Abstract
The tandem BRCA1 C-terminal (BRCT) domains are phospho-serine/threonine recognition modules essential for the function of BRCA1. Recent studies suggest that acetyl-CoA carboxylase 1 (ACC1), an enzyme with crucial roles in de novo fatty acid biosynthesis and lipogenesis and essential for cancer cell survival, may be a novel binding partner for BRCA1, through interactions with its BRCT domains. We report here the crystal structure at 3.2 Å resolution of human BRCA1 BRCT domains in complex with a phospho-peptide from human ACC1 (p-ACC1 peptide, with the sequence 1258-DSPPQ-pS-PTFPEAGH-1271), which provides molecular evidence for direct interactions between BRCA1 and ACC1. The p-ACC1 peptide is bound in an extended conformation, located in a groove between the tandem BRCT domains. There are recognizable and significant structural differences to the binding modes of two other phospho-peptides to these domains, from BACH1 and CtIP, even though they share a conserved pSer-Pro-(Thr/Val)-Phe motif. Our studies establish a framework for understanding the regulation of lipid biosynthesis by BRCA1 through its inhibition of ACC1 activity, which could be a novel tumor suppressor function of BRCA1.
Breast and ovarian cancers are among the most commonly diagnosed cancers in women. Hereditary predisposition to these cancers account for 5-10% of the cases, and roughly half of these familial cases can be traced to germline mutations in the BRCA1 (breast cancer associated 1) gene (1, 2). The 220-kDa BRCA1 protein is a tumor suppressor and is involved in multiple cellular functions, such as DNA repair, cell cycle checkpoint control, transcription and ubiquitination (3-5).
A large portion of BRCA1-linked cancer mutations is located in its two tandem repeats of the BRCA1 C-terminal (BRCT) domains (4, 6). Besides BRCA1, many other proteins are known to contain a single or multiple BRCT domains, with 90-100 amino acid residues in each domain (7). The tandem BRCT domains function as phospho-serine/threonine recognition modules (8-10) and mediate protein-protein interactions (11). The BRCT domains of BRCA1 have been demonstrated to bind the phosphorylated DNA helicase BACH1, the phosphorylated transcriptional corepressor CtIP, and phosphorylated Abraxas (5, 8, 10, 12-14).
Crystal structures of free human BRCA1 BRCT domains and their complex with phospho-peptides p-BACH1, p-CtIP and an artificial phospho-peptide optimized for binding are available (15-19). In addition, the structures of the BRCT domains from p53-binding protein 1 (53BP1), mediator of DNA damage checkpoint protein 1 (MDC1), and BRCA1 associated RING domain protein 1 (BARD1) have also been determined (20-23). The BRCT domains belong to the α/β class and the two domains are packed in a head-to-tail manner. The phospho-peptide is bound at the interface between the two BRCT domains (7, 24), and cancer-causing mutations in the BRCT domains of BRCA1 can disrupt the binding of the phospho-peptides (16-19, 25).
Most recently, acetyl-CoA carboxylase 1 (ACC1) has been identified as a novel binding partner for the BRCT domains of BRCA1 in a phosphorylation dependent manner (26-28). The site of phosphorylation in ACC1 that is essential for this interaction has been mapped to Ser1263 (27), in the central region of ACC1 and outside its biotin carboxylase and carboxyltransferase domains. Interestingly, the phospho-serine sites in BACH1, CtIP and ACC1 share a common motif, pSer-Pro-Thr/Val-Phe, and peptide binding studies revealed that the Phe residue (at the +3 position) is strongly preferred by the BRCA1 BRCT domains (16).
