Abstract
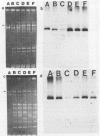
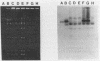
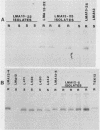
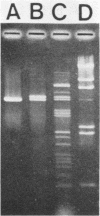
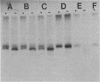
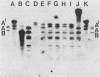
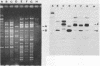
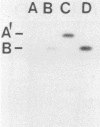
Bacteriophage-resistant dairy streptococci were obtained following conjugal transfer of pTR2030 from a lactose-negative donor, Streptococcus lactis TEK12, to lactose-positive recipient strains, Streptococcus cremoris LMA13 and 924 and S. lactis LMA12. Fast acid-producing, phage-resistant transconjugants were selected by challenge with homologous phage on fast-slow differential agar or lactose indicator agar. Acquisition of pTR2030 by the transconjugants was confirmed by DNA-DNA hybridization. Resistance of transconjugants to homologous phage was complete. Curing or deletion of pTR2030 in the transconjugants confirmed that phage resistance was due to pTR2030 acquisition and not to coincident background mutation. Phage-sensitive pTR2030 deletion derivatives of LMA12 transconjugants were isolated in vivo. The HindIII fragment B of pTR2030 was subcloned into pBR322 to yield a recombinant plasmid, pMET2, useful as a source of pTR2030 DNA. A specific, chemically synthesized oligomer useful as a pTR2030 probe was derived from the sequence of a small portion of pTR2030. The conjugal strategy presented here was effective in yielding fast acid-producing, phage-resistant S. cremoris and S. lactis strains without the use of antibiotic resistance markers and without interfering with the acid-producing ability of the recipient strain.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984 Jan;47(1):68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purrello M., Balazs I. Direct hybridization of labeled DNA to DNA in agarose gels. Anal Biochem. 1983 Feb 1;128(2):393–397. doi: 10.1016/0003-2697(83)90391-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing W. D., Klaenhammer T. R. Conjugal Transfer of Bacteriophage Resistance Determinants on pTR2030 into Streptococcus cremoris Strains. Appl Environ Microbiol. 1986 Jun;51(6):1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]