Abstract
Kinesin is a processive motor protein: A single molecule can walk continuously along a microtubule for several micrometers, taking hundreds of 8-nm steps without dissociating. To elucidate the biochemical and structural basis for processivity, we have engineered a heterodimeric one-headed kinesin and compared its biochemical properties to those of the wild-type two-headed molecule. Our construct retains the functionally important neck and tail domains and supports motility in high-density microtubule gliding assays, though it fails to move at the single-molecule level. We find that the ATPase rate of one-headed kinesin is 3–6 s−1 and that detachment from the microtubule occurs at a similar rate (3 s−1). This establishes that one-headed kinesin usually detaches once per ATP hydrolysis cycle. Furthermore, we identify the rate-limiting step in the one-headed hydrolysis cycle as detachment from the microtubule in the ADP⋅Pi state. Because the ATPase and detachment rates are roughly an order of magnitude lower than the corresponding rates for two-headed kinesin, the detachment of one head in the homodimer (in the ADP⋅Pi state) must be accelerated by the other head. We hypothesize that this results from internal strain generated when the second head binds. This idea accords with a hand-over-hand model for processivity in which the release of the trailing head is contingent on the binding of the forward head. These new results, together with previously published ones, allow us to propose a pathway that defines the chemical and mechanical cycle for two-headed kinesin.
Conventional kinesin is a motor protein that transports membrane-bound vesicles and organelles along microtubules in neurons and other cells (1–3). An important functional property of kinesin is that it is processive: Individual motor molecules can move continuously along the surface of a microtubule for several microns without dissociating (4–6). Processivity is likely to be an adaptation to ensure that organelles or vesicles, even small ones that contain few motors on their surfaces, are transported reliably over long distances.
Processivity implies that a kinesin molecule has only a small probability of dissociating from the microtubule during each cycle of ATP hydrolysis. On average, a single kinesin molecule moves along the surface of a microtubule through a distance of 5 μm in microtubule gliding assays (4), 1.4 μm in bead assays (5), and 0.6–1.3 μm in fluorescence assays (6, 7). Because the movement takes place in 8-nm steps (8, 9), the spacing of the adjacent tubulin dimers that form kinesin’s consecutive binding sites (10), and because each step is associated with one cycle of ATP hydrolysis (11–13), kinesin is expected to hydrolyze some 80–600 ATP molecules before it dissociates from the microtubule. This accords with radionucleotide studies (14). These observations show that the probability of kinesin dissociating during any one ATP hydrolysis cycle is only ≈1%. Because kinesin remains processive even against high loads of up to several piconewtons (9, 15, 16), kinesin must spend <1 μs in a detached state per hydrolysis cycle (16), and it is likely that kinesin maintains continuous attachment to the microtubule.
Processivity relies on kinesin having two motor domains. Kinesin is a homodimer. Each subunit contributes one globular motor domain, also called a head, and two subunits dimerize via amphipathic regions that form a series of coiled coils (17–20). When one of the heads is removed by coexpressing the full-length subunit with a truncated subunit whose head has been deleted, no motility is observed in single-molecule motility assays (21). Failure of this one-headed heterodimer to move is not attributable to loss of motor activity per se because the construct moves in high-density motility assays in which five or more motors cooperate to move a microtubule (21). Kinesin head fragments, monomeric constructs that contain only the motor domain, also fail to move in single-molecule assays (6, 22, 23); the one exception (24) may have dimerized under the motility conditions.
One model for processivity, the “hand-over-hand” model, postulates that the motor maintains continuous attachment to the microtubule because the release of the bound head is contingent on the binding of the other, so there is always one head bound (4). This model is supported by the observation from microtubule gliding assays that the rate at which the one-headed heterodimer detaches from a microtubule (0.3 s−1) is much smaller than the rate at which the individual heads of a homodimer detach from the microtubule during movement [50 s−1 when the dimeric motor is moving at 800 nm/s (21)]; a second head is needed to accelerate release of the first head.
However, kinetic experiments on monomeric kinesins, constructs lacking the dimerization and tail domains, argue against the hand-over hand model. At high ATP concentration, the one-headed monomers detach more quickly (not less quickly) than the individual heads in the corresponding homodimers [range of 22–46 s−1 for the monomer and 20–44 s−1 for the homodimer from the ATPase rate per head (25–30)]. This suggests an alternative, “kinetic” model for processivity whereby the motor remains attached not because the heads are coordinated but because the rate of binding of the free head is simply much faster than the rate of release of the bound head.
To resolve this issue, we have measured the ATPase activities and the rates of detachment from microtubules of individual one-headed heterodimers and two-headed homodimers bound to 200-nm-diameter silica beads. The results support a coordinated, hand-over-hand model for two-headed kinesin that is consistent with a large body of biochemical, structural, and motility studies.
Materials and Methods
ATPase assays.
