Abstract
Proteins with multiple cysteine residues often require disulfide isomerization reactions before they attain their correct conformation. In prokaryotes this reaction is catalysed mainly by DsbC, a protein that shares many similarities in structure and mechanism to the eukaryotic protein disulfide isomerase. This review discusses the current knowledge about disulfide isomerization in prokaryotes.
Keywords: protein folding, disulfide bond formation, isomerization
1. Introduction
Disulfide bonds are important in the folding of secreted proteins. The formation of disulfides can conceptually be divided into two steps, the oxidation of cysteine residues to form disulfides, and the isomerization of incorrect disulfides to the correct disulfide bonded pattern. For proteins with more than two cysteines, the number of possible incorrect disulfide bonding patterns increases very rapidly with cysteine number, whereas the number of correct disulfide bonding patterns remains constant at one. Thus, disulfide isomerases play vital roles in the folding of proteins, particularly for proteins with multiple cysteine residues. In the early 1960s, Anfinsen hypothesized the need for a protein to catalyze disulfide isomerization and discovered an endoplasmic reticulum protein, later called protein disulfide isomerase (PDI), involved in this process [1, 2]. More recently, prokaryotes were shown to contain a related protein called DsbC that is involved in disulfide isomerization. Both PDI and DsbC act by thiol-disulfide exchange with secreted proteins, and function to catalyze protein folding [3]. This review focuses on disulfide isomerization in prokaryotes, particularly in E. coli since it is the best studied. The process of disulfide oxidation is only described as it is relevant to disulfide isomerization.
2. DsbA is the main oxidase in the periplasm of E. coli
Oxidative protein folding is catalyzed by the Dsb proteins present in the periplasmic space of prokaryotes [4]. DsbA serves as the direct donor of disulfides to secreted proteins, whereas DsbC serves as the isomerase of incorrect disulfide bonds. Strains that are mutant in the dsbA gene show a severe defect in disulfide bond formation.
DsbA acts by donating its disulfide to secreted proteins, thereby catalyzing their oxidative folding. DsbA oxidizes reduced polypeptides extremely rapidly, with rate constants of about 107 M−1s−1 at pH 7.5. This is about a million times faster than normal values for disulfide exchange reactions between proteins [43]. DsbA is also one of the most oxidizing thiol disulfide oxidoreductases known, with a redox potential of −121 mV [5]. Given the very oxidizing redox potential of DsbA, it is not surprising that it can form incorrect disulfides. One reasonable model of DsbA function is that it oxidizes adjacent cysteines (consecutive disulfides) as they are secreted into the periplasmic space. Proteins secreted by the commonly used “sec” secretion apparatus are secreted from the N to C terminus. Thus, one simple model would be that DsbA may only be able to form correct disulfides if they all form between cysteines that are adjacent in the sequence [6]. Any other disulfide configuration (non-consecutive disulfide) would then require the disulfide isomerase DsbC. Thus, the paradigm that has been guiding the disulfide bond formation field for some time is that DsbA functions entirely as an oxidizing thiol-disulfide oxidoreductase, whereas DsbC functions as a disulfide isomerase. This is in contrast to the situation in the eukaryotic field, in which a single protein disulfide isomerase is thought to play roles in both isomerization and oxidation.
3. DsbB is a membrane protein that reoxidizes DsbA
The oxidation of a substrate protein by DsbA reduces DsbA. In order to be catalytic, DsbA must be reoxidized; this reoxidation is performed by the membrane protein DsbB. DsbB generates protein disulfides de novo, via the reduction of quinones [8, 9]. DsbB is thus the primary source of disulfides in prokaryotic organisms. Strains that have mutations in the dsbB gene showed accumulation of reduced DsbA in the periplasm and exhibited the same pleiotropic phenotypes as dsbA mutants. DsbB is reoxidized by components of the respiratory chain, which accept electrons from DsbB. Under aerobic conditions, DsbB transfers the electrons to oxidized ubiquinone. The reduced ubiquinone then becomes reoxidized by the terminal cytochrome oxidases, which finally transfer the electrons onto molecular oxygen [8, 10]. Under anaerobic conditions, DsbB transfers electrons to menaquinone, which then passes them to the final electron acceptors fumarate or nitrate [8]. DsbB has four membrane-spanning segments and two pairs of essential cysteines, Cys41-Cys44, and Cys104-Cys130. One of each pair is located in each of the two periplasmic domains [11]. These cysteines are involved in a disulfide cascade reaction within DsbB that ends up transferring a disulfide to DsbA [9].
