Abstract
Members of the myc family of nuclear protooncogenes play roles in cell proliferation, differentiation, and apoptosis. Moreover, inappropriate expression of c-myc genes contributes to the development of many types of cancers, including B cell lymphomas in humans. Although Myc proteins have been shown to function as transcription factors, their immediate effects on the cell have not been well defined. Here we have utilized a murine model of lymphomagenesis (Eμ-myc mice) to show that constitutive expression of a c-myc transgene under control of the Ig heavy-chain enhancer (Eμ) results in an increase in cell size of normal pretransformed B lymphocytes at all stages of B cell development. Furthermore, we show that c-Myc-induced growth occurs independently of cell cycle phase and correlates with an increase in protein synthesis. These results suggest that Myc may normally function by coordinating expression of growth-related genes in response to mitogenic signals. Deregulated c-myc expression may predispose to cancer by enhancing cell growth to levels required for unrestrained cell division.
It has been known for some time that cell cycle progression is tightly coupled to an accumulation of cell mass (i.e., cell growth) (1). However, the molecules that control cell growth and the mechanisms through which growth and proliferation are coupled are only beginning to be defined (2). One group of genes whose function may be important for both growth and proliferation is the members of the myc protooncogene family, c-, N-, and L-myc.
The proteins encoded by myc family genes are members of the basic-helix-loop-helix-zipper (bHLHZ) class of transcription factors. Dimerization of Myc protein with its obligate partner Max results in formation of a heterodimer with sequence-specific DNA-binding activity (3, 4). Myc-Max heterodimers appear to activate transcription when bound to promoter-proximal sites on DNA (5, 6). In addition, Myc is known to repress transcription of specific genes (7). Although max expression is constitutive, myc expression is highly regulated at transcriptional, posttranscriptional, translational, and posttranslational levels (8–11). In general, c-Myc expression is associated with proliferation and is down-regulated in quiescent and differentiated cells. After serum or mitogen stimulation of quiescent cells, myc levels peak within several hours, followed by a decline to a low basal level maintained by synthesis and degradation (12–15) and dependent on the continued presence of growth factors (16). Ectopic expression of c-myc in normally quiescent cells can potentiate entry into S phase, and cells that constitutively express myc have reduced growth factor requirements, shortened doubling times, and in some cases have circumvented cell cycle exit (17–20). Conversely, failure to induce c-myc in response to mitogenic signaling abrogates cell cycle progression (21) whereas a fibroblast cell line bearing targeted homozygous c-myc deletions has significantly decreased proliferation rates and lengthened G1 and G2 phases (22). These observations together have led to the notion that Myc-Max functions to modulate expression of genes promoting cell cycle progression (for reviews, see refs. 23–25).
Deregulated expression of myc family genes, through gene amplification, viral promoter insertion, chromosomal translocation, or promoter mutation, has long been associated with neoplastic disease in a wide range of vertebrates including humans (for reviews, see refs. 26–29). Among the most striking examples of the importance of myc in cancer development are the chromosomal translocations involving c-myc and Ig heavy- or light-chain loci that are characteristic of Burkitt’s lymphoma in humans and plasmacytomas in mice and rats. A powerful model system for B cell lymphoma (Eμ-myc mice) entails expression of c-myc in murine lymphoid cells as a transgene under control of the Ig heavy-chain enhancer (30) (for review see ref. 31). Eμ-myc mice develop clonal B cell lymphomas with a mean latency of 12–16 wk of age. Before transformation, B cell progenitors appear relatively normal in these mice. However, an expanded pre-B cell compartment exhibiting increased apoptosis is observed and was suggested to serve as a pool of cells from which secondary cooperating mutations would generate frank lymphomas (32). Indeed, subsequent experiments demonstrated that lymphomagenesis involving myc can be accelerated by activation of a number of cooperating oncogenes including v-abl, bmi1, N-ras, cyclin D1, and pim1 (33–38).
Despite the apparent importance of Myc in proliferation and tumor evolution, we lack a clear understanding of Myc’s primary function in normal and transformed cells. Although it is tempting to conclude that Myc simply modulates expression of target genes involved in cell cycle progression, few such targets have been identified or validated (see refs. 24 and 25 for reviews). In addition, myc has been found to be expressed in nondividing cell types (39–41), and myc knockout mice display extensive cell division before death (42, 43). Furthermore, rat1 cells in which c-myc has been somatically deleted and which do not express other myc family genes are nonetheless capable of proliferation, although at a reduced rate (22). Thus, it is conceivable that Myc may regulate aspects of cell physiology distinct from cell cycle progression. In fact, a recent study using hypomorphic mutations as well as overexpression of Drosophila myc (dmyc) has concluded that whereas dmyc has little effect on cell division, it has a profound influence on cell growth (44). To determine whether Myc could also influence growth in mammalian cells, we used the Eμ-myc mouse model system to examine the effects of ectopic myc expression on cell size and protein synthesis.