Single-headed kinesin heterodimer was made by co-expressing the full-length Drosophila kinesin heavy chain with a truncated heavy chain that is missing the N-terminal 340 amino acids (21). This construct includes the native tail, rod, and dimerization domains but is missing one head domain. Motors were purified as described (21), the concentration of active protein was determined by radionucleotide binding (13), and microtubule-stimulated ATPase rates were determined by using malachite green to assay phosphate liberation (13). The ATPase rates were measured in BRB80 (80 mM Pipes/1 mM EDTA/1 mM MgCl2, pH 6.9) augmented with 1 mM MgATP, 10 μM taxol, 1 mg/ml casein, motors (typically 1–2 nM), and microtubules (at the indicated tubulin dimer concentration). Single-headed ATPase data are from four to seven experiments on at least two different kinesin preparations. ATPase data were fit to the Michaelis-Menten equation by using igor (Wavemetrics, Lake Oswego, OR), with 1/SEM2 weighting. Two-hundred-nanometer-diameter silica beads (SS02N, Bangs Laboratories, Fischers, IN) were sonicated in the presence of casein (typically 6 nM beads, 6 mg/ml casein in BRB80 buffer) for 3 hours to ensure a homogeneous solution of casein-coated beads. To adsorb motors, kinesin and beads (typically 3 nM of each) were rapidly mixed and incubated for 30 minutes on ice. Motor activity, as determined by radionucleotide binding, was unaffected by adsorption to beads, as shown previously (13).
Visualization of Beads in Binding and Motility Assays.
Microtubules (typically 400 nM tubulin dimer in BRB80 plus 10 μM taxol) were infused into flow cells constructed with silanized coverslips (13) and were allowed to adsorb to the surface for 3 minutes. The free microtubules then were washed out with BRB80 containing 2 mg/ml BSA and 10 μM taxol. Three minutes later, the motor-bead solution (BRB80 containing 10 pM motor-beads, 1 mg/ml casein, 10 μM taxol, and 1 mM ATP, unless otherwise stated) was introduced. All stock nucleotide solutions contained equimolar magnesium. The level of ATP contamination in the ADP stock was determined to be ≤0.2% by thin layer chromatography [cellulose polyethyleneimine F (J. T. Baker, Phillipsburg, NJ), developed in 0.6 M potassium phosphate buffer (pH 3.4) and visualized with short wavelength UV light].
Microtubules and beads were visualized with a Zeiss Axiovert microscope using differential interference contrast optics, were filmed with a Dage-MTI NC-67MD camera (Michigan City, IN), and images were processed with a Hamamatsu (Bridgewater, NJ) image processor. Experiments were videotaped for later analysis. The attachment and detachment of kinesin-coated beads were measured by frame-by-frame analysis of the video records, samples of which are published as supplemental data on the PNAS web site, www.pnas.org. The high diffusion coefficient of the beads (≈2 μm2/s) permitted detection of binding events of duration as short as 33 ms, corresponding to one full video frame, during which time a free bead undergoes a rms displacement of ≈600 nm. Rate constants for the detachment of motors from microtubules in ATP and ADP were calculated by fitting a single-exponential function to the duration histogram; data were fit to the square root of the bin counts as appropriate for counting statistics. The slow off rates in adenosine 5′-(β,γ-imino)-triphosphate (AMP-PNP) or no nucleotide were calculated by dividing the total number of releases by the total observation time for all beads. Detachment data in Fig. 2 c and d were fit to the Michaelis-Menten equation.
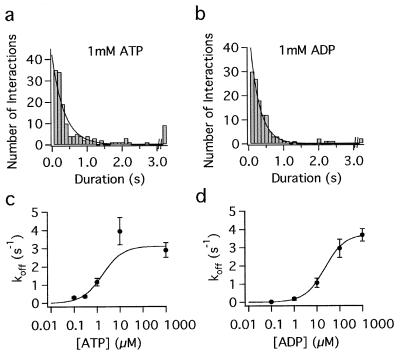
Figure 2.
Duration of attachments of one-headed kinesin (1 motor/bead) to microtubules in 1 mM ATP (a) and 1 mM ADP (b). The last bin denotes all events >3 s in duration. Lower panels show the dependence of the detachment rate on the ATP concentration (c) and ADP concentration (d). Nucleotide concentrations for half maximal off rates were 1.7 μM ATP and 22 μM ADP, respectively.
Results
The ATPase Rate of One- and Two-Headed Kinesins.
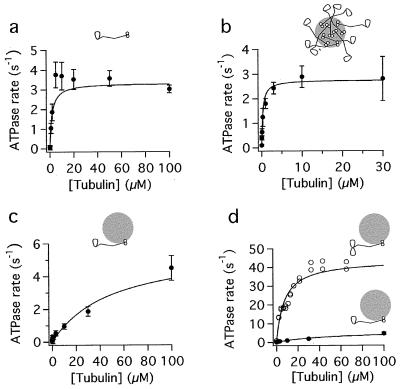
The steady-state ATPase rate of the one-headed heterodimers was measured by incubating the motors with various concentrations of polymerized tubulin in the presence of saturating ATP (1 mM) and monitoring the formation of the inorganic phosphate using a colorimetric assay. Measurements were made under three conditions: motors free in solution, motors adsorbed to beads at a high motor:bead ratio, and motors adsorbed to beads at 1 motor/bead.