4. DsbC, a periplasmic isomerase
The role of the disulfide isomerase proteins DsbC and possibly DsbG is the isomerization of incorrect disulfides [3, 12, 13]. These members of the Dsb family were identified by two different genetic approaches [3, 13]. DsbC and DsbG share 24% sequence identity with each other and about 10% sequence identity to thioredoxin [3, 14].
DsbC can rearrange disulfides in vitro and in vivo. The addition of catalytic amounts of DsbC to fully reduced and denatured BPTI in vitro promotes the formation of correct disulfides in the presence of GSH/GSSG and leads to active BPTI [15]. In this case glutathione oxidizes BPTI and potentially incorrect disulfides are isomerized by DsbC. In an in vivo study BPTI was oxidized in the presence of DsbA, leading to mispaired disulfides, which were corrected in a DsbC dependent manner. This is consistent with results that we have obtained with RNaseA. DsbA can oxidize reduced RNaseA to the point that no free thiols are available, but this does not result in significant RNaseA activity; only upon addition of DsbC is RNaseA activity recovered [16]. We interpreted this to mean that the DsbA oxidizes the protein to completion but incorporates at least one incorrect disulfide, and DsbC then reshuffles the disulfides to the correct configuration. As shown in the figure there are several possible mechanisms by which DsbC may isomerizes incorrect disulfide bonds. DsbC attacks incorrect disulfides forming a mixed disulfide between the more N terminal cysteine of DsbC and the protein substrate (step 1 in the figure). This disulfide may be immediately transferred back to the protein substrate resulting in a different and in this case correct disulfide bond pattern (step 2 in the figure). This mechanism is called the shuffling model for the purposes of this review. Alternatively the mixed disulfide may be resolved such that DsbC is oxidized and the substrate is reduced (step 3 in the figure) The reduced substrate may then be reoxidized by DsbA or DsbC (step 4 or 5 in the figure). This mechanism is called the reduction/reoxidation model. This may result in a correct pattern of disulfides. Since correct disulfides are, in general, stable and buried they will not be attacked by reduced DsbC so steps 2 and 5 can be considered essentially irreversable. Incorrect disulfides in contrast are expected to be exposed and susceptible to attack and will undergo another round of isomerization. Seps 1, 3 and 4 can be considered to be reversable. It should be noted that often, particularly in the case where consecutive disulfides are correct, DsbA will correctly form disulfides in the first place, eliminating the need for isomerization.
Figure.
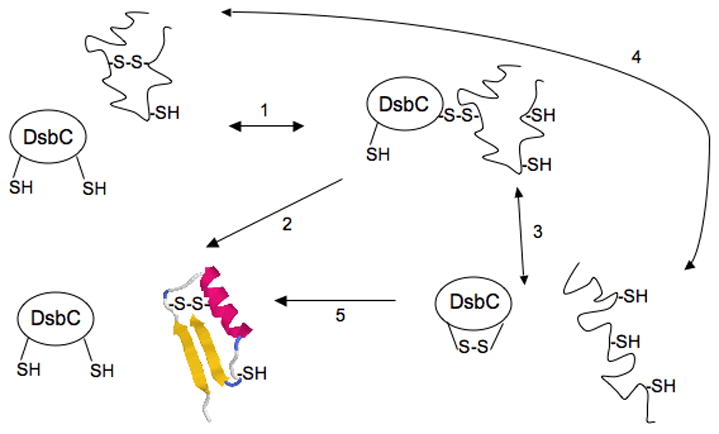
Some evidence against the shuffling model and in favor of the reduction/reoxidation mechanism are the observations by Rietsch et al [33]. They showed that DsbC mutants that lack the more C terminal cysteine are strongly deficient in the expression of urokinase, implying that they are deficient in isomerization. They also showed that in a DsbA, DsbD double mutant, DsbC’s active site cysteines are in a reduced state, while in a DsbD mutant DsbC is in the oxidized state. This seems to indicate that the oxidation of DsbC’s active site cysteines is dependent on DsbA. Since DsbA and DsbC do not directly interact, the oxidation of DsbC likely takes place through DsbA oxidizing protein substrates which are then reduced by DsbC, favoring the reduction/isomerization model.