Materials and Methods
Mice.
C57BL/6 Eμ-myc transgenic mice were obtained from The Jackson Laboratory (30) and were maintained under pathogen-free conditions. Transgenic mice were maintained on C57BL/6 background and were genotyped by PCR according to instructions from the supplier. Mice were analyzed at 4–5 wk of age, before onset of pathology associated with B cell transformation.
Flow Cytometry.
Cells were isolated from bone marrow, spleen, and thymus as previously described (45) and resuspended in Hanks’ Balanced Salt Solution (GIBCO/BRL) plus 3% fetal calf serum (HyClone). Erythrocytes were depleted by ammonium chloride lysis (46). Staining for surface markers by using flow cytometry was performed as previously described (45), and 10,000 gated events were collected utilizing a FACsCaliber machine (Becton Dickinson). Data were then analyzed by using cellquest software (Becton Dickinson) and repromac software, version 2.07 (Truefacts Software, Seattle, WA). For DNA content analysis, 2 × 106 cells were washed twice in 10 ml of sample buffer (0.1% glucose in PBS). Cell pellets were resuspended in ice-cold 70% ethanol while slowly vortexing, fixed overnight or longer, and then resuspended in 1 ml of propidium iodide (PI) staining solution [50 μg/ml propidium iodide/100 units/ml RNase A (Sigma), in sample buffer], and incubated at room temperature in the dark for 2 hr. Fifty-thousand gated events were collected on a FACsCaliber machine and analyzed by using cellquest software. For cell sorting, cells were stained with PI (as described above), and 50,000 G2-phase cells were sorted by using a FACs Vantage machine (Becton Dickinson) and analyzed by using cellquest software.
Cell Volume.
Cell volume measurements were performed by using a Coulter Model Z2 (Coulter) (47). Cells were diluted in Isoton II (Beckman Coulter) at 100,000 cells/ml in 10 ml. A 1-ml sample was analyzed according to manufacturer’s instructions. Fifty-thousand sorted cells were resuspended in 6 ml of Isoton II, and 1 ml was analyzed.
B Cell Purification.
Total splenocytes were isolated by crushing spleens between the frosted ends of glass microscope slides (Fisher Scientific) into Hanks’ Balanced Salt Solution plus 3% FCS. The resulting cells, depleted of erythrocytes, were incubated in 1:5,000 anti-Thy1.2 (Sigma) in PBS for 30 min on ice, followed by the addition of guinea pig complement (GIBCO/BRL) at 1:10 dilution for 1 hr at 37°C. Live cells were enriched by separation over 100% FCS. Splenic B cells are typically 90% pure as determined by flow cytometry.
Protein Analysis.
Purified B cells (2 × 106) were incubated for 30 min in methionine-free medium DMEM (GIBCO/BRL) supplemented with 10% fetal calf serum. [35S]methionine (16 μCi) was added to the cells and the incubation continued for 30 min. Cells were lysed in 50 μl of RIPA buffer (48) and frozen at −70°C. One microliter of thawed lysate was spotted onto a Hybond glass filter for liquid scintillation counting. For immunoblots, 2 × 107 cells were lysed in 30 μl of sample buffer (1% SDS/5% glycerol/25 mM Tris, pH6.5/0.005% Bromphenol blue) and 30 μl of PBS, boiled for 5 min, and sonicated for 30 sec. Cell equivalents (4 × 106) were separated on 10% SDS/PAGE, transferred to nitrocellulose by semidry transfer, and blocked overnight in 5% skim milk. Blots were probed with anti-c-Myc (9E10), or anti-Max (49), as previously described. For protein quantitation, 1 × 106 purified cells were lysed in TNT and protein concentration determined by the Bradford method (45).
Results
Expression of an Eμ-myc Transgene in B Lymphocyte Progenitors Results in Impaired B Lymphocyte Development.