In solution (i.e., in the absence of beads), the maximum microtubule-stimulated ATPase rate of one-headed kinesin was 3.3 ± 0.2 ATP s−1 (Fig. 1a; Table 1). This measurement could underestimate the true ATPase for two possible reasons. First, the protein may be in a partially inhibited conformation—in solution, two-headed kinesin is folded up (31) and has a low ATPase activity compared with the unfolded conformation (32). Second, the protein preparation used for these assays also contains headless protein formed by the dimerization of the truncated chains (21). This motorless, rod-tail protein may inhibit the one-headed heterodimers, just as the tail domain inhibits the ATPase activity of two-headed kinesin (32), although the inhibition is expected to be only slight because the concentration of the headless protein in the ATPase assays (≤10 nM) is lower than the inhibitory constant for the tails [≈100 nM (32)].
Figure 1.
Microtubule-stimulated ATPase rates of one-headed kinesin heterodimer and two-headed kinesin homodimer. (a) One-headed kinesin in solution. (b) One-headed kinesin adsorbed to beads at 500 motors/bead. (c) One-headed kinesin adsorbed to beads at 1 motor/bead. (d) Comparison of one-headed heterodimer (filled circles) and two-headed kinesin homodimer (open circles) adsorbed at 1 motor/bead. Note: beads are not drawn to scale.
Table 1.
Summary of ATPase parameters
| Motor | Condition | Km, μM tubulin | Vmax, ATP/s per head |
|---|---|---|---|
| One-headed | In solution | 1.8 ± 0.5 | 3.3 ± 0.2 |
| One-headed | 1 motor/bead | 50 ± 24 | 5.8 ± 1.9 |
| One-headed | 500 motors/bead | 0.29 ± 0.06 | 2.8 ± 0.2 |
| Two-headed | 1 motor/bead | 7.4 | 44 |
Shown are Michaelis-Menten parameters Vmax (maximum microtubule-stimulated ATPase rate) and Km (tubulin concentration, in polymerized form, for half-maximal ATPase rate). Two-headed data are from Coy et al. (13).
To circumvent these problems, one-headed motors were adsorbed to 200-nm silica beads before measuring the ATPase activity. Adsorption to beads did not result in any loss of radionucleotide binding activity of the motor domain, consistent with the protein binding to the bead surface via the rod or tail domains rather than the motor domain. Control experiments with two-headed protein showed that adsorption to beads fully activates the microtubule-stimulated ATPase to a rate of 44 s−1 per head [corresponding to 1 ATP hydrolyzed per 8-nm step (Table 1; refs. 13 and 32)]. When one-headed kinesin was bound to beads under high-density conditions (≈500 motors/bead), the ATPase rate was 2.8 ± 0.2 s−1 (Fig. 1b; Table 1). This is similar to the ATPase rate of the protein in solution.
The bead experiment also suffers from a potential problem: It is possible that, at this high density of motors on the beads, not all of the kinesin molecules are able to interact with microtubules (even at high concentrations of polymerized tubulin), and so again the ATPase rate might be underestimated. We therefore repeated these experiments with a low density of kinesin on the bead surfaces. Under conditions in which there were on average one kinesin heterodimer per 200-nm bead, the maximum microtubule-stimulated ATPase rate was 5.8 ± 1.9 s−1 (Fig. 1c; Table 1).
The one-motor-per-bead experiment suffers from a different sort of problem. The problem is that the Km is high: 50 μM polymerized tubulin is necessary to half-maximally activate the ATPase rate. Because of the high viscosity of the microtubule-containing solutions, we could not use more than 100 μM (≈10 mg/ml) polymerized tubulin in the ATPase assays, and so it was difficult to completely saturate the ATPase rate, although there were sufficient signs of saturation in the data that the curve-fitting program converged and gave a reliable estimate for the maximum microtubule-activated ATPase. Nevertheless, if the true Km were much greater than 50 μM, then the true maximally activated ATPase rate would be much higher than our estimated value. To rule out this possibility, we calculated the Km by an independent technique—by comparing the relative microtubule affinities of one- and two-headed kinesin observed in the bead binding assay. If the Km for microtubules is regulated by the fraction of time the motor spends bound to microtubules versus free in solution, then the ratio of dissociation constants (Kd = koff/kon) for the two motors should equal the ratio of their Kms. The binding of beads coated with one-headed kinesin to microtubules was directly observed by differential interference contrast microscopy. The binding rate was equal to 25 ± 10 (μM beads)−1⋅(μm microtubules)−1⋅s−1, similar to the corresponding rate for beads coated with two-headed kinesin [22 ± 5 (μM beads)−1⋅(μm microtubules)−1⋅s−1]. On the other hand, the rates at which the beads coated with one- and two-headed motors detached from the microtubules in saturating ATP were 2.9 ± 0.4 s−1 and 0.66 ± 0.16 s−1, respectively (Table 2; see below). Hence, we expect the Km for one-headed kinesin to be ≈4× greater than that for two-headed kinesin [≅(2.9/25)/(0.66/22)]. The Km for two-headed kinesin is reliably determined because the microtubule-stimulated ATPase is well saturated (Fig. 1d): Its value is 7.4 μM. Therefore, the expected Km for one-headed kinesin is ≈30 μM. This is in good agreement with the measured value of 50 μM and implies a maximum ATPase rate of 3–4 s−1. Thus the ATPase rate of 5.8 s−1 is not an underestimate, and there is little suggestion that the full-length one-headed kinesin is partially inhibited in solution like the two-headed protein.