Compared to PDI, a well-studied eukaryotic isomerase, DsbC is not as catalytically active, with PDI being 10-fold faster in regard to the disulfide rearrangement in bovine pancreatic trypsin inhibitor (BPTI) [15]. Compared to DsbA, however, DsbC is much more active as an isomerase in the in vitro refolding of Ragi bifunctional inhibitor (RBI), a protein with five nonconsecutive disulfides. DsbC seems to be particularly efficient in rearranging buried, nonnative disulfides that have been formed by treatment of RBI with oxidants such as GSSG [17].
In vivo DsbC has been shown to increase the yield of overall correctly disulfide bonded E. coli and eukaryotic proteins with non-consecutive disulfide bonds. For example, the formation of active murine urokinase containing 12 disulfides is decreased 100-fold in a dsbC− strain compared to the wild-type strain, which indicates that DsbC is required for formation of the correct disulfide bonds [18]. Joly and Swartz investigated the expression of a variety of eukaryotic proteins in a dsbC− strain [19]. The influence of DsbC on the formation of active eukaryotic proteins strongly depends on the model protein used, and whether the disulfides are consecutive or nonconsecutive. For melanoma growth stimulating activity (MGSA) and insulin-like growth factor 1 (IGF-1), proteins that require nonconsecutive disulfides, the absence of DsbC reduced the amount of expressed protein. However, human growth hormone (hGH) and anti-CD18 antibody, proteins that contain consecutive disulfides, showed similar expression levels in a dsbC− and a wild-type strain [19]. The assembly of pertussis toxin, which requires 11 consecutive disulfides, requires DsbA but not DsbC [20]. Periplasmic proteins whose disulfide bonding pattern are known to be decreased in amount in dsbC− strains include RNaseI, and phytase, both of which contain a non-consecutive disulfide [21]. This ability of DsbC to specifically assist in the expression of proteins with nonconsecutive disulfides strongly suggests that DsbC acts as an isomerase.
Perhaps the strongest evidence for a specific requirement of DsbC in folding proteins with nonconsecutive disulfides comes from experiments conducted with phytase. Phytase is an E. coli periplasmic protein that contains three consecutive disulfide bonds and one nonconsecutive one. Its expression is DsbC-dependent, unless the nonconsecutive disulfide is removed by mutation, when it becomes DsbC-independent [6]. It has been proposed that DsbA acts immediately on proteins as they are translocated into the periplasm. Evidence for this also comes from a mutant in alkaline phosphatase (AP) in which the most amino-terminal cysteine was replaced with serine. Alkaline phosphatase normally contains two consecutive disulfides, one that links the first and second cysteines in the protein, and one that links the third and fourth. Upon removal of the first cysteine in AP, DsbA forms an incorrect disulfide. This disulfide is slowly resolved in a DsbC-dependent reaction. Interestingly, more active alkaline phosphatase results from expression of this mutant AP in a dsbA null strain than is formed in a dsbA+ strain. This implies that DsbA may be too oxidizing for the expression of proteins with nonpaired cysteines [7]. However, a recent report found that RNAseI is oxidized correctly by DsbA, even though it has a nonconsecutive disulfide[44].