ACC1 is present in the cytosol in all tissues but is enriched in liver, adipocytes and other lipogenic tissues. It catalyzes the first and rate-limiting step of de novo fatty acid biosynthesis (29, 30). Many lines of evidence suggest that up-regulation of fatty acid biosynthesis is required for carcinogenesis, as more than 90% of the lipids are synthesized through de novo lipogenesis in cancer cells (31). Studies utilizing the RNAi technique or chemical inhibition have shown that ACC1 is essential for cancer cell survival (32-34). BRCA1 significantly reduces the rate of fatty acid biosynthesis when complexed with ACC1, suggesting a novel function for BRCA1 in its tumor suppressor activity (28, 35).
We report here the crystal structure of human BRCA1 BRCT domains in complex with a phospho-peptide from human ACC1 (the p-ACC1 peptide). There are recognizable and significant structural differences to the binding modes of the p-BACH1 and p-CtIP peptides, even though they share a conserved pSer-Pro-Thr/Val-Phe motif. The observed interactions for the p-ACC1 peptide are consistent with the Kd value for the complex as determined from isothermal titration calorimetry experiments. Our studies provide structural evidence for direct interactions between BRCA1 and ACC1, which may be the molecular basis for the regulation of lipid biosynthesis by BRCA1.
Materials and Methods
Protein production and crystallization
The BRCT domains of human BRCA1 (residues 1646-1859) were sub-cloned from a GST-BRCT template into the pET28a vector. The BRCT domains were over-expressed in E. coli BL21/Star (DE3) cells (Novagen) and purified using nickel agarose and gel-filtration chromatography. The purified BRCT domains were concentrated to 20-30 mg/ml in a buffer containing 20 mM Tris (pH 8.5), 200 mM NaCl, 5% (v/v) glycerol and 2 mM DTT. The N-terminal His-tag was not removed for crystallization.
Crystals of human BRCT domains in complex with the phosphorylated human ACC1 peptide 1258-DSPPQ-pS-PTFPEAGH-1271 (purchased from Elim Biopharmaceuticals Inc., CA) were obtained by the hanging-drop vapor diffusion method at 21°C. The protein solution at 15 mg/ml was mixed and incubated with the phospho-peptide at a molar ratio of 1:1.5 on ice for 45 minutes before crystallization. 2 μl of the protein/peptide solution was mixed with 1 μl of the reservoir solution containing 100 mM sodium acetate (pH 4.5), 25-27% (v/v) PEG400, and 200 mM calcium acetate. The crystals were cryo-protected with the reservoir solution supplemented with 10% (v/v) glycerol.
Structure Determination
X-ray diffraction data were collected at the X4A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the HKL package (Table 1) (36). The crystals belong to space group P212121, with cell parameters of a=83.3 Å, b=181.5 Å, and c=194.6 Å. The diffraction intensity of most crystals falls sharply at around 7 Å resolution. After screening through many crystals, the best crystal diffracted to 3.2 Å resolution.
Table 1. Summary of crystallographic information.
| Structure | BRCT/p-ACC1 complex |
|---|---|
| Resolution range (Å) | 30-3.2 |
| Number of observations | 141,866 |
| Rmerge 1(%) | 11.8 (39.1) |
| I/σI | 7.8 (2.8) |
| Observation redundancy | 3.2 (2.2) |
| Number of reflections | 42,426 |
| Completeness (%) | 92 (72) |
| R factor2(%) | 25.7 (36.8) |
| Free R factor2(%) | 30.8 (38.7) |
| rms deviation in bond lengths (Å) | 0.008 |
| rms deviation in bond angles (°) | 1.2 |
| Ramachandran plot statistics | |
| Most favored region | 80.9% |
| Additional allowed region | 16.7% |
| Generously allowed region | 2.4% |
| Disallowed region | 0% |
| Average B value (Å2) | 88 |
. The numbers in parentheses are for the highest resolution shell.
. 5% of the reflections were used for the free R calculation.
