), and substitution of aspartate(I) by alanine in Nav1.4 did not change the reversal potential relative to the WT channel (43.1 mV), while a similar mutation of glutamate(II) shifted it negatively to 33.6 mV (Penzotti et al., 1998; Hilber et al., 2005). Selectivity can apparently be achieved by the side chains of only two amino acid residues, glutamate(II) and lysine(III).
Glutamate(II) most likely repels aspartate(I) in the WT channel, excluding aspartate from participating in the selectivity process. There is experimental support for the suggestion that aspartate(I) is physically located away from the critical selectivity region of glutamate-lysine, probably below it. Chiamvimonvat et al. (1996) reported that the cysteine mutant of aspartate(I) could not be modified by any of the methanesulfonate derivatives they tested (MTSEA and MTSET), although cysteine mutants of glutamate(II) and lysine(III) could be, implying that aspartate(I) is deeper in the pore or shielded in some way. Their Cd2+ block is also supportive of this idea. IC50 values for Cd2+ block of the mutants D400C and E755C were 192 and 0.03 μM, correspondingly. However, when glutamate(II) is removed, the new configuration may reorient aspartate(I) to interact with lysine(III) to allow it to participate actively in selectivity.
Impressions from the MD Analysis
Two concepts have traditionally been used in considering the structural basis of selectivity in cation channels; one assumes substantial dehydration of the cation and the other assumes minimal dehydration. For the first approach, the channel is considered to be a narrow and rigid cylindrical pore with an inner diameter close to that of the bare, unhydrated permeating ion (Mullins, 1959; Bezanilla and Armstrong, 1972; Hille, 1973), so that the large energy requirement for dehydration of the cation is compensated by interaction with oxygens lining the pore walls. The x-ray structure of the K+-selective channel (KcsA; Doyle et al., 1998) is consistent with this idea. The carbonyl oxygens may be somewhat flexible and mutually interact (Noskov et al., 2004), but almost complete dehydration of the cation is still required. Alternatively, Eisenman (1962) proposed another possible mechanism for cationic selectivity that did not require structural rigidity of the selectivity filter or substantial dehydration of the cation. He considered an ion-exchange equilibrium reaction in ion-selective glass and its energetic and thermodynamic factors. In this concept cation selectivity results from a balance between the hydration energy of an ion and its electrostatic interaction with a “binding site”, implying a strong field strength site for the Na+ channel and a selectivity sequence of Li+ > Na+ > K+ > Rb+ > Cs+. The Na+ channel selectivity filter is formed by flexible side chains, and it seems to present an example of the Eisenman mechanism that permits such side chain flexibility and allows the cation to remain mostly hydrated.
The Eisenman concept of a strong-field-strength anionic site in the selectivity region of a channel favoring the smaller Na+ over the larger K+ is supported by the presence of several carboxylates in the Na+ channel selectivity ring. However, charge density at the interactive surface of Na+ is only about twice that of K+ (spherical approximation), but the channel selectivity is 10–30 to 1 (Hille, 2001). Mutational experiments (DAAA, AEAA, AAEA, DEAA, DQEA, NEEA, DECA, DEDA, EEEA, DEEE, EEEE) demonstrate clearly that one or more carboxylates in the Na+ channel selectivity filter without an accompanying lysine do not confer Na+ selectivity (Chiamvimonvat et al., 1996; Favre et al., 1996; Schlief et al., 1996). In addition, the four carboxylates of the Ca2+ channel allow permeation of univalent cations in the absence of divalent cations, but the channel does not distinguish between Na+ and K+. The simplest application of the Eisenman concept is therefore insufficient to achieve selectivity in the Na+ channel, because the inclusion of a lysine is absolutely required for Na+/K+ selectivity. It is not simply the positive charge, because arginine substitution (DERA) yields a nonselective channel. Although arginine has the same charge as lysine, it is distributed over a large guanidinium group, emphasizing that for selectivity, short-range local field intensity on the scale of a few Å is important. Flexibility of the positively charged residue and its precise structural and chemical interaction with the carboxylate of glutamate(II) is also essential because replacing lysine(III) with cysteine and rendering the site positive by addition of an amino group by MTSEA interaction fails to restore selectivity (Perez-Garcia et al., 1997). Furthermore, the lysine environment is important, since moving it to a different location in the selectivity ring (DKEA) or adding a third carboxylate (DEKD) reduces selectivity and conductance (Schlief et al., 1996).