The abundance of a number of proteins, including MepA, End1, YebF, and Ivy, is decreased in the absence of DsbC, suggesting that these E. coli proteins are in vivo substrates for DsbC [22]. A larger number of proteins are decreased in amount in dsbA− strains, implying that DsbA, as the presumptive oxidase, is more important for the folding of proteins than is DsbC, the presumptive isomerase. Darby and Creighton measured the rates at which a 28-mer model peptide reacts with DsbC and compared it with the rates at which DsbA reacts with the same peptide. To function as an isomerase, DsbC needs to attack incorrect disulfide bonds. The initial rate at which DsbC reacts with the model peptide’s disulfide is about 6 × 103 more rapid than the same reaction of the oxidized peptide with reduced DsbA. For isomerization to occur, a free thiol needs to attack this mixed disulfide, resulting in a different disulfide. This reaction occurs 26- to 43-fold more rapidly with DsbC than it does with DsbA. These kinetic differences help explain the greater isomerase activity of DsbC than DsbA [23].
The crystal structure of oxidized DsbC [24] reveals two 23 kDa monomers of DsbC forming a V shaped homodimer [24]. Each monomer of DsbC consist of two domains: a C-terminal thioredoxin-like domain containing the active site Cys98-Gly-Tyr-Cys102 motif, a structural disulfide, and an N-terminal dimerization domain connected via a flexible linker helix. Mutations that remove the active site cysteines, as expected, show that they are required for the thiol-disulfide exchange activities of DsbC, but not the chaperone activity. Mutations that remove the structural disulfide increase the sensitivity of DsbC to denaturants and decrease its folding rate [25]. Dimerization of DsbC acts to form a large cleft built of uncharged and hydrophobic residues, which may be the binding site for substrate proteins of DsbC. The active site cysteines of each monomer are oriented facing each other. The thioredoxin-like domains are separated from the dimerization domain by a linker alpha helix. Relative movement of the actives sites and the presumptive peptide binding cleft at the junction of the arms of the V may be important in DsbC’s isomerase or chaperone activities. It has been suggested that the cleft may act to both bind and partially shield incompletely folded proteins while the protein explores various conformations and disulfide bonding patterns [24]. DsbC reacts with peptides up to a million-fold faster than it reacts with glutathione, and the mixed disulfide formed between the peptide and DsbC is 10,000 times more stable than glutathione-DsbC mixed disulfides. Noncovalent binding interactions that occur between the peptide and DsbC likely contribute to these very rapid kinetics of substrate utilization and the increased stability of the mixed disulfide [23]. In reduced DsbC, the catalytic domains are shifted and twisted up to 13 Å around an alpha helical hinge located between the catalytic and dimerization domains. This apparent flexibility of DsbC may play a role in accommodating substrates in the cleft between the catalytic domains.
The function of DsbC as isomerase and chaperone is strictly dependent on it’s dimerization [26]. A screen to find mutants of dsbC that complement a dsbA deficiency revealed that mutations in the dimerization domain of DsbC lead to monomeric DsbC that can act as an oxidase and, in addition, can interact with DsbB to become reoxidized [27]. The dimerization of DsbC therefore, is a structural safeguard against the interaction with DsbB, protecting the chaperone and isomerase functions of DsbC. A heterodimer that has had one of the CXXC active sites inactivated by carboxymethylation has no isomerase activity [26]. This suggests strongly that both active sites of the dimer are required for isomerization.
Deletion mutations in the alpha helical linker domain that connects the dimerization domain to the catalytic domain allow DsbC to be oxidized by DsbB and also decrease DsbC’s isomerization activity [29, 30]. In wild-type DsbC, the active site cysteines are facing each other; these deletion mutations probably function by rotating the catalytic domains with respect to each other. The ability of the mutants to oxidize proteins was inversely related to their ability to isomerize incorrect disulfide bonds. A number of orientations (including an inward pointing orientation) of the active sites appear to be associated with high isomerase activity; other orientations leave DsbC accessible to DsbB, and DsbC thus acquires oxidase activity. All of the alpha helical linker truncations tested by Segatori et al. are capable of serving as both an oxidant and an isomerase [29, 30]. This mimics the situation present in eukaryotes, in which protein disulfide isomerase is thought to serve both roles.