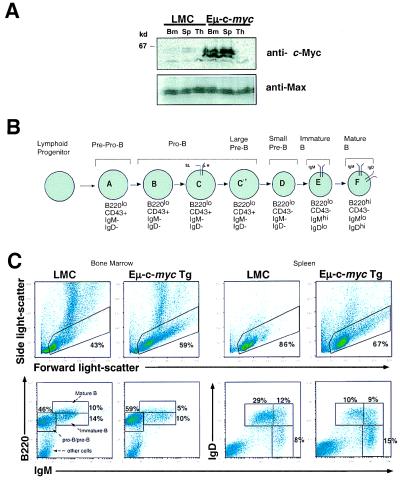
To examine the effects of c-Myc on development and cell growth of primary cells, we utilized transgenic mice that express c-myc under control of the Ig heavy-chain enhancer transcriptional element (Eμ-myc). Previous analyses of lymphoid cells derived from Eμ-myc mice showed that Eμ-myc-expressing B cells in young mice are relatively normal in terms of marker expression, growth factor requirement, and immune response (32; 50–52). We first examined B cells from Eμ-myc mice for c-Myc and Max protein by immunoblotting with antibodies against c-Myc and Max (49, 53). As expected, Eμ-myc mice, when compared with wild-type littermates, exhibited increased levels of c-Myc, but not Max, in B cells derived from bone marrow as well as in mature B cells from peripheral lymphoid tissue such as the spleen. Thymocytes from Eμ-myc mice did not express high levels of c-Myc (Fig. 1A and ref. 32).
Figure 1.
Expression of Eμ-c-myc transgene results in impaired B cell development. (A) Bone marrow cells (Bm), splenocytes (Sp), or thymocytes (Th) were isolated from Eμ-c-myc and littermate control mice (LMC). Cells (4 × 106) were lysed in sample buffer (see Materials and Methods) and proteins separated by SDS/PAGE. Samples were transferred to nitrocellulose membranes and protein visualized by probing blots with anti-c-Myc or anti-Max. (B) A model for characterizing B lymphocyte development utilizing monoclonal antibodies to surface markers and flow cytometry (modified from ref. 54). (C) Total bone marrow cells were stained with phycoerythrin (PE)-conjugated anti-B220 and FITC-conjugated anti-IgM. Total splenocytes were stained with FITC-conjugated anti-IgD and PE-conjugated anti-IgM. Cells were then visualized by flow cytometry, gated according to forward and side light scatter (lymphocyte gate), and staged according to a general scheme for B lymphocyte development as described by Hardy et al. (see diagram and ref. 54). The forward and side light-scatter gate excluded small apoptotic cells and granular cells, whereas large cells were included (Top). The expression of Eμ-c-myc transgene results in an increase in representation of pro-B and pre-B cells in the bone marrow (Bottom Left) and immature B cells in the spleen (Bottom Right) relative to littermate control mice.
To characterize the effects of c-myc overexpression on B cell development in more detail, we utilized the model of Hardy et al. (54), in which B cells are staged developmentally according to size and surface markers (Fig. 1B). The earliest B cells that can be identified in bone marrow or fetal liver (Hardy fraction A) are characterized by expression of B220 (CD45R) in the absence of detectable surface or cytoplasmic IgM. In response to cytokines such as IL-7 and kit-ligand, these cells subsequently mature to the progenitor B cell stage (large pro-B; Hardy Fractions B and C), where they initiate V(D)J rearrangements of their Ig heavy-chain genes leading to expression of cytoplasmic μ heavy chain. The μ chain then appears at the cell surface along with a “surrogate” light chain composed of the Vpre-B and λ5 polypeptides, as a component of the pre-B cell receptor (large pre-B; Hardy Fraction C′). Thereafter, signals derived from the pre-B cell receptor complex stimulate Ig light-chain rearrangement and further maturation (small pre-B; Hardy Fraction D), eventually giving rise to B220loIgMhiIgDlo immature B cells (Hardy Fraction E). These cells then populate peripheral lymphoid tissues wherein they mature further to become B220hiIgMloIgDhi long-lived recirculating B cells (Hardy Fraction F).
As shown in Fig. 1C (Top Left), expression of Eμ-c-myc transgene results in an increase in the representation of total bone marrow cells that fall within a forward and side light-scatter lymphocyte gate. Analysis of total bone marrow cells that fall within the lymphocyte gate reveals an increase in representation of developing B220+IgM− progenitor B (pro-B) and precursor B (pre-B) cells, at the expense of more mature IgM+ cells in the bone marrow (Fig. 1C Bottom Left and ref. 30). In addition, the representation of splenocytes from Eμ-c-myc mice that fall within the light-scatter lymphocyte gate is slightly decreased relative to littermate control mice (Fig. 1C Top Right). Analysis of these cells from Eμ-c-myc mice reveals an increase in representation of IgMhiIgDlo immature B cells at the expense of longer-lived IgMloIgDhi mature B cells relative to littermate control mice (Fig. 1C Bottom Right). Hence, the decrease in representation of splenic B cells in young Eμ-c-myc mice (<5 wk of age) is likely because of impaired B cell development and increased apoptosis in response to inappropriate c-myc expression (52), as has been previously shown (55). Overexpression of c-myc eventually results in clonal transformation of pre-B cells and occasionally mature B cells, with a median age of 12–16 wk before transformation (ref. 51 and data not shown).
c-myc Transgene Expression Results in Increased Growth of Primary B Lymphocytes at All Stages of B Cell Development.