A model of selectivity based on a balance of electrostatic and steric interactions has been presented by Nonner et al. (2000) and Boda et al. (2007). In their model, the selectivity region is a small rigid confined volume containing four oxygens, each with one half of a unitary negative charge, a positively charged ammonium group, and a Na+ or a K+ ion. The oxygens correspond to the two flexible carboxylate side chains of DEKA, and the ammonium to the butyl ammonium group of lysine. These charged moities are freely mobile in the defined small volume and interact in response to electrostatic and van der Waals forces. Selectivity in this model results from the size difference between cations. They conclude that any small pore with a −1 permanent charge and side chains that occupy a significant small volume is a Na+-selective channel. However, experimental studies argue against this proposal. Experimental removal of two of the oxygens (AEKA or DAKA) or addition of two more oxygens (DEKD) changes selectivity modestly or not at all. Their model has no specific role for lysine other than provision of a positive charge. However, lysine is essential for selectivity, since its replacement with the larger arginine (DERA) abolishes selectivity. The experimental results and our calculations are more consistent with a defined cation binding site, modified by short range electrostatic forces from nearby tethered charges.
Noskov and Roux (2007) have studied a flexible four-carbonyl model of the K+ channel pore (“toy model”) to explore K+/Na+ selectivity. If one carbonyl is changed to a carboxylate, so that the site is now composed of one carboxylate, three carbonyls, and two water molecules, binding is highly selective for Na+. The closest Na+ channel mutants to this modeled structure are DAAA and AEAA, both of which are experimentally completely nonselective for univalent cations and permeable even for Ca2+ (Favre et al., 1996). Perhaps the channel fails to be selective because the Na+ channel selectivity ring is sufficiently rigid that it cannot collapse around the cation in the way that it occurs in the “toy model”. Their model also provides no role for lysine, which is essential for selectivity in the Na+ channel.
How does the essential lysine achieve selectivity? Our calculations show that it adds a precise repulsive electrostatic force that must be subtracted from the Na+-carboxylate oxygen interaction energy and from the K+-carboxylate oxygen interaction energy. Although it reduces the Na+-carboxylate interaction energy, it nulls the K+ interaction energy and makes Rb+ interaction impossible, so that Na+ binding is greatly favored. The binding site for cations is normally blocked by the side chain of lysine(III), and only strong alkali metal cations (Li+, Na+) can compete for interaction with the carboxylates of the selectivity filter. The repulsive energy of lysine required for WT selectivity is precise, because reducing it by replacing lysine with the larger arginine (AERA) abolishes selectivity. The location of a carboxylate on both sides of the lysine (DKEA and DEKD/E) does have a measurable modest effect on selectivity, perhaps because lysine interaction with the critical carboxyl is reduced by its interaction with the carboxylate on its other side. A reasonable test of this suggestion would be determination of selectivity of the mutations AKEA and DKAA, which would be unchanged from WT only if position within the ring is not important at all. The dependence of selectivity on extent of separation of the carboxylate and amino group could be considered in mutant configurations such as ADKA and EAKA.
Several features of selectivity of the WT Na+ channel selectivity filter are not well explained by these MD calculations. Mutational data are clear that a selectivity ring with only glutamate(II) and lysine(III) are sufficient for WT selectivity, and the MD calculations for the WT DEKA ring do show that Na+ binds preferentially to glutamate(II), but the role of aspartate(I) is not resolved. The reason that the model makes sense when only one carboxylate is present is that the model of the DEKA used for these calculations does not fully represent the true structure of the selectivity region of the Na channel, which is probably more complicated than the symmetrical arrangement we have proposed on the basis of the KcsA crystal structure. Perhaps in the WT channel, aspartate(I) contributes mainly by maintaining a negative potential at the selectivity ring (McNulty et al., 2007) and only affects selectivity in the absence of glutamate(II).
In addition to the carboxylates in the selectivity ring, the outer vestibule has four others located perhaps 9–10 Å above the selectivity ring. Their neutralization by mutation or pH titration (Chiamvimonvat et al., 1996; Khan et al., 2002) has minimal or no effect on selectivity, but it has large effect on permeation rate, presumably because of a substantial negative electrostatic field (McNulty et al., 2007). Similarly, mutational addition of charge ∼9–10 Å below the selectivity ring did not affect selectivity, but it did alter permeation rate (McNulty et al., 2007).