Further evidence that the presence of two active sites is important in facilitating isomerase action is the observation that the addition of a dimerization domain onto monomeric thioredoxin-like proteins generates a protein with significant isomerase activity in vivo and in vitro. Dimerization of DsbA, thioredoxin, or monomeric PDI domains by the fusion of these proteins to the dimerization domain of DsbC results in a protein that is capable of supporting the formation of active vtPA in the periplasm and RNaseA in vitro, indicating that these artificial dimeric proteins have acquired disulfide isomerization activity [29, 30]. Formation of a mixed disulfide between these dimeric disulfide isomerases and their substrates will greatly increase the local concentration of the active sites relative to the substrate. In contrast, monomeric disulfide oxidoreductases like thioredoxin and DsbA are very inefficient disulfide isomerases. This higher isomerase activity of DsbC compared to DsbA is proposed to be due to a number of factors in addition to the increased effective concentration of active sites upon formation of mixed disulfides; these factors include a higher peptide affinity and a slower and less random oxidation of substrates by DsbC.
It appears that the equilibrium redox potential alone does not determine whether a protein primarily functions in the cell as an oxidase or an isomerase, since DsbC and DsbA both have very similar and strongly oxidizing redox potentials, and both react very rapidly with folding proteins and peptides [15]. The ability of mutants in DsbA to catalyze the folding of multidisulfide proteins does not correlate with their redox potential [31]. To function as an isomerase, DsbC is kept in the reduced form in vivo by DsbD [ 18, 19, 28, 32, 33]. In contrast, DsbA functions as an oxidase in part because it is kept in the oxidized state by DsbB. Large kinetic barriers separate the oxidation pathway from the isomerization pathway. Nonphysiological interactions between DsbC-DsbB, DsbA-DsbD, and DsbC-DsbA are three to seven orders of magnitude slower than the physiological interactions [34].
DsbC mutants are copper sensitive. Copper is a redox metal, which catalyzes disulfide bond formation and appears to introduce incorrect disulfides more frequently than DsbA does. Thus, the copper sensitivity of dsbC− strains may arise from the inability of the cell to rearrange copper-catalyzed incorrect disulfide bonds. These results suggest that the primary role of DsbC may be to rearrange incorrect disulfide bonds that are formed during certain forms of oxidative stress [35]. The structure of DsbC in complex with a fragment of DsbD revealed that the large cleft can adopt two conformations: an open and a closed conformation [36]. This flexibility presumably allows DsbC to adopt the cleft for different substrates. The interface between the DsbC-DsbD alpha subunit involves only residues from the thioredoxin domain of DsbC. The contact residues are highly conserved, which helps explain how a variety of DsbC homologues isolated from gamma proteobacteria are capable of complementing E. coli DsbC.
In addition to functioning as an isomerase, DsbC also has chaperone activity and can assist the refolding of single chain Fv fragments [37], lysozyme, and glyceraldehyde-3-phosphate-dehydrogenase (GAPDH). For the latter two proteins, this chaperone activity is independent of the presence of DsbC’s active site cysteines [25]. The dual mode of action of DsbC is probably the key to its functionality. As a chaperone, DsbC recognizes misfolded proteins, then the isomerase function resolves incorrectly formed disulfides. DsbC’s ability to bind to peptides probably contributes to its chaperone activity. A mixed disulfide between a peptide derived from BPTI and DsbC is 100-fold more stable than between the peptide and DsbA [23].