Table 2.
Rate of detachment of kinesin from microtubules
| Nucleotide condition | One-headed rate, s−1 | Two-headed rate, s−1 |
|---|---|---|
| 1 mM ATP | 2.9 ± 0.4 | 48 ± 2 |
| 1 mM ADP | 3.7 ± 0.4 | 1.01 ± 0.28 |
| 1 mM ADP + 10 mM Pi | 3.8 ± 0.6 | 1.67 ± 0.50 |
| No nucleotide | 0.019 ± 0.007 | 0.0009 ± 0.0002 |
| 1 mM AMP-PNP | 0.009 ± 0.002 | 0.0010 ± 0.0004 |
All rates were measured for kinesin bound to beads (one motor/bead). In the case of two-headed kinesin in ATP, the detachment rate was calculated by dividing the bead velocity [770 nm/s (13)] by the step size of 16.2 nm per head; the observed rate for release of these beads from microtubules were 0.66 ± 0.16 s−1. 1 mM ADP samples contain ≈2 μM contaminating ATP, and no nucleotide contains 100 nM ADP and ≈1 nM ATP.
The Dissociation Rates of One- and Two-Headed Kinesins from Microtubules.
A key prediction of the hand-over-hand model is that a single head should stay attached to a microtubule until the other head binds. In other words, one-headed kinesin should detach only very slowly from microtubules. To test this prediction, the attachment and detachment of beads coated with one-headed kinesin at low density was observed by video-enhanced differential interference contrast microscopy. The distribution of attached times in the presence of 1 mM ATP is shown in Fig. 2a. Fitting an exponential to these durations gave a detachment rate of 2.9 ± 0.4 s−1.
This detachment rate (2.9 s−1) is ≈9× greater than the detachment rate that we measured from observing microtubules dissociate from glass surfaces sparsely coated with the same one-headed kinesin [0.31 s−1 (21)]. We believe that much of this discrepancy can be accounted for by the rebinding of the slowly diffusing microtubules to the surface: As pointed out by Block et al. (5), such rebinding probably accounts for the higher processivity observed in microtubule gliding assays [5 μm (4)] than in bead assays [1.4 μm (5)].
The Hydrolysis Cycle for One-Headed Kinesin.
Because the detachment rate of one-headed kinesin (≈3 s−1) is similar to its ATPase rate (3–6 s−1), one-headed kinesin usually detaches during each hydrolysis cycle, although it is possible that occasionally two ATPs are hydrolyzed while the enzyme remains attached to the microtubule. To determine which nucleotide state one-headed kinesin is in when it detaches, we measured detachment rates in the absence of nucleotides, in the presence of AMP-PNP (which is thought to trap the motor in an ATP-like state), and in the presence of various concentrations of ADP, ATP, and/or Pi (Fig. 2; Table 2). The detachment rates in the absence of nucleotide and in the presence of 1 mM AMP-PNP (0.019 s−1 and 0.009 s−1, respectively; Table 2) were very low, too low to account for the detachment rate in the presence of ATP (3 s−1). By contrast, the detachment rates in the presence of 1 mM ADP (3.7 s−1; Fig. 2 b and d; Table 2), as well as in 1 mM ADP and 10 mM Pi (3.8 s−1; Table 2), were similar to the detachment rate in the presence of ATP. Hence, detachment occurs when kinesin is in either the ADP or in the ADP⋅Pi state.
Detachment in the ADP state is ruled out by nucleotide release experiments from other laboratories. These experiments, described in the next paragraph, show that kinesin releases ADP very quickly when it is bound to a microtubule; the rate constant is 50–300 s−1 (26, 30). This fast release of ADP implies that, before one-headed kinesin in the ADP state could detach from the microtubule (at 3.7 s−1), the ADP itself would unbind (at 50–300 s−1). At the high ATP concentration used for the hydrolysis assays, ATP would then rapidly bind, and the result would be many cycles of ATP hydrolysis per dissociation (14 or 81, corresponding to 50/3.7 and 300/3.7 respectively), contrary to our observed one-to-one ratio (or perhaps two-to-one ratio) of ATP hydrolyzed per dissociation. Hence, the only remaining possibility is that one-headed kinesin detaches from the microtubule in the ADP⋅Pi state.
The rapid release of ADP from the microtubule⋅kinesin⋅ADP complex follows from experiments showing that, when two-headed kinesin binds to a microtubule, ADP is released from the first head at a rate of 50 s−1 (26) to 300 s−1 (30). We expect one-headed kinesin to release its ADP at a similar rate because the crystal structure of the kinesin homodimer (33) shows that the two heads make minimal contact with each other. Thus, provided that both heads are not bound to the microtubule (in which case the molecule may be strained), we do not expect that the presence of a second head should significantly affect the nucleotide release kinetics of the other head. Though we have not directly measured the ADP off rate from our one-headed motor because of the difficulty of obtaining sufficient protein for enzyme kinetic experiments, we do have indirect experiments that suggest that the off rate is indeed fast. The ADP concentration necessary for a half-maximal detachment rate is 22 μM (Fig. 2d), and, if the ADP on-rate is similar to the ATP on-rate (≈2 μM−1⋅s−1), as found in kinetic experiments on kinesin homodimer (34), then the 22 μM ADP concentration corresponds to an ADP off rate of 44 s−1, in agreement with the transient kinetic data.