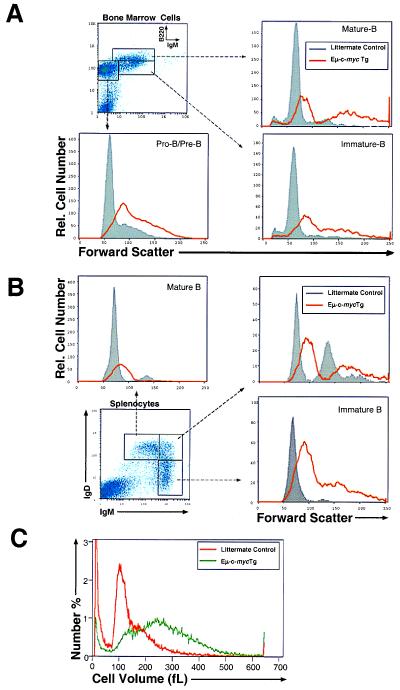
We observed that overexpression of c-myc in Eμ-myc transgenic mice results in an increase in cell size (higher forward light scatter) in cells derived from both bone marrow and spleen (Fig. 1C Top), as has been previously shown (32, 51). However, in earlier studies, it was not determined whether the increase in cell size simply reflected a greater representation of larger less mature B cell progenitors (large pro-B and large pre-B cells; Hardy Fraction A-C′), or a greater representation of cells that had entered the S and G2 phases of the cell cycle. To examine these possibilities, we analyzed the size of B cell progenitors isolated from young prepubescent (<5 wk of age) Eμ-myc mice at various stages of B cell development utilizing fluorochrome-labeled antibodies and flow cytometry. As shown in Fig. 2A, bone marrow-derived B220loIgM− pro-B/pre-B cells, B220loIgM+ immature B, and B220hiIgM+ mature B cells from Eμ-myc mice are all significantly larger then cells of the same developmental stage isolated from littermate control mice, as shown by forward light-scatter characteristics (FSC). In addition, IgMhiIgDlo immature B cells, IgMhiIgDhi transitional cells, and IgMloIgDhi long-lived mature B cells isolated from the spleen of Eμ-myc mice are also significantly larger than corresponding B cells of the same developmental stage from littermate control mice (Fig. 2B). Hence, the increase in cell size resulting from c-myc expression is not caused by an overrepresentation of larger less mature progenitors. Rather, overexpression of c-Myc protein results in an increase in size of B lymphocytes regardless of their developmental stage. Importantly, these results also demonstrate that the increase in cell growth observed in Eμ-myc transgenic mice is not caused by isolation of a transformed population of B cells, because B cells of all developmental stages are uniformly enlarged, and the cells were isolated from young mice before pathology onset. Furthermore, the increase in cell size is not caused by a secondary effect of c-Myc on B cell activation, because pro-B and pre-B cells (which lack antigen receptor expression) are uniformly enlarged.
Figure 2.
Expression of Eμ-c-myc transgene results in an increase in B cell size (growth) during all stages of B cell development. (A) Total bone marrow cells isolated from Eμ-c-myc transgenic or littermate control mice were stained with PE-conjugated anti-B220 and FITC-conjugated anti-IgM. Cells were visualized by flow cytometry. Shown are the FSC of gated B220loIgM− pro-B/pre-B cells, B220loIgM+ immature B cells and B220hiIgM+ mature B cells. Cells with higher FSC are larger than cells with lower FSC. (B) Total splenocytes were stained with FITC-conjugated anti-IgD and PE-conjugated anti-IgM. Shown are the FSC of IgMhiIgDlo immature B cells, IgMhiIgDhi, and IgMloIgDhi mature B cells as visualized by flow cytometry. (C) Purified splenic B lymphocytes from either Eμ-c-myc transgenic or littermate control mice were analyzed for cell volume utilizing a Coulter Z2 counter (47). Shown is a representative single-parameter histogram.