5. DsbG, a second disulfide isomerase?
Although multigene families are less common in prokaryotes than they are in eukaryotes, they do occasionally occur, the disulfide isomerase family is one example. In general individual members of multigene families are assumed to have different substrate specificities, patterns of expression, or localizations. In some cases overlapping substrate specificity and expression patterns may lead to redundancy. A second putative isomerase in the periplasm of E. coli, DsbG, was discovered using a DTT based screen and because of its 24% sequence identity to DsbC [12]. Originally DsbG was described as a disulfide oxidase [12], but it is now thought that DsbG acts as a second isomerase in the periplasm of E. coli [14]. In vitro, DsbG has chaperone and isomerase activity [14, 38]. Overexpression of DsbG, like DsbC overexpression, enhances the proper expression of single chain Fv antibody fragments, and the effect of DsbC and DsbG appear to be additive. However, DsbG does not catalyze insulin reduction, a standard thiol-disulfide redox reaction, and fails to catalyze the oxidative refolding of RNaseA [14]. The structures of DsbC and DsbG closely resemble each other [39]. However, DsbG is significantly larger because the helix that links the dimerization and catalytic domains is about 2.5 turns longer in DsbG. This acts to substantially increase the dimensions of the substrate binding cleft. In addition, DsbG possesses some conserved acidic residues in the generally hydrophobic substrate binding cleft that are not present in DsbC. These differences have led to the hypothesis that DsbC and DsbG vary in substrate specificity. DsbC is proposed to interact with unfolded hydrophobic polypeptide chains, whereas DsbG is proposed to interact with larger, at least partially folded substrates with charged surfaces [39]. Although a number of E. coli substrates are known for DsbC, none have yet been detected for DsbG. DsbG is normally expressed at lower levels than is DsbC. Since no protein that is abundant under normal laboratory growth conditions is present in dramatically reduced concentrations in DsbG null mutants it seems likely that DsbG is acting on proteins that are not normally abundant. In vivo, overexpression of DsbG in dsbC− strains allows the production of comparable levels of BPTI as in dsbC+ E. coli strains, suggesting that DsbG also functions in vivo as an isomerase [14]. Mutations in DsbG have been isolated that allow it to complement the copper sensitivity of dsbC null strains [40].
That simple overexpression of the isomerases DsbC and DsbG assists in the folding of a number of eukaryotic proteins indicates that the periplasm of E. coli can be altered to improve the folding of eukaryotic disulfide containing proteins. Most peptides and proteins with therapeutic applications contain disulfides. The proper formation of these disulfides is one of the rate limiting steps in the commercial production of these proteins. Because the purification of the correctly disulfide bonded form of the protein as the active pharmaceutical ingredient turns out to be very complicated when multiple possibilities of disulfide bonding are possible, these therapeutic proteins are very expensive. Further optimization of the periplasm for thiol-disulfide exchange is clearly of great interest for the commercial production of disulfide containing proteins. It has been observed that DsbC variants that contain alterations in the dipeptide sequence between the active site cysteine residues increase the yield of mouse urokinase up to fivefold [31]. Further, optimization of the periplasm for thiol-disulfide exchange is clearly of great interest for the commercial production of disulfide containing proteins. Selection for chromosomal mutations that enhance the folding of tissue plasminogen activator yielded mutants that up-regulate DsbC due to mutations that result in reduced function of RNaseE. This suggests a critical role of RnaseE in regulating DsbC expression [41].
6. Comparison between the prokaryotic disulfide isomerases DsbC and DsbG, and eukaryotic PDI
The recently solved crystal structure of eukaryotic PDI reveals a number of striking similarities to DsbC and DsbG, including domain arrangement, active site location, and surface features [42]. Interestingly, these structural similarities are in large part due to convergent evolution. Nevertheless, the overall effect is very similar. Both prokaryotic and eukaryotic disulfide isomerases have a V or U shape, with two active-site CXXC motifs embedded in a thioredoxin-like fold that face each other across the U. The eukaryotic PDI achieves this by covalently linking four thioredoxin domains, with the N-and C-terminal domains possessing the active-site CXXC motifs, which face each other across the interior of the U; the two internal domains are catalytically inactive. In contrast, DsbC and DsbG utilize a thioredoxin unrelated domain to dimerize two monomers, each of which possesses a single CXXC active site containing a thioredoxin-like domain. It appears that two thioredoxin-like domains in close spatial proximity on opposite sides of a large cleft are required for disulfide isomerase activity. Individual catalytic domains of PDI, though redox active, are essentially inactive as isomerases, are essentially inactive as isomerases, and heterodimers of DsbC in which one active CXXC motif is carboxymethylated is also inactive as an isomerase in vivo(26). The two active sites of PDI presumably act in a concerted fashion on its substrates.