In summary, selectivity in the Na+ channel depends on both composition and conformation of the selectivity filter residues, derived from its four nonidentical domains. We propose a plausible energetic mechanism for selectivity with electrostatic competition between alkali cations and the glutamate(II)-lysine(III) residue pair. The extensive experimental data and our MD calculations allow us to draw several conclusions:
(1) The primary interaction site for cations in the Na+ channel selectivity filter is the carboxylate of glutamate(II). It is normally blocked by a hydrogen bond to the side chain of lysine(III).
(2) Only strong alkali metals (Li+, Na+) can compete with this interaction between the carboxylate(II) and the amino group(III), displacing lysine laterally toward alanine(IV). These ions need to lose only one or two waters of hydration for this interaction.
(3) Larger cations (K+, Rb+) do not compete successfully with the amino group of lysine(III), excluding those ions from the carboxylate of glutamate(II) and preventing permeation.
(4) The repulsive energy between alkali cations and lysine(III) is precise, because reduction of the electrostatic interaction by replacing lysine with arginine (with its distributed positive charge) fails to maintain selectivity.
(5) The influence of conformation dependence of the selectivity filter residues remains to be defined experimentally by studying mutations with paired carboxylate and lysine in different positions in the ring, such as AKEA, DKAA, ADKA, and EAKA.
Acknowledgments
We appreciate the advice of Dr. Dorothy Hanck.
This work was supported by USPHS NHLBI RO1 65661.
Olaf S. Andersen served as editor.
Abbreviation used in this paper: MD, molecular dynamics.
References
- Bezanilla, F., and C.M. Armstrong. 1972. Negative conductance caused by entry of sodium and cesium ions into potassium channels of squid axons. J. Gen. Physiol. 60:588–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda, D., W. Nonner, M. Valisko, D. Henderson, B. Eisenberg, and D. Gillespie. 2007. Steric selectivity in Na channels arising from protein polarization and mobile side chains. Biophys. J. 93:1960–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiamvimonvat, N., M.T. Perez-Garcia, R. Ranjan, E. Marban, and G.F. Tomaselli. 1996. Depth asymmetries of the pore-lining segments of the Na channel revealed by cysteine mutagenesis. Neuron. 16:1037–1047. [DOI] [PubMed] [Google Scholar]
- Doyle, D.A., J.M. Cabral, R.A. Pfuetzner, A. Kuo, J.M. Gulbis, S.L. Cohen, B.T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Eisenman, G. 1962. Cation selective glass electrodes and their mode of operation. Biophys. J. 2(Suppl. 2):259–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre, I., E. Moczydlowski, and L. Schild. 1996. On the structural basis for ionic selectivity among Na, K, and Ca in the voltage-gated sodium channel. Biophys. J. 71:3110–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, H.R., and P. Seetharamulu. 1986. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. USA. 83:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, S.H., H. Terlau, W. Stühmer, K. Imoto, and S. Numa. 1992. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 356:441–443. [DOI] [PubMed] [Google Scholar]
- Hilber, K., W. Sandner, T. Zarrabi, E. Zebedin, O. Kudlacek, H.A. Fozzard, and H. Todt. 2005. Selectivity filter residues contribute unequally to pore stabilization in voltage-gated sodium channels. Biochemistry. 44:13874–13882. [DOI] [PubMed] [Google Scholar]
- Hille, B. 1971. The permeability of the sodium channel to organic cations in myelinated nerve. J. Gen. Physiol. 58:599–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 1973. Potassium channels in myelinated nerve. Selective permeability to small cations. J. Gen. Physiol. 61:669–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 1975. Ionic selectivity, saturation, and block in sodium channels. A four-barrier model. J. Gen. Physiol. 66:535–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Inc., Sunderland, MA. 814 pp.