To examine whether the increase in FSC observed in Eμ-myc mice actually corresponds to an increase in cell volume, we purified B lymphocytes from young Eμ-myc transgenic mice and analyzed cell volume via the Coulter principle (47). As shown in Fig. 2C, mature B lymphocytes from 4- to 5-wk-old Eμ-myc mice are significantly larger (1.6-fold average increase in volume) than B lymphocytes from littermate control mice, consistent with the changes observed in FSC as measured by flow cytometry.
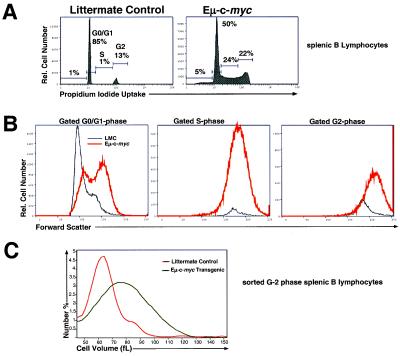
Increased Growth of B Lymphocytes Induced by c-Myc Occurs Independently of Cell Cycle Alterations.
To determine whether the increase in cell size induced by the c-myc transgene is caused by an increase in representation of larger S- and/or G2-phase cells, we measured cell size of gated populations of purified splenic B cells that lie within different phases of the cell cycle. As shown in Fig. 3A, expression of the c-myc transgene results in an increase in cells that have entered the S and G2 phases of the cell cycle, as has been previously shown (32). However, this increase in cycling cells does not account for the uniform increase in cell size observed in Eμ-myc mice, because gated cells that fall within the G0/G1 or G2 phases of the cell cycle are uniformly enlarged relative to littermate control B cells (Fig. 3B). To further confirm the validity of our forward light-scatter results, we sorted the largest cells (G2 phase) from both Eμ-myc and littermate control mice and measured cell volume by using a Coulter counter. As expected, the Coulter results mirrored the increase in cell size observed by forward light-scatter analysis (Fig. 3C). For the S-phase populations, the relative shift in cell size is small; however, it has been consistently observed. Hence, overexpression of c-Myc protein results in an increase in cell growth, regardless of the stage of the cell cycle.
Figure 3.
B cells from Eμ-c-myc mice are enlarged during all phases of the cell cycle. (A) Purified splenic B lymphocytes isolated from Eμ-c-myc or littermate control mice were stained with PI, and cell cycle status was determined by flow cytometry. Shown is a representative single-parameter flow cytometric histogram. (B) FSC were determined for purified splenic B cells that fell within G0/G1, S, or G2 gates, as outlined by using PI staining in A. The biphasic G0/G1 peak likely represents separation of G0- and G1-phase cells. G0-, G1-, and G2-phase cells from Eμ-c-myc mice exhibit higher FSC than similar phase cells from littermate control mice. (C) Splenic B lymphocytes were purified, fixed in ethanol, and stained with PI (see Materials and Methods). G2-phase cells (50,000 cells) from Eμ-c-myc and littermate control mice were sorted by flow cytometry, and cell volume was determined by using the Coulter principle. (Ethanol fixation results in a significant, but proportional, reduction in cell size.)
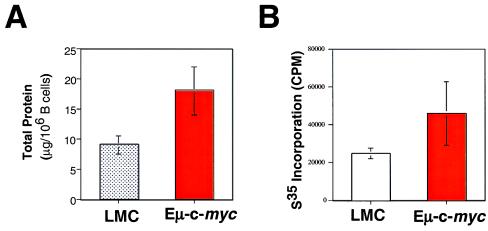
Increased Cell Growth of B Lymphocytes Induced by c-Myc Correlates with an Increase in Protein Synthesis.
To determine whether the increase in cell growth induced by overexpression of c-myc correlates with an increase in protein synthesis, we measured steady-state protein content and protein biosynthetic rates from purified B lymphocytes from either Eμ-myc or littermate control mice. B cells from Eμ-myc mice contained 2-fold greater levels of total protein when compared with littermate control mice (Fig. 4A). This increase in total protein correlates with a 2-fold increase in protein biosynthesis, as determined by incorporation of [35S]methionine (Fig. 4B). Taken together, these results show a correlation between Myc overexpression, cell size, and increased protein synthesis and accumulation.
Figure 4.
Expression of Eμ-c-myc transgene results in increased protein synthesis. (A) Purified splenic B lymphocytes (2 × 106 cells) isolated from Eμ-c-myc or littermate control mice were lysed in TNT buffer with protease inhibitors, and total protein was determined by the Bradford method. Error bars denote standard error for triplicate samples from a representative experiment. (B) Purified splenic B lymphocytes (2 × 106 cells) from the above mice were cultured for 30 min in methionine-free medium, followed by 30 min in [35S]methionine-containing medium. Cells were washed and lysed in RIPA buffer (48). Lysate was spotted onto glass filters, and [35S]methionine incorporation was determined by using a liquid scintillation counter.