Interaction of Kinesin⋅ADP with Microtubules.
Until now, kinesin has generally been thought to detach in the ADP state (26, 35–37), although detachment in the ADP⋅Pi state has been considered (29, 38). This prevailing view has been based on the weak binding of kinesin·ADP to microtubules (26, 35, 39). We performed two control experiments to confirm the slow detachment of kinesin·ADP from microtubules and to establish that spontaneous detachment in the ADP state is too slow to be part of the one- or two-headed hydrolysis cycle. First, to make sure that the slow dissociation in the ADP state was not a peculiarity of our one-headed construct, we measured the dissociation of individual two-headed molecules from microtubules in the presence of 1 mM ADP (in the absence of ATP). Consistent with the one-headed results, the dissociation rate was 1 s−1 (Table 2), too slow to be on the pathway for a motor that takes 100 steps per second. Second, in contrast with Romberg and Vale (35), we observed microtubules interacting for several seconds with individual, two-headed motors bound to the surfaces of flow cells coated with motors at low density in the presence of 1 mM ADP; at high motor density the microtubules bound almost irreversibly, even at 10 mM ADP. Thus, the ADP state has high enough affinity for microtubules to maintain the motor-microtubule contact in motility assays. The slow detachment of kinesin·ADP from microtubules is also supported by transient kinetic experiments in the literature (see Discussion).
Discussion
We have shown that one-headed kinesin heterodimer has a low ATPase rate (3–6 s−1), which is similar to the rate at which it detaches from microtubules in the presence of ATP (3 s−1). Because the corresponding rates of the two-headed motor are much greater (44 s−1 and 48 s−1, respectively, from Tables 1 and 2), we infer that the second head must accelerate the ATPase rate and the detachment of the other head at least 10-fold. These observations support a model in which the hydrolysis cycles of the two heads are tightly coupled and confirm a key prediction of the hand-over-hand model.
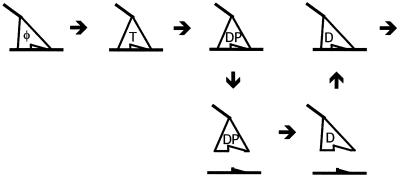
From single-molecule binding assays under various nucleotide conditions, we have argued that the one-headed heterodimer dissociates from the microtubule in the ADP⋅Pi state. This leads to a model for the one-headed hydrolysis cycle shown in Fig. 3. A corollary of this model is that the release of phosphate must be slow when the motor is attached to the microtubule for otherwise the detachment pathway would be bypassed, leading to many ATP hydrolyzed per dissociation (which is inconsistent with our results). This proposed cycle is analogous to myosin’s, but shifted one-quarter of a cycle: Myosin releases phosphate while attached to actin whereas kinesin releases phosphate after detachment from the microtubule. The slow release of phosphate while kinesin is bound to the microtubule could be attributable to a steric block of the exit path of phosphate—perhaps, like myosin (40), kinesin has a “back door” through which phosphate leaves and which is obstructed by the microtubule—or it could be attributable to an allosteric interaction between the microtubule and nucleotide binding sites.
Figure 3.
Model for the one-headed kinesin hydrolysis pathway at high ATP concentration and low ADP and Pi concentrations. T, ATP; D, ADP; P, Pi; φ, no nucleotide. For simplicity, nucleotide states not on the principal pathway and reverse rate constants are not shown. Our results argue that detachment occurs in the ADP⋅Pi state. Results from Hackney (41) show that, in the absence of microtubules, reversal of ATP hydrolysis is slow compared with phosphate release and that the rebinding of kinesin⋅ADP to the microtubule is necessary to accelerate ADP release.
The model for the one-headed cycle gives insight into which step may be regulated by the second head. If the detachment of kinesin⋅ADP⋅Pi is the rate-limiting step in the one-headed cycle, then this step must be accelerated, at least 10-fold, by the second head. The atomic structure of the homodimer shows that there are minimal contacts between the two heads, so it is natural to think that the acceleration of the release of the first head is attributable to intramolecular strain generated when the second head binds to the microtubule. According to this scheme, the ADP⋅Pi state acts as a checkpoint: The first head must wait in this attached state for the second head to bind before proceeding to the next mechanical step (detachment) and the subsequent chemical step (phosphate release). In this way, spontaneous detachment is kept low to ensure that two-headed kinesin has a high degree of processivity.
Before accepting these conclusions, however, it is necessary to explain why the one-headed monomers mentioned in the Introduction have such different enzyme activities from the one-headed heterodimer studied here. To facilitate the discussion of the one-headed monomer, we will first present our new model for the two-headed hydrolysis cycle (Fig. 4). The model includes a number of steps that should not be in dispute and should allow us to focus on those steps that are currently controversial.