Acknowledgments
We thank our anonymous reviewers for useful suggestions. This work was supported by Grants from the NIH to JCAB. JCAB is a Howard Hughes Investigator
Abbreviations
- BPTI
bovine pancreatic trypsin inhibitor
- PDI
protein disulfide isomerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldberger RF, Epstein CJ, Anfinsen CB. Purification and properties of a microsomal enzyme system catalyzing the reactivation of reduced ribonuclease and lysozyme. J Biol Chem. 1964;239:1406–10. [PubMed] [Google Scholar]
- 2.Anfinsen CB, Haber E. Studies on the reduction and re-formation of protein disulfide bonds. J Biol Chem. 1961;236:1361–3. [PubMed] [Google Scholar]
- 3.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. Embo J. 1994;13:2013–20. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–9. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Zapun A, Bardwell JC, Creighton TE. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry. 1993;32:5083–92. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 6.Berkmen M, Boyd D, Beckwith J. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J Biol Chem. 2005;280:11387–94. doi: 10.1074/jbc.M411774200. [DOI] [PubMed] [Google Scholar]
- 7.Sone M, Akiyama Y, Ito K. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J Biol Chem. 1997;272:10349–52. doi: 10.1074/jbc.272.16.10349. [DOI] [PubMed] [Google Scholar]
- 8.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–27. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. Embo J. 1999;18:1192–8. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader MW, Xie T, Yu CA, Bardwell JC. Disulfide bonds are generated by quinone reduction. J Biol Chem. 2000;275:26082–8. doi: 10.1074/jbc.M003850200. [DOI] [PubMed] [Google Scholar]
- 11.Jander G, Martin NL, Beckwith J. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. Embo J. 1994;13:5121–7. doi: 10.1002/j.1460-2075.1994.tb06841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen CL, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–32. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 13.Shevchik VE, Condemine G, Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. Embo J. 1994;13:2007–12. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessette PH, Cotto JJ, Gilbert HF, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–92. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 15.Zapun A, Missiakas D, Raina S, Creighton TE. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry. 1995;34:5075–89. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 16.Bader M, Muse W, Zander T, Bardwell J. Reconstitution of a protein disulfide catalytic system. J Biol Chem. 1998;273:10302–7. doi: 10.1074/jbc.273.17.10302. [DOI] [PubMed] [Google Scholar]
- 17.Maskos K, Huber-Wunderlich M, Glockshuber R. DsbA and DsbC-catalyzed oxidative folding of proteins with complex disulfide bridge patterns in vitro and in vivo. J Mol Biol. 2003;325:495–513. doi: 10.1016/s0022-2836(02)01248-2. [DOI] [PubMed] [Google Scholar]
- 18.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:13048–53. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joly JC, Swartz JR. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–72. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 20.Stenson TH, Weiss AA. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infect Immun. 2002;70:2297–303. doi: 10.1128/IAI.70.5.2297-2303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiniker A, Bardwell JC. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J Biol Chem. 2004;279:12967–73. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 22.Vertommen D, Depuydt M, Pan J, Leverrier P, Knoops L, Szikora JP, Messens J, Bardwell JC, Collet JF. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol. 2007 doi: 10.1111/j.1365-2958.2007.06030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darby NJ, Raina S, Creighton TE. Contributions of substrate binding to the catalytic activity of DsbC. Biochemistry. 1998;37:783–91. doi: 10.1021/bi971888f. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy AA, Haebel PW, Torronen A, Rybin V, Baker EN, Metcalf P. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol. 2000;7:196–9. doi: 10.1038/73295. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Wang CC. Disulfide-dependent folding and export of Escherichia coli DsbC. J Biol Chem. 2001;276:1146–51. doi: 10.1074/jbc.M004929200. [DOI] [PubMed] [Google Scholar]