The structure was solved by the molecular replacement method with the program Phaser (37), and the structure of the free BRCT domains was used as the model (15). Based on the Vmcalculation, there could be between 7 and 14 molecules per asymmetric unit of the crystal. A total of 8 molecules were located by Phaser. Careful examination of crystal packing and unsuccessful attempts at finding the 9th molecule excluded the possibility that there could be more molecules in the asymmetric unit. The Vm of the crystal is 3.8 Å3/Dalton, corresponding to a solvent content of 67% and consistent with the poor diffraction quality of the crystal. After the first round of rigid-body refinement, with the program Refmac (38), the Fo-Fc map clearly indicated the electron density for 5 out of 8 bound peptides. The models for the BRCT domains were adjusted with the program O (39), and 8 peptide models were built into the electron density after the second round of refinement. Eight-fold non-crystallographic symmetry restraint (with weight of 0.5) was applied throughout the refinement, and TLS parameters were introduced in the later stages of the refinement. The crystallographic information is summarized in Table 1.
Isothermal Titration Calorimetry (ITC)
The ITC experiment was performed on an OMEGA Titration Calorimeter (MicroCal, Inc., Northampton, MA). 4 μl aliquots of the p-ACC1 peptide at 4 mM concentration were titrated against BRCT domains at 0.1 mM concentration in a solution containing 20 mM sodium phosphate (pH 7.4) and 300 mM NaCl at 25 °C. The titration data were recorded and analyzed using the Origin software (MicroCal Software, Northampton, MA) supplied with the instrument. The experiment was repeated once to ensure reproducibility.
Results and Discussion
Overall structure of the BRCT-phosphorylated ACC1 peptide complex
The crystal structure of the human BRCA1 BRCT domains (residues 1648-1859) in complex with a phosphorylated peptide from human ACC1 (residues 1258-1271, with the sequence DSPPQ-pS1263-PTFPEAGH, to be referred to as the p-ACC1 peptide) has been determined at 3.2 Å resolution (Fig. 1A). The crystallographic asymmetric unit contains eight copies of the BRCT/p-ACC1 complex, and the 8-fold non-crystallographic symmetry improves the overall quality of the electron density map and the refined structure despite the lower diffraction resolution (Table 1). None of the residues in the His-tag are observed in the electron density. The atomic coordinates and the observed structure factors have been deposited into the PDB (accession code 3COJ).
Figure 1.
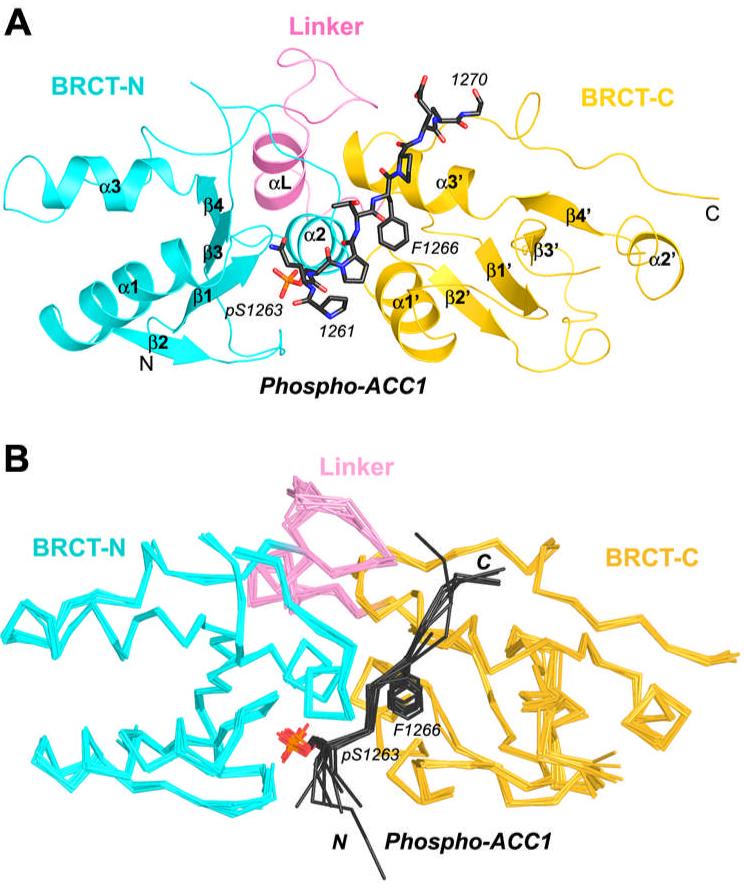
Overall structure of human BRCA1 BRCT domains in complex with a human ACC1 phospho-peptide. (A). Overview of the BRCT/p-ACC1 complex. The BRCT-N and BRCT-C domains are colored in cyan and yellow, respectively. The linker segment is colored in magenta. The p-ACC1 peptide is drawn as a stick model in black. (B). Superimposition of the eight BRCT/p-ACC1 complexes in the crystallographic asymmetric unit. The BRCT domains and p-ACC1 peptides are shown as Cα trace, and the side chain of the pSer1263 residue of p-ACC1 is drawn as stick models. All structure figures in this paper are produced with Pymol (44).
