Abstract
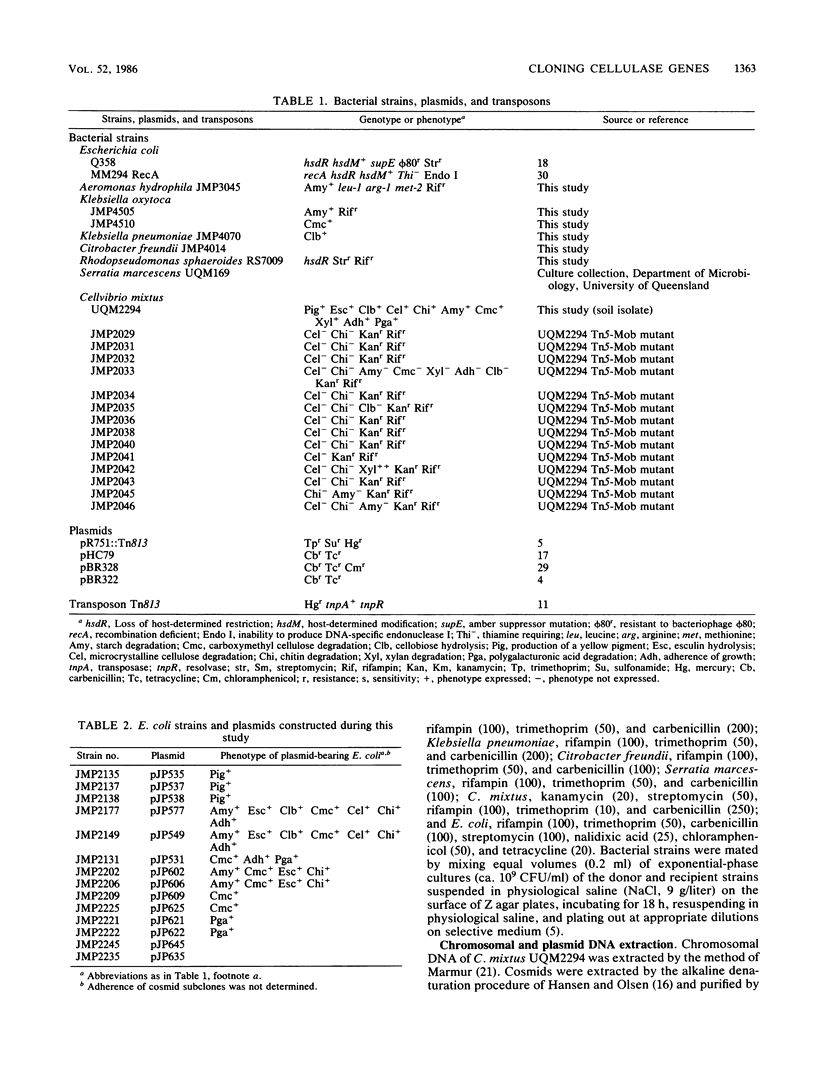
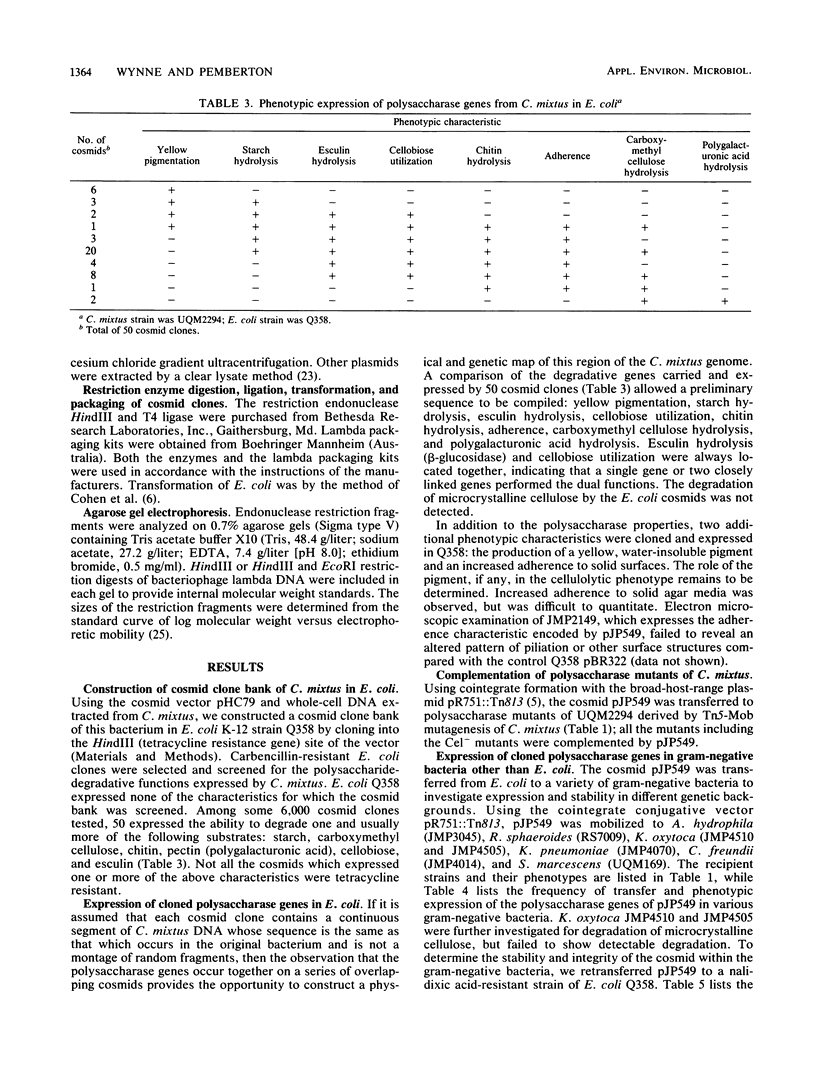
The soil isolate Cellvibrio mixtus UQM2294 degraded a variety of polysaccharides including microcrystalline cellulose. Among 6,000 cosmid clones carrying C. mixtus DNA, constructed in Escherichia coli with pHC79, 50 expressed the ability to degrade one or more of the following substrates: carboxymethyl cellulose, chitin, pectin (polygalacturonic acid), cellobiose, and starch. These degradative genes are encoded in a single 94.1-kilobase segment of the C. mixtus genome; a preliminary order of the genes is starch hydrolysis, esculin hydrolysis, cellobiose utilization, chitin hydrolysis, carboxymethyl cellulose hydrolysis, and polygalacturonic acid hydrolysis. A restriction endonuclease cleavage map was constructed, and the genes for starch, carboxymethyl cellulose, cellobiose, chitin, and pectin hydrolysis were subcloned.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS G. H., PARK R. W. A taxonomic study of certain bacteria currently classified as Vibrio species. J Gen Microbiol. 1962 Jan;27:101–119. doi: 10.1099/00221287-27-1-101. [DOI] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Postgate J. R. Transfer of nitrogen-fixation genes by conjugation in Klebsiella pneumoniae. Nature. 1971 Nov 5;234(5323):47–48. doi: 10.1038/234047a0. [DOI] [PubMed] [Google Scholar]
- Gilardi G. L. Characterization of nonfermentative nonfastidious gram negative bacteria encountered in medical bacteriology. J Appl Bacteriol. 1971 Sep;34(3):623–644. doi: 10.1111/j.1365-2672.1971.tb02326.x. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 1985 Mar;4(3):781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Molecular cloning of eucaryotic genes required for excision repair of UV-irradiated DNA: isolation and partial characterization of the RAD3 gene of Saccharomyces cerevisiae. J Bacteriol. 1982 Oct;152(1):323–331. doi: 10.1128/jb.152.1.323-331.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Cooke S., Bowen A. R. Gene transfer mechanisms among members of the genus Rhodopseudomonas. Ann Microbiol (Paris) 1983 Jul-Aug;134B(1):195–204. doi: 10.1016/s0769-2609(83)80105-7. [DOI] [PubMed] [Google Scholar]
- Sashihara N., Kudo T., Horikoshi K. Molecular cloning and expression of cellulase genes of alkalophilic Bacillus sp. strain N-4 in Escherichia coli. J Bacteriol. 1984 May;158(2):503–506. doi: 10.1128/jb.158.2.503-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Gilberg W. Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene. 1980 Dec;12(3-4):235–241. doi: 10.1016/0378-1119(80)90105-5. [DOI] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis L. The genetics of dissimilarity pathways in Pseudomonas. Annu Rev Microbiol. 1975;29:505–524. doi: 10.1146/annurev.mi.29.100175.002445. [DOI] [PubMed] [Google Scholar]
- Whittle D. J., Kilburn D. G., Warren R. A., Miller R. C., Jr Molecular cloning of a Cellulomonas fimi cellulose gene in Escherichia coli. Gene. 1982 Feb;17(2):139–145. doi: 10.1016/0378-1119(82)90066-x. [DOI] [PubMed] [Google Scholar]