- 26.Sun XX, Wang CC. The N-terminal sequence (residues 1–65) is essential for dimerization, activities, and peptide binding of Escherichia coli DsbC. J Biol Chem. 2000;275:22743–9. doi: 10.1074/jbc.M002406200. [DOI] [PubMed] [Google Scholar]
- 27.Bader MW, Hiniker A, Regeimbal J, Goldstone D, Haebel PW, Riemer J, Metcalf P, Bardwell JC. Turning a disulfide isomerase into an oxidase: DsbC mutants that imitate DsbA. Embo J. 2001;20:1555–62. doi: 10.1093/emboj/20.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collet JF, Riemer J, Bader MW, Bardwell JC. Reconstitution of a disulfide isomerization system. J Biol Chem. 2002;277:26886–92. doi: 10.1074/jbc.M203028200. [DOI] [PubMed] [Google Scholar]
- 29.Segatori L, Murphy L, Arredondo S, Kadokura H, Gilbert H, Beckwith J, Georgiou G. Conserved role of the linker alpha-helix of the bacterial disulfide isomerase DsbC in the avoidance of misoxidation by DsbB. J Biol Chem. 2006;281:4911–9. doi: 10.1074/jbc.M505453200. [DOI] [PubMed] [Google Scholar]
- 30.Segatori L, Paukstelis PJ, Gilbert HF, Georgiou G. Engineered DsbC chimeras catalyze both protein oxidation and disulfide-bond isomerization in Escherichia coli: Reconciling two competing pathways. Proc Natl Acad Sci U S A. 2004;101:10018–23. doi: 10.1073/pnas.0403003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bessette PH, Qiu J, Bardwell JC, Swartz JR, Georgiou G. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J Bacteriol. 2001;183:980–8. doi: 10.1128/JB.183.3.980-988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. Embo J. 1995;14:3415–24. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–8. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozhkova A, Stirnimann CU, Frei P, Grauschopf U, Brunisholz R, Grutter MG, Capitani G, Glockshuber R. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. Embo J. 2004;23:1709–19. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiniker A, Collet JF, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280:33785–91. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- 36.Haebel PW, Goldstone D, Katzen F, Beckwith J, Metcalf P. The disulfide bond isomerase DsbC is activated by an immunoglobulin-fold thiol oxidoreductase: crystal structure of the DsbC-DsbDalpha complex. Embo J. 2002;21:4774–84. doi: 10.1093/emboj/cdf489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsumoto K, Umetsu M, Yamada H, Ito T, Misawa S, Kumagai I. Immobilized oxidoreductase as an additive for refolding inclusion bodies: application to antibody fragments. Protein Eng. 2003;16:535–41. doi: 10.1093/protein/gzg064. [DOI] [PubMed] [Google Scholar]
- 38.Shao F, Bader MW, Jakob U, Bardwell JC. DsbG, a protein disulfide isomerase with chaperone activity. J Biol Chem. 2000;275:13349–52. doi: 10.1074/jbc.275.18.13349. [DOI] [PubMed] [Google Scholar]
- 39.Heras B, Edeling MA, Schirra HJ, Raina S, Martin JL. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc Natl Acad Sci U S A. 2004;101:8876–81. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiniker A, Ren G, Heras B, Zheng Y, Laurinec S, Jobson RW, Stuckey JA, Martin JL, Bardwell JC. Laboratory evolution of one disulfide isomerase to resemble another. Proc Natl Acad Sci U S A. 2007;104:11670–5. doi: 10.1073/pnas.0704692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan X, Gao J, Jain C, Cieslewicz MJ, Swartz JR, Georgiou G. Genetic analysis of disulfide isomerization in Escherichia coli: expression of DsbC is modulated by RNase E-dependent mRNA processing. J Bacteriol. 2004;186:654–60. doi: 10.1128/JB.186.3.654-660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 43.Stafford SJ, Lund PA. Mutagenic studies on human protein disulfide isomerase by complementation of Escherichia coli dsbA and dsbC mutants. FEBS Lett. 2000;466:317–22. doi: 10.1016/s0014-5793(99)01728-7. [DOI] [PubMed] [Google Scholar]
- 44.Darby NJ, Raina S, Creighton TE. Contributions of Substrate Binding to the Catalytic Activity of DsbC. Biochemistry. 1998;37:783–791. doi: 10.1021/bi971888f. [DOI] [PubMed] [Google Scholar]
- 45.Messens J, Collet J, Van Belle K, Brosens E, Loris R, Wyns The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem. 2007;282:31302–7. doi: 10.1074/jbc.M705236200. [DOI] [PubMed] [Google Scholar]


