Abstract
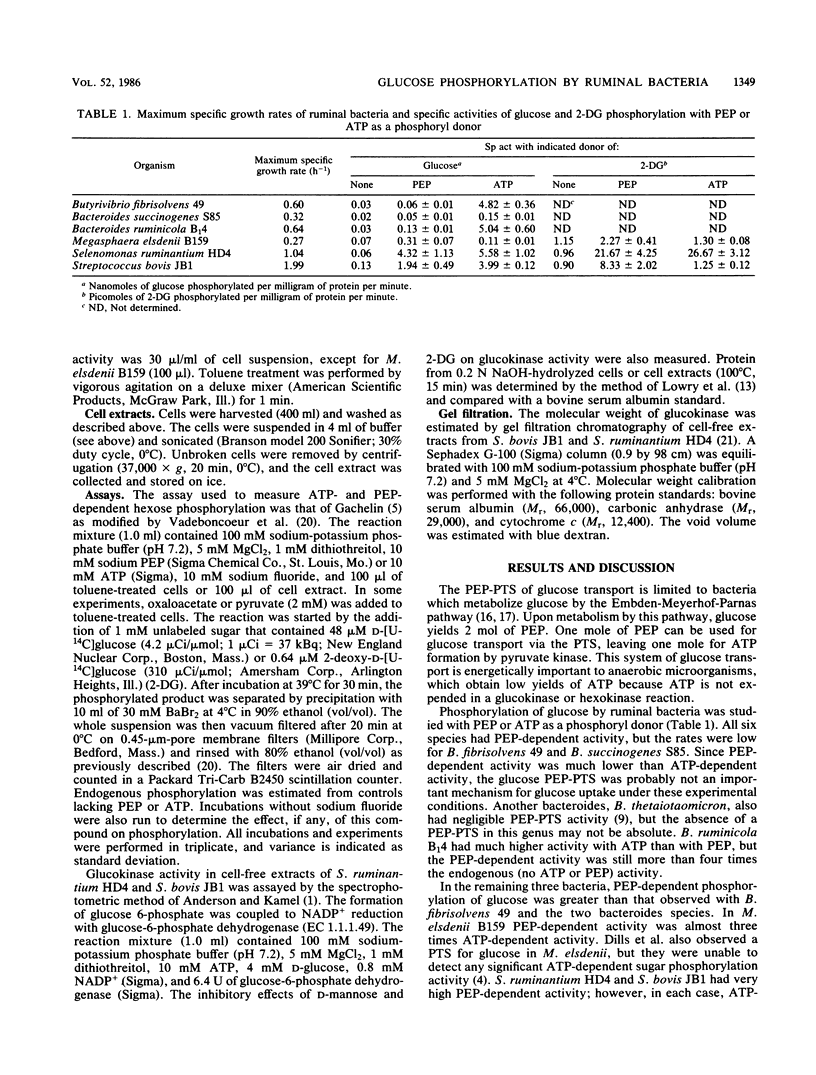
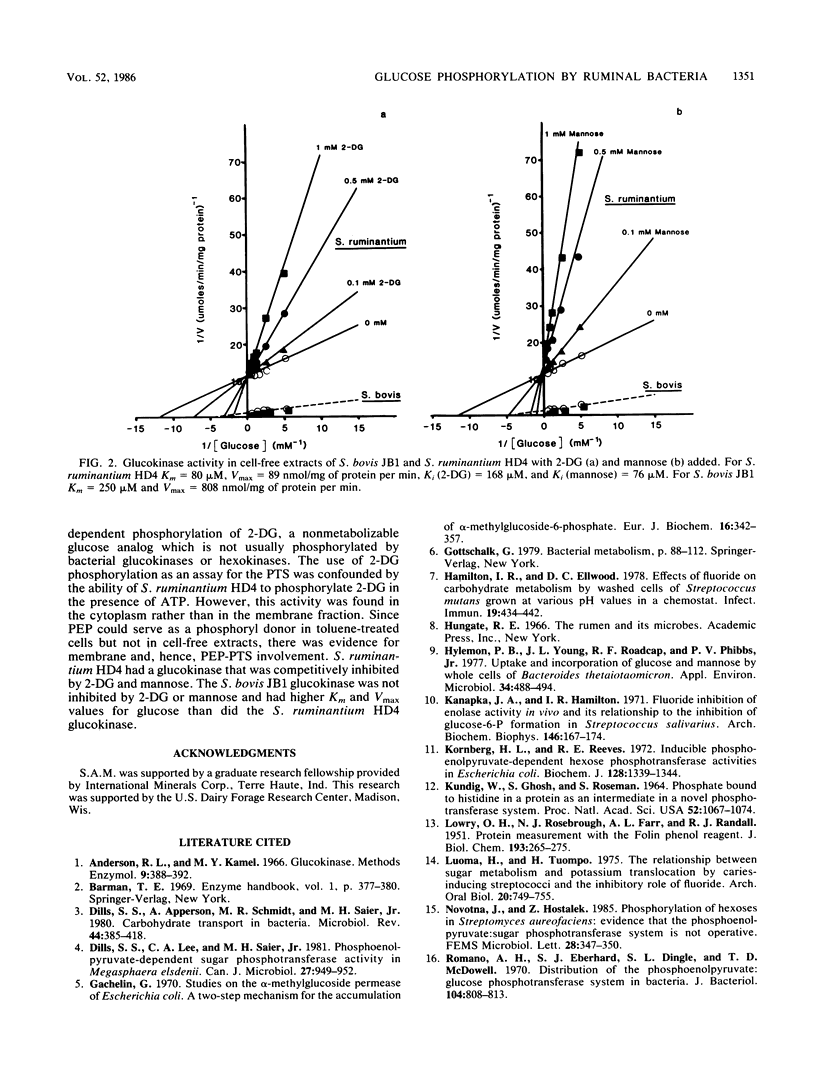
Six species of ruminal bacteria were surveyed for the phosphoenolpyruvate (PEP)-dependent phosphorylation of glucose. Selenomonas ruminantium HD4, Streptococcus bovis JB1, and Megasphaera elsdenii B159 all showed significant activity, but Butyrivibrio fibrisolvens 49, Bacteroides succinogenes S85, and Bacteroides ruminicola B1(4) showed low rates of PEP-dependent phosphorylation and much higher rates in the presence of ATP. S. ruminantium HD4, S. bovis JB1, and M. elsdenii B159 also used PEP to phosphorylate the nonmetabolizable glucose analog 2-deoxy-D-glucose (2-DG). Rates of 2-DG phosphorylation with ATP were negligible for S. bovis JB1 and M. elsdenii B159, but toluene-treated cells of S. ruminantium HD4 phosphorylated 2-DG in the presence of ATP as well as PEP. Cell-free extracts of S. ruminantium HD4 used ATP but not PEP to phosphorylate glucose and 2-DG. Since PEP could serve as a phosphoryl donor in toluene-treated cells but not in cell-free extracts, there was evidence for membrane and hence phosphotransferase system involvement in the PEP-dependent activity. The ATP-dependent phosphorylating enzymes from S. ruminantium HD4 and S. bovis JB1 had molecular weights of approximately 48,000 and were not inhibited by glucose 6-phosphate. Based on these criteria, they were glucokinases rather than hexokinases. The S. ruminantium HD4 glucokinase was competitively inhibited by 2-DG and mannose, sugars that differ from glucose in the C-2 position. Since 2-DG was a competitive inhibitor of glucose, the same enzyme probably phosphorylates both sugars. The S. bovis JB1 glucokinase was not inhibited by either 2-DG or mannose and had a higher Km and Vmax for glucose.