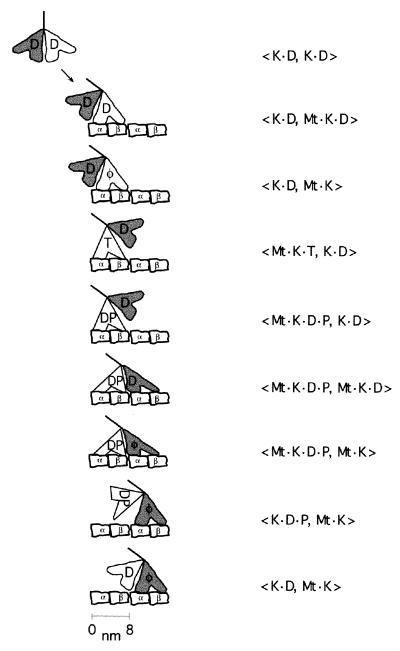
Figure 4.
Proposed two-headed hydrolysis cycle for kinesin. K, kinesin; Mt, microtubule. α and β are tubulin subunits; nucleotide abbreviations are same as in Fig. 3. The order of terms in the brackets denotes the position of the two heads along the microtubule from the minus- to the plus-end. See text for a detailed justification for this sequence of steps. Structural correlates for various states are as follows. (i) The 〈K⋅D, K⋅D〉 state corresponds to the crystal structure of homodimeric kinesin in the absence of microtubules (33). (ii) The 〈K⋅D, Mt⋅K⋅D〉 and the 〈K⋅D, Mt⋅K〉 states correspond to cryoEM reconstructions of microtubules decorated with kinesin in saturating ADP (47) and no nucleotide (47, 48), respectively, both of which show the detached head trailing the attached head in a poor orientation for microtubule binding. (iii) The 〈Mt⋅K⋅T, K⋅D〉 and 〈Mt⋅K⋅D⋅P, K⋅D〉 states correspond to the cryo-electron microscopic structures in AMP-PNP (48, 49) and in 1 mM ADP and 10 mM phosphate buffer (48, 50), respectively, in which the free head is situated closer to the next tubulin binding site.
Kinetic Model for Kinesin’s Two-Headed ATPase.
Kinesin in solution predominantly has ADP bound in both heads (〈K⋅D, K⋅D〉) because of the very slow release of ADP (≈0.01 s−1) (41).The binding of one head to the microtubule (〈K⋅D, Mt⋅K⋅D〉) greatly accelerates the release of ADP from this head [50–300 s−1 (26, 30)] to give the 〈K⋅D, Mt⋅K〉 state. In the absence of nucleotides, the second, free head releases its ADP only very slowly [range of 0.01–0.5 s−1 (26, 30, 42)]. However, the binding of ATP to the attached head, to give the 〈Mt⋅K⋅T, K⋅D〉 state, greatly accelerates the release of the second ADP (42) to a rate of 110–300 s−1 (26, 30). The nonhydrolyzable ATP analogue AMP-PNP also accelerates the release of the second ADP; this suggests that the free head in the 〈Mt⋅K⋅T, K⋅D〉 state also can bind to the microtubule and release its ADP. However, the release of the second ADP in AMP-PNP is only ≈30 s−1 (26, 30), an order of magnitude slower than the release in ATP and slower than the hydrolysis rate (26, 30). The simplest interpretation is that the nucleotide is hydrolyzed first to give 〈Mt⋅K⋅D⋅P, K⋅D〉, then the second head binds to give 〈Mt⋅K⋅D⋅P, Mt⋅K⋅D〉, and then ADP is released to give 〈Mt⋅K⋅D⋅P, Mt⋅K〉. Up to this stage, there should be little disagreement because the facts have all been confirmed in at least two laboratories.
If we assume, as argued above, that when one head is free the other head behaves like the one-headed protein, then our results imply that the bound head in the 〈Mt⋅K⋅D⋅P, K⋅D〉 state detaches much more slowly (≈4 s−1) than the free head attaches [≈300 s−1 (30)]. This ensures a high degree of processivity (≈75 = 300/4). Furthermore, if detachment of the head is a prerequisite for phosphate release, as we believe for the one-headed heterodimer, then the futile cycle 〈Mt⋅K⋅D⋅P, Mt⋅K〉 → 〈Mt⋅K⋅D, Mt⋅K〉 → 〈Mt⋅K, Mt⋅K〉, which completes a hydrolysis cycle without producing a step, is averted.
After detachment of the K⋅D⋅P head to form 〈K⋅D⋅P, Mt⋅K〉, the phosphate is assumed to release rapidly (the rate must be greater than the two-headed ATPase rate) to give 〈K⋅D, Mt⋅K〉. This completes one ATPase cycle, during which the two-headed motor has moved 8 nm to the next tubulin dimer. A hydrolysis cycle recently published by Cross et al. (37) includes a similar checkpoint at the phosphate release step; however, in that model, phosphate release occurs before detachment of the bound head.
Processivity, Stoichiometry, and the Discrepant One-Headed Motors.