The eight BRCT domains share very similar conformation, with rms deviation of 0.5 Å for their equivalent Cα atoms (Fig. 1B). The central portion of the eight p-ACC1 peptides, in the pS-P-T-F motif, also has essentially the same conformation. Variations in the conformation of these peptides at their N- and C-termini are partly due to crystal packing differences, and some of these terminal residues are disordered (Fig. 1B). Residue His1271 of the p-ACC1 peptide is not observed in any of the complexes, and residues Asp1258-Pro1260 are observed in only one of the complexes (Fig. 1B).
Each BRCT domain contains a central 4-stranded β-sheet (β1-β4 for the N-terminal domain, BRCT-N, and β1’-β4’ for BRCT-C) surrounded by two helices (α1 and α3 for BRCT-N) on one face and one helix (α2) on the other (Fig. 1A). The two domains are packed against each other through the interactions between helix α2 from BRCT-N and helices α1’ and α3’ from BRCT-C (Fig. 1A). The linker segment connecting the two domains also helps mediate interactions between them (Fig. 1A).
Binding mode of the p-ACC1 peptide
The 2Fo-Fc electron density illustrates unambiguously an extended conformation for the p-ACC1 peptide in the complex (Fig. 2A). The peptide is nestled at the interface between the two BRCT domains (Figs. 1A, 2B). Its N-terminal half (residues Pro1261-pSer1263) is bound by the BRCT-N domain, whereas its C-terminal half (Pro1264-Gly1270) is bound by the BRCT-C domain. The β1, β1-α1 loop, β3-α2 loop and α2 from the BRCT-N domain, and β1’-α1’ loop and α3’ from the BRCT-C domain constitute the phospho-peptide binding surface (Figs. 1A, 3A). The interactions between BRCT-N and the p-ACC1 peptide are hydrophilic in nature, primarily with the pSer1263 residue (Fig. 3B). In comparison, the interactions between BRCT-C and the p-ACC1 peptide are more hydrophobic in nature, primarily with the Phe1266 side chain. Besides pSer1263 and Phe1266, the C-terminal portion of the p-ACC1 peptide also contributes to the interactions with the BRCT domains (Figs. 3A, 3B).
Figure 2.