- Kim, M.S., T. Morii, X. Sun, K. Imoto, and Y. Mori. 1993. Structural determinants of ion selectivity in brain calcium channels. FEBS Lett. 318:145–148. [DOI] [PubMed] [Google Scholar]
- Khan, A., L. Romantseva, A. Lam, G.M. Lipkind, and H.A. Fozzard. 2002. Role of outer A. ring carboxylates of the rat skeletal muscle sodium channel pore in proton block. J. Physiol. 543:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., J.W. Kyle, D.A. Hanck, G.M. Lipkind, and H.A. Fozzard. 2006. Isoform-dependent interaction of voltage-gated sodium channels with protons. J. Physiol. 576:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind, G., and H.A. Fozzard. 1994. A structural model of the tetrodotoxin and saxitoxin binding site of the Na+ channel. Biophys. J. 66:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind, G.M., and H.A. Fozzard. 2000. KcsA crystal structure as framework for a molecular model of the Na channel pore. Biochemistry. 39:8161–8170. [DOI] [PubMed] [Google Scholar]
- McNulty, M.M., G.B. Edgerton, R.D. Shah, D.A. Hanck, H.A. Fozzard, and G.M. Lipkind. 2007. Charge at the lidocaine binding site residue Phe-1759 affects permeation in human cardiac voltage-gated sodium channels. J. Physiol. 581:741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, L.J. 1959. Penetration of some cations into muscle. J. Gen. Physiol. 42:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonner, W., L. Catacuzzeno, and B. Eisenberg. 2000. Binding and selectivity in L-type calcium channels: a mean spherical approximation. Biophys. J. 79:1976–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov, S.Y., S. Berneche, and B. Roux. 2004. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834. [DOI] [PubMed] [Google Scholar]
- Noskov, S.Y., and B. Roux. 2007. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzotti, J.L., H.A. Fozzard, G.M. Lipkind, and S.C. Dudley Jr. 1998. Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the outer vestibule of the Na channel. Biophys. J. 75:2647–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garcia, M.T., N. Chiamvimonvat, R. Ranjan, J.R. Balser, G.F. Tomaselli, and E. Marban. 1997. Mechanisms of sodium/calcium selectivity in sodium channels probed by cysteine mutagenesis and sulfhydryl modification. Biophys. J. 72:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran, A., L. Schild, and E. Moczydlowski. 1991. Divalent cation selectivity for external block of voltage-gated sodium channels prolonged by batrachotoxin: Zn2+ induces discrete substates in cardiac Na channels. J. Gen. Physiol. 97:89–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom, M.S.P., G.R. Smith, C. Adcock, and P.C. Biggin. 1997. The dielectric properties of water within model transbilayer pores. Biophys. J. 73:2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlief, T., R. Schonherr, K. Imoto, and S.H. Heinemann. 1996. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur. Biophys. J. 25:75–91. [DOI] [PubMed] [Google Scholar]
- Simonson, T., and D. Perahia. 1995. Internal and interfacial dielectric properties of cytochrome c from molecular dynamics in aqueous solution. Proc. Natl. Acad. Sci. USA. 92:1082–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C., R. Sankararamakrishnan, S. Subramanian, and E. Jakobsson. 1996. Solvation, water permeation, and ionic selectivity of a putative model for the pore region on the voltage-gated sodium channel. Biophys. J. 71:2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.-M., I. Favre, L. Schild, and E. Moczydlowski. 1997. On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. J. Gen. Physiol. 110:693–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau, H., S.H. Heinemann, W. Stühmer, M. Pusch, F. Conti, K. Imoto, and S. Numa. 1991. Mapping the site of block by tetrodotoxin and saxitoxin of the sodium channel. FEBS Lett. 293:93–96. [DOI] [PubMed] [Google Scholar]
- Tsushima, R.G., R.A. Li, and P.H. Backx. 1997. Altered ionic selectivity of the sodium channel revealed by cysteine mutations within the pore. J. Gen. Physiol. 109:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, S., and S.B. Rempe. 2007. Tuning ion coordination architectures to enable selective partitioning. Biophys. J. 93:1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, T., M. Janecki, E. Marban, and G.F. Tomaselli. 1997. Topology of the P segments in the sodium channel revealed by cysteine mutagenesis. Biophys. J. 73:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., P.T. Ellinor, W.A. Sather, J.F. Zhang, and R.W. Tsien. 1993. Molecular determinants of Ca selectivity and ion permeation in L-type Ca channels. Nature. 366:158–161. [DOI] [PubMed] [Google Scholar]
- Zhu, S.-B., and W. Robinson. 1992. Molecular dynamics computer simulation of an aqueous NaCl solution: structure. J. Chem. Phys. 97:4336–4348. [Google Scholar]