The kinetics of the one-headed kinesin monomers do not fit this scheme. The rapid detachment of the monomers in the presence of ATP supports the kinetic model for processivity rather than the hand-over-hand model. Furthermore, the ATPase rates of the monomers are greater than those of the corresponding homodimers (25, 27–29), suggesting that the nucleotide hydrolysis cycles of the two heads are not tightly coupled as they are in the model presented here.
We have two arguments that suggest that the monomers have altered kinetic properties that make them poor models for a one-headed protein. First, the rapid detachment of the monomer from microtubules in the presence of ADP [80 s−1 (25); 50 s−1 (27)] is not consistent with the slow detachment of 〈Mt⋅K⋅D, K⋅D〉 from the microtubule. We find that two-headed kinesin in saturating ADP detaches from microtubules at the slow rate of 1 s−1. This slow detachment of 〈Mt⋅K⋅D, K⋅D〉 is also inferred from experiments in which ADP is rapidly added to 〈Mt⋅K, K⋅D*〉 and the release of ADP from the tethered head (denoted by D*) is monitored. At a high ADP concentration (to ensure rapid binding of ADP to the attached head) and high polymerized tubulin concentration (to ensure rapid rebinding to microtubules after detachment), the ADP* releases at a rate of only 2–4 s−1 (26, 30). But, if 〈Mt⋅K⋅D, K⋅D*〉 detached at a rate of 80 s−1, like the monomer, then the ADP* would release at a much higher rate, ≈40 s−1 (= 80/2 because there is only a 50% chance of the reattachment of the previously free head).
The second problem with the monomer is that, to account for the high degree of processivity for two-headed kinesin (<1% chance of detachment per cycle), a very high, possibly unrealistic, rate of binding of the free head must be assumed. The key branch-point in the cycle is 〈Mt⋅K⋅D⋅P, K⋅D〉. The release of phosphate from the attached head before the attachment of the free head leads to a futile hydrolysis cycle because it returns the motor to 〈K⋅D, Mt⋅K⋅D〉 without a forward step. If the rate at which the Mt⋅K⋅D head detaches from the microtubule [50–80 s−1 (25, 27)] is similar to the rate at which the ADP releases from that head [50–300 s−1 (26, 30)], then there is a 50% chance that 〈Mt⋅K⋅D, K⋅D〉 will detach, leading to loss of processive movement. Hence, to have a 1% chance of detachment per cycle, binding of the second head would need to be 50-fold faster than the rate of phosphate release from the attached head. If phosphate release is as fast as suggested from experiments on kinesin monomers [>200 s−1 (25, 27)], then the free head must attach at the improbably high rate of >10,000 s−1 to account for the observed processivity.
We believe that a simpler explanation is that truncation (perhaps the absence of the neck or dimerization domain) has created an uncoupled ATPase: The checkpoint that prevents phosphate from releasing while the motor is attached has been short-circuited, and the motor is able to rapidly detach in the ADP state. The net effect is an uncoupling of the chemical and mechanical cycles. Such uncoupling of motor and ATPase activities of truncated constructs was observed by Stewart et al. (43), who found that, as more and more of the dimerization domain was removed, the ATPase rate increased as the speed of movement decreased. It is significant that no group has succeeded in making any of the monomers used in the biochemical assays move, even in multiple-motor microtubule gliding assays. Motion is only observed when an artificial tail is fused with the motor domain (6, 22–24, 43, 44). By contrast, motility of the one-headed heterodimer is readily observed in high density assays (21). The simplest explanation is that the structural integrity of the neck/dimerization domain is necessary for the correct coupling of the nucleotide- and microtubule-binding sites in the motor domain.
The Force-Generating Step.
Our model provides a simple structural explanation for why kinesin slows down under high load. The rate-limiting step at high load must be associated with a large conformational change that brings the load (i.e., the dimerization domain) toward the plus end of the microtubule; only in this way can a force parallel to the axis of the microtubule couple to the reaction. We also know that the rate-limiting step is associated with a movement away from the surface of the microtubule because in the presence of a high parallel load, application of a perpendicular load that pulls the motor away from the surface of the microtubule increases the speed (45). It is apparent from Fig. 4 that the step that satisfies both these conditions is the release of the trailing head. If this is the case, then the physical reason why the motor slows down at high load is that the external force counteracts the force generated by the forward head, thereby canceling the accelerated detachment of the trailing head. Such a mechanism will help maintain kinesin’s processivity under high parallel loads because the motor will be pulled back into a state with both heads bound to the microtubule. Because slowed detachment in turn slows phosphate release, our model predicts that high loads will slow the ATPase rate, as argued by Visscher et al. (46), so that tight coupling between ATP turnover and the stepping rate will also be maintained under load.
Supplementary Material
Acknowledgments
We thank Drs. Albert Gordon, Ellen Lumpkin, Ravi Sawhney, Peter Detwiler, and Mr. Andy Hunter for comments on earlier versions of this manuscript. This work was supported by a grant from the U.S. Public Health Service (AR40593) to J.H.