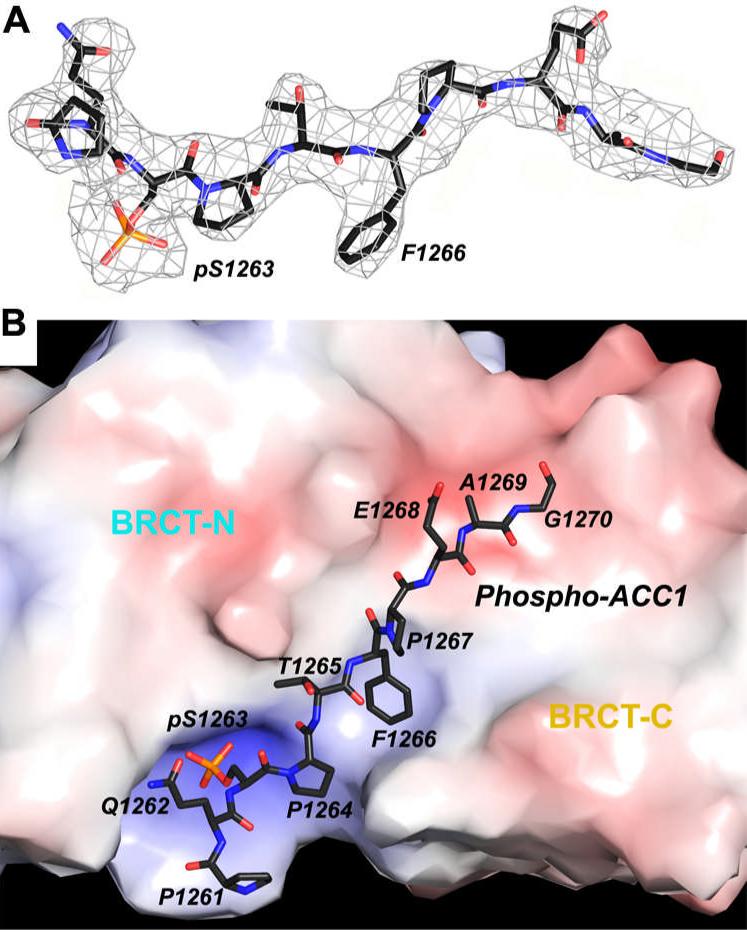
The p-ACC1 peptide is bound at the interface between the two BRCT domains. (A). The final 2Fo-Fc electron density map for the p-ACC1 peptide at 3.2 Å resolution, contoured at 1σ level. (B). The electrostatic surface view of the p-ACC1 binding site. The p-ACC1 peptide is shown as a stick model, and the two BRCT domains are labeled. The surface is colored by the electrostatic potential (red-negative, blue-positive), starting at 10 kT/e.
Figure 3.
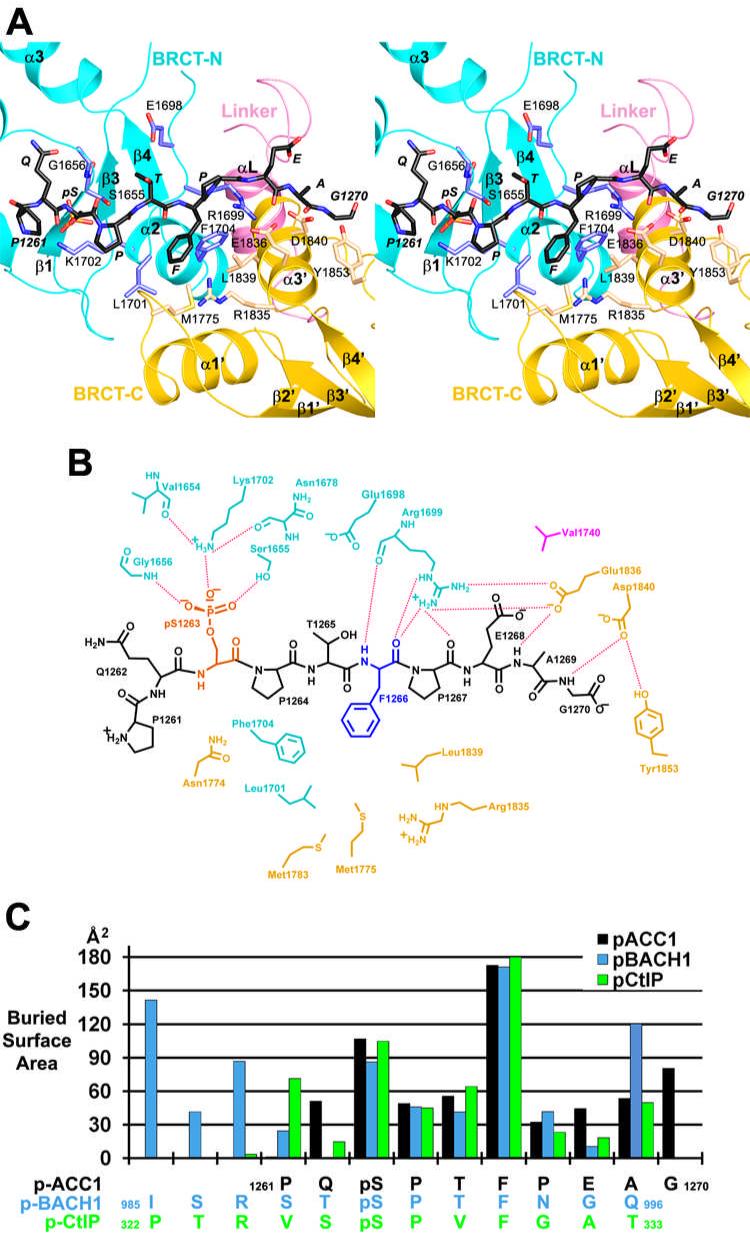
The binding mode of the p-ACC1 peptide. (A). Stereo drawing of the p-ACC1 binding mode. The BRCT-N domain is colored in cyan (main-chain) and light blue (side-chain). The BRCT-C domain is colored in yellow (main-chain) and wheat (side-chain). The linker is colored in magenta. The p-ACC1 peptide is shown as a stick model and labeled in italic. (B). Schematic drawing of the interactions between the p-ACC1 peptide and the BRCT domains. The dashed lines represent hydrogen-bonding and ion-pair interactions. (C). Contribution of individual residues in the phospho-peptide to the surface area burial in the BRCT/p-ACC1, BRCT/p-BACH1 and BRCT/p-CtIP complexes.
The pSer1263 residue of the p-ACC1 peptide protrudes into a positively-charged shallow pocket in the BRCT-N domain (Fig. 2B). One of the terminal oxygen atoms on the phosphate has ionic interactions with the side chain ammonium ion of residue Lys1702, which is held in place by two hydrogen-bonds with the main-chain carbonyls of Val1654 and Asn1678 (Figs. 3A, 3B). Lys1702 is