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dills S. S., Apperson A., Schmidt M. R., Saier M. H., Jr Carbohydrate transport in bacteria. Microbiol Rev. 1980 Sep;44(3):385–418. doi: 10.1128/mr.44.3.385-418.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dills S. S., Lee C. A., Saier M. H., Jr Phosphoenolpyruvate-dependent sugar phosphotransferase activity in Megasphaera elsdenii. Can J Microbiol. 1981 Sep;27(9):949–952. doi: 10.1139/m81-148. [DOI] [PubMed] [Google Scholar]
- Gachelin G. Studies on the alpha-methylglucoside permease of Escherichia coli. A two-step mechanism for the accumulation of alpha-methylglucoside 6-phosphate. Eur J Biochem. 1970 Oct;16(2):342–357. doi: 10.1111/j.1432-1033.1970.tb01088.x. [DOI] [PubMed] [Google Scholar]
- Hamilton I. R., Ellwood D. C. Effects of fluoride on carbohydrate metabolism by washed cells of Streptococcus mutans grown at various pH values in a chemostat. Infect Immun. 1978 Feb;19(2):434–442. doi: 10.1128/iai.19.2.434-442.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon P. B., Young J. L., Roadcap R. F., Phibbs P. V., Jr Uptake and incorporation of glucose and mannose by whole cells of Bacteroides thetaiotaomicron. Appl Environ Microbiol. 1977 Nov;34(5):488–494. doi: 10.1128/aem.34.5.488-494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUNDIG W., GHOSH S., ROSEMAN S. PHOSPHATE BOUND TO HISTIDINE IN A PROTEIN AS AN INTERMEDIATE IN A NOVEL PHOSPHO-TRANSFERASE SYSTEM. Proc Natl Acad Sci U S A. 1964 Oct;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanapka J. A., Hamilton I. R. Fluoride inhibition of enolase activity in vivo and its relationship to the inhibition of glucose-6-P formation in Streptococcus salivarius. Arch Biochem Biophys. 1971 Sep;146(1):167–174. doi: 10.1016/s0003-9861(71)80053-x. [DOI] [PubMed] [Google Scholar]
- Kornberg H. L., Reeves R. E. Inducible phosphoenolpyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem J. 1972 Aug;128(5):1339–1344. doi: 10.1042/bj1281339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Luoma H., Tuompo H. The relationship between sugar metabolism and potassium translocation by caries-inducing streptococci and the inhibitory role of fluoride. Arch Oral Biol. 1975 Nov;20(11):749–755. doi: 10.1016/0003-9969(75)90047-3. [DOI] [PubMed] [Google Scholar]
- Romano A. H., Eberhard S. J., Dingle S. L., McDowell T. D. Distribution of the phosphoenolpyruvate: glucose phosphotransferase system in bacteria. J Bacteriol. 1970 Nov;104(2):808–813. doi: 10.1128/jb.104.2.808-813.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A. H., Trifone J. D., Brustolon M. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in fermentative bacteria. J Bacteriol. 1979 Jul;139(1):93–97. doi: 10.1128/jb.139.1.93-97.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr Bacterial phosphoenolpyruvate: sugar phosphotransferase systems: structural, functional, and evolutionary interrelationships. Bacteriol Rev. 1977 Dec;41(4):856–871. doi: 10.1128/br.41.4.856-871.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Staley J. T. Phosphoenolpyruvate:sugar phosphotransferase system in Ancalomicrobium adetum. J Bacteriol. 1977 Aug;131(2):716–718. doi: 10.1128/jb.131.2.716-718.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadeboncoeur C., Proulx M., Trahan L. Purification of proteins similar to HPr and enzyme I from the oral bacterium Streptococcus salivarius. Biochemical and immunochemical properties. Can J Microbiol. 1983 Dec;29(12):1694–1705. doi: 10.1139/m83-260. [DOI] [PubMed] [Google Scholar]