Abbreviation
- AMP-PNP
adenosine 5′-(β,γ-imino)-triphosphate
References
- 1.Coy D L, Howard J. Curr Opin Neurobiol. 1994;4:662–667. doi: 10.1016/0959-4388(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 2.Bloom G S, Endow S A. Protein Profile. 1995;2:1109–1171. [PubMed] [Google Scholar]
- 3.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 4.Howard J, Hudspeth A J, Vale R D. Nature (London) 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 5.Block S M, Goldstein L S, Schnapp B J. Nature (London) 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 6.Vale R D, Funatsu T, Pierce D W, Romberg L, Harada Y, Yanagida T. Nature (London) 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romberg L, Pierce D W, Vale R D. J Cell Biol. 1998;140:1407–1416. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svoboda K, Schmidt C F, Schnapp B J, Block S M. Nature (London) 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 9.Kojima H, Muto E, Higuchi H, Yanagida T. Biophys J. 1997;73:2012–2022. doi: 10.1016/S0006-3495(97)78231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray S, Meyhofer E, Milligan R A, Howard J. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua W, Young E C, Fleming M L, Gelles J. Nature (London) 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 12.Schnitzer M J, Block S M. Nature (London) 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 13.Coy D L, Wagenbach M, Howard J. J Biol Chem. 1999;276:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 14.Hackney D D. Nature (London) 1995;377:448–450. doi: 10.1038/377448a0. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda K, Block S M. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 16.Meyhofer E, Howard J. Proc Natl Acad Sci USA. 1995;92:574–578. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J T, Laymon R A, Goldstein L S. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- 18.de Cuevas M, Tao T, Goldstein L S. J Cell Biol. 1992;116:957–965. doi: 10.1083/jcb.116.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang T G, Suhan J, Hackney D D. J Biol Chem. 1994;269:16502–16507. [PubMed] [Google Scholar]
- 20.Howard J. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- 21.Hancock W O, Howard J. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berliner E, Young E C, Anderson K, Mahtani H K, Gelles J. Nature (London) 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Hirokawa N. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y, Toyoshima Y Y, Iwane A H, Morimoto S, Higuchi H, Yanagida T. Proc Natl Acad Sci USA. 1997;94:7275–7280. doi: 10.1073/pnas.94.14.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y Z, Taylor E W. J Biol Chem. 1997;272:717–723. doi: 10.1074/jbc.272.2.717. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y Z, Taylor E W. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 27.Jiang W, Hackney D D. J Biol Chem. 1997;272:5616–5621. doi: 10.1074/jbc.272.9.5616. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Stock M F, Li X, Hackney D D. J Biol Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- 29.Moyer M L, Gilbert S P, Johnson K A. Biochemistry. 1998;37:800–813. doi: 10.1021/bi9711184. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert S P, Moyer M L, Johnson K A. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- 31.Hackney D D, Levitt J D, Suhan J. J Biol Chem. 1992;267:8696–8701. [PubMed] [Google Scholar]
- 32.Coy D L, Hancock W O, Wagenbach M, Howard J. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 33.Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow E M, Mandelkow E. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y Z, Taylor E W. Biochemistry. 1995;34:13233–13241. doi: 10.1021/bi00040a039. [DOI] [PubMed] [Google Scholar]
- 35.Romberg L, Vale R D. Nature (London) 1993;361:168–170. doi: 10.1038/361168a0. [DOI] [PubMed] [Google Scholar]
- 36.Cross R A. Curr Biol. 1997;7:R631–R633. doi: 10.1016/s0960-9822(06)00320-4. [DOI] [PubMed] [Google Scholar]
- 37.Cross, R. A., Crevel, I., Carter, N. J., Alonso, M. C., Hirose, K. & Amos, L. A. (1999) Proc. R. Soc. London Ser. B, in press. [DOI] [PMC free article] [PubMed]
- 38.Gilbert S P, Webb M R, Brune M, Johnson K A. Nature (London) 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crevel I M, Lockhart A, Cross R A. J Mol Biol. 1996;257:66–76. doi: 10.1006/jmbi.1996.0147. [DOI] [PubMed] [Google Scholar]
- 40.Yount R G, Lawson D, Rayment I. Biophys J. 1995;68:44S–47S. [PMC free article] [PubMed] [Google Scholar]
- 41.Hackney D D. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hackney D D. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart R J, Thaler J P, Goldstein L S. Proc Natl Acad Sci USA. 1993;90:5209–5213. doi: 10.1073/pnas.90.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J T, Saxton W M, Stewart R J, Raff E C, Goldstein L S. Science. 1990;249:42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]
- 45.Gittes F, Meyhofer E, Baek S, Howard J. Biophys J. 1996;70:418–429. doi: 10.1016/S0006-3495(96)79585-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visscher K, Schnitzer M J, Block S M. Nature (London) 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 47.Hirose K, Lowe J, Alonso M, Cross R A, Amos L A. Mol Biol Cell. 1999;10:2063–2074. doi: 10.1091/mbc.10.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnal I, Wade R H. Structure (London) 1998;6:33–38. doi: 10.1016/s0969-2126(98)00005-7. [DOI] [PubMed] [Google Scholar]
- 49.Hirose K, Lockhart A, Cross R A, Amos L A. Proc Natl Acad Sci USA. 1996;93:9539–9544. doi: 10.1073/pnas.93.18.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozielski F, Arnal I, Wade R H. Curr Biol. 1998;8:191–198. doi: 10.1016/s0960-9822(98)70083-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.