Discussion
Cell proliferation is a coordinated process whereby cells duplicate their contents and increase their mass and size (cell growth) before initiating cell division. Although growth and division generally appear coupled, proliferation mutants identified in yeast suggest that these events are indeed separable. For example, one class of yeast mutants blocks cell cycle progression while allowing cell growth to continue, whereas a second class abrogates both cell growth and cell cycle progression (56, 57). The first class of mutants is known to affect cell cycle proteins, whereas the second affects general biosynthesis. These genetic studies in yeast suggest that cell growth is dominant to, and limiting for, cell cycle progression (58). Although there is increasing evidence that in metazoan cells growth and proliferation are also coupled and/or coordinated, the key molecules involved in regulating growth are only beginning to be identified (see ref. 2 for review).
Utilizing a murine model of Burkitt’s lymphoma (30), we show that expression of the c-myc protooncogene, driven by the Ig heavy-chain enhancer (Eμ) in otherwise normal B lymphocyte progenitors results in a significant increase in cell growth and protein synthesis. These changes correlate with impaired B cell development, an increase in cell cycle entry, and predisposition to B cell lymphoma. However, it is unlikely that the observed growth enhancement is secondary to the increased proliferation or developmental alterations because the cells were larger in all phases of the cell cycle and at all stages of B cell development (including stages lacking antigen receptors). In addition B cells exhibiting increased size are not transformed and were detected before onset of lymphomas, suggesting that these cells are not highly abnormal (our data, ref. 32; refs. 50–52). Importantly, the fact that we could detect a substantial cell size increase suggests that c-myc overexpression uncouples growth from proliferation, otherwise the effect on cell size would have been offset by increased division. Therefore, Myc’s predominant effect appears to be on cell growth. Our findings extend previous work showing that B cells from Eμ-myc mice are larger, but which did not examine size as a function of development or cell cycle (32, 51). Our results are also consistent with recent findings indicating that Myc overexpression induces growth, but not cell cycle progression, in an immortalized human B cell line (M. Schuhmacher, M. S. Staege, A. Pajic, U. H. Weidle, G. W. Bornkamm, D. Eick and F. Kohlhuber, personal communication).
We postulate that deregulated c-myc in B lymphocytes predisposes to transformation at least in part by enhancing cell growth. Because in c-myc-deregulated cells growth may no longer be limiting for cell division, there may exist strong selective pressure for secondary activating mutations in genes involved in cell cycle progression. Thus ability of deregulated c-myc to drive cell growth may underlie the requirement for c-Myc to collaborate in cell transformation with oncogenes such as ras or pim-1. Consistent with this notion, activating mutants of Ras and Raf oncoproteins, which collaborate with c-Myc to transform primary B lymphocytes, do not stimulate cell growth at levels sufficient to drive development of T or B lymphocytes on recombinase-activating gene-deficient backgrounds (59, 60). These results also demonstrate that the ability of deregulated c-myc to potently drive cell growth is not a property common to all oncoproteins.
Although our data show that deregulated expression of c-myc stimulates cell growth in otherwise normal B lymphocytes, we cannot conclude from this work that the normal function of endogenous c-Myc is to regulate growth. Nonetheless, several lines of evidence suggest that this is the case. Firstly, in Drosophila, decreased expression of dMyc (the ortholog of vertebrate Myc) in dmyc mutant wing imaginal disc cells retards cell proliferation and reduces cell size, whereas dmyc overexpression results in increased cell growth rate and cell size without affecting cell division (44). Secondly, c-myc null fibroblasts (which also lack expression of L- and N-myc) exhibit significantly decreased accumulation of protein and RNA as well as reduced proliferation, which probably accounts for the fact that no change in cell size was detected (22). Thirdly, overexpression of a mad1 transgene (whose protein product competes with Myc/Max heterodimers for E-box binding) selectively in lymphocytes results in impaired lymphocyte development, proliferation, and slightly reduced cell size (B.M.I., unpublished data).
In mammalian cells, the control of cell growth is largely determined by the availability of growth factors produced by other cells. These factors generally activate intracellular signaling pathways that in part stimulate the protein synthesis machinery so that the rate of macromolecular production exceeds the rate of degradation. c-myc is normally up-regulated in lymphocytes after crosslinking of T or B cell antigen receptors by MHC-peptide complexes or free antigen, respectively. With appropriate costimulation, these responses generally result in increased cell growth and proliferation (for review, see ref. 61). It is tempting to speculate that the sensitivity of a particular cell type to transformation by Myc may be determined in part by the intrinsic capability of a cell to synthesize macromolecules. B lymphocytes have engaged their protein synthesis machinery in preparation for rapid clonal proliferation and antibody synthesis in response to infection by extracellular pathogens. Hence, constitutive activation of Myc may readily push a metabolically “poised” B lymphocyte into growth factor or mitogen-independent cell overgrowth and proliferation, perhaps accounting for the high sensitivity of B cells to myc transformation (27).
How does c-Myc stimulate cell growth? As mentioned above, Myc functions with its dimerization partner Max as a transcription factor, and about 30 putative target genes have been identified to date (for recent review, see ref. 25). Although a number of these candidate gene targets have been linked to cell cycle control, immortalization, adhesion, metastasis, and stress response, the majority of potential targets can be construed as being involved in cell growth and metabolism. Several of these latter genes, such as CAD, ODC, DHFR, and TK, are closely linked to DNA metabolism, whereas others are involved in glycolysis (LDH-A), iron metabolism (H-ferritin and IRP-2) (62), and protein synthesis (eIF4E, eIF2α) (see ref. 25 and references therein). The preponderance of vertebrate Myc target genes related to growth and metabolism has been previously noted and has led to the suggestion that at least part of Myc’s function may be directed toward cell growth (24, 25, 63). It is also likely that other, yet to be identified, target genes mediate growth. Utilizing DNA array technology to compare global RNA expression patterns in B cells from Eμ-c-myc and control mice should make it possible to identify growth-specific target genes. Identification of such targets may permit the design of metabolic inhibitors that could specifically subvert the growth stimulating effects of c-Myc in tumor cells.
Acknowledgments
We are grateful to Ivan Gomez and Leni Sue Carlos for expert technical assistance, to the Breeden laboratory for use of their Coulter counter, and to Peter Gallant and Bruce Edgar for helpful discussions. We also thank Jerry Adams, Alan Harris, Paul Neiman, Laura Johnston, Cynthia Yost, and Carla Grandori for critically reviewing this manuscript. This work was supported by a National Institutes of Health (NIH) Mentored Clinical Investigator Award 1 K08 AJ01445-01 (to B.M.I.) and NIH/National Cancer Institute grants RO1CA20525 and HL54881 (to R.N.E.). R.N.E. is a Research Professor of the American Cancer Society.
Abbreviations
- FSC
forward light scatter characteristics
- PI
propidium iodide
References
- 1.Killander D, Zetterberg A. Exp Cell Res. 1965;38:272–284. doi: 10.1016/0014-4827(65)90403-9. [DOI] [PubMed] [Google Scholar]
- 2.Neufeld T P, Edgar B A. Curr Opin Cell Biol. 1998;10:784–790. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 4.Prendergast G C, Lawe D, Ziff E B. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 5.Kretzner L, Blackwood E M, Eisenman R N. Nature (London) 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 6.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Nature (London) 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Nerlov K, Prendergast G, MacGregor D, Ziff E B. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer C A, Groudine M. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 9.West M J, Stoneley M, Willis A E. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 10.Salghetti S E, Kim S Y, Tansey W P. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sears R, Leone G, DeGregori J, Nevins J R. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 12.Kelly K, Cochran B H, Stiles C D, Leder P. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 13.Campisi J, Gray H E, Pardee A B, Dean M, Sonenshein G. Cell. 1984;36:241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg M E, Ziff E B. Nature (London) 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 15.Hann S R, Thompson C B, Eisenman R N. Nature (London) 1985;314:366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- 16.Waters C M, Littlewood T D, Hancock D C, Moore J P, Evan G I. Oncogene. 1991;6:797–805. [PubMed] [Google Scholar]