conserved among BRCA1 homologs as well as other BRCT domains, such as BARD1, 53BP1 and MDC1 (18), underlining its importance for recognizing the phosphate group. The other two terminal oxygen atoms of the phosphate are hydrogen-bonded to the side-chain hydroxyl group of Ser1655 and the main-chain amide of Gly1656 (Figs. 3A, 3B). This pattern of recognizing the phosphate group is also observed in other protein modules, including the 14-3-3 and FHA families (24). The two residues N-terminal to pSer1263 in the p-ACC1 peptide (Pro1261 and Gln1262) make little contact to the BRCT domains (Fig. 3B).
The side chain of Phe1266 in the p-ACC1 peptide is located in a deep, hydrophobic pocket at the interface between the two BRCT domains (Figs. 1A, 2B), involving residues Met1775 and Leu1839 from the BRCT-C domain, and Leu1701 and Phe1704 from the BRCT-N domain (Figs. 3A, 3B). The main-chain amide of Phe1266 is recognized by a hydrogen-bond with the main-chain carbonyl of Arg1699 in the BRCT-N domain (Fig. 3B), whereas the main-chain carbonyl group of Phe1266 is hydrogen-bonded to the side-chain guanidinium group this Arg residue (Fig. 3A). This extensive network of interactions with the main-chain and side-chain of Phe1266 explains the strong preference for a phenylalanine residue at this position in the phospho-peptide (16).
The C-terminal region of the p-ACC1 peptide makes contact to the BRCT-C domain through several main-chain interactions (Fig. 3B). For example, the main-chain carbonyl group of Pro1267 is hydrogen-bonded to the side-chain guanidinium