- 17.Palmieri S, Kahn P, Graf T. EMBO J. 1983;2:2385–2389. doi: 10.1002/j.1460-2075.1983.tb01750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stern D, Roberts A, Roche N S, Sporn M B, Weinberg R A. Mol Cell Biol. 1986;6:870–877. doi: 10.1128/mcb.6.3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karn J, Watson J V, Lowe A D, Green S M, Vedeckis W. Oncogene. 1989;4:773–787. [PubMed] [Google Scholar]
- 20.Eilers M, Schirm S, Bishop J M. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roussel M F, Cleveland J L, Shurtleff S A, Sherr C J. Nature (London) 1991;353:361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- 22.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 23.Henriksson M, Luscher B. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 24.Grandori C, Eisenman R N. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 25.Dang C V. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cory S. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- 27.Magrath I. Adv Cancer Res. 1990;55:134–251. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 28.Tonini G P. In: Encyclopedia of Cancer. Bertino J R, editor. San Diego: Academic; 1997. pp. 1212–1239. [Google Scholar]
- 29.McCormack S J, Lippman M E, Dickson R B. In: Encyclopedia of Cancer. Bertino J R, editor. San Diego: Academic; 1997. pp. 165–172. [Google Scholar]
- 30.Adams J M, Harris A W, Pinkert C A, Corcoran L M, Alexander W S, Cory S, Palmiter R D, Brinster R L. Nature (London) 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 31.Cory S, Adams J. Ann Rev Immunol. 1988;6:25–48. doi: 10.1146/annurev.iy.06.040188.000325. [DOI] [PubMed] [Google Scholar]
- 32.Langdon W Y, Harris A W, Cory S, Adams J M. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 33.van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 34.Rosenbaum H, Harris A W, Bath M L, McNeall J, Webb E, Adams J M, Cory S. EMBO J. 1990;9:897–905. doi: 10.1002/j.1460-2075.1990.tb08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 36.Haupt Y, Alexander W S, Barri G, Klinken S P, Adams J M. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 37.Haupt Y, Harris A W, Adams J M. Oncogene. 1992;7:981–986. [PubMed] [Google Scholar]
- 38.Bodrug S E, Warner B J, Bath M L, Lindemann G J, Harris A W, Adams J M. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Downs K M, Martin G R, Bishop J M. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 40.Craig R W, Buchan H L, Civin C I, Kastan M B. Cell Growth Differ. 1993;4:349–357. [PubMed] [Google Scholar]
- 41.Wakamatsu Y, Watanabe Y, Shimono A, Kondoh H. Neuron. 1993;10:1–9. doi: 10.1016/0896-6273(93)90236-k. [DOI] [PubMed] [Google Scholar]
- 42.Davis A C, Wims M, Spotts G D, Hann S R, Bradley A. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 43.Stanton B R, Perkins A S, Tessarollo L, Sassoon D A, Parada L F. Genes Dev. 1993;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 44.Johnston L A, Prober D A, Edgar B A, Eisenman R N, Gallant P. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iritani B M, Forbush K A, Farrar M A, Perlmutter R M. EMBO J. 1997;16:7019–7031. doi: 10.1093/emboj/16.23.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishell R I, Shiigi S M. Selected Methods in Cellular Immunology. San Francisco: Freeman; 1980. [Google Scholar]
- 47.Said S, Tamura T, Gerdes A M. BioTechniques. 1998;25:522–525. doi: 10.2144/98253pf02. [DOI] [PubMed] [Google Scholar]
- 48.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 49.Blackwood E M, Luscher B, Eisenman R N. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 50.Vaux D L, Adams J M, Alexander W S, Pike B L. J Immunol. 1987;139:3854–3860. [PubMed] [Google Scholar]
- 51.Harris A W, Pinkert C A, Crawford M, Langdon W Y, Brinster R L, Adams J M. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langdon W Y, Harris A W, Cory S. Oncogene Res. 1988;3:271–279. [PubMed] [Google Scholar]
- 53.Hann S R, Eisenman R N. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Askew D, Ashmun R, Simmons B, Cleveland J. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 56.Hartwell L H. J Mol Biol. 1971;14:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 57.Nurse P, Thuriaux P, Nasmyth K 1, Nasmyth K 1. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 58.Johnston G C, Pringle J R, Hartwell L H. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 59.Swat W, Shinkai Y, Cheng H L, Davidson L, Alt F W. Proc Natl Acad Sci USA. 1996;93:4683–4687. doi: 10.1073/pnas.93.10.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iritani B M, Alberola-Ila J, Forbush K A, Perlmutter R M. Immunity. 1999;10:713–722. doi: 10.1016/s1074-7613(00)80070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melchers F. Curr Top Microbiol Immunol. 1997;224:19–30. doi: 10.1007/978-3-642-60801-8_2. [DOI] [PubMed] [Google Scholar]
- 62.Wu K-J, Polack A, Dalla-Favera R. Science. 1999;283:676–679. doi: 10.1126/science.283.5402.676. [DOI] [PubMed] [Google Scholar]
- 63.Polymenis M, Schmidt E V. Curr Opin Genet Dev. 1999;9:76–80. doi: 10.1016/s0959-437x(99)80011-2. [DOI] [PubMed] [Google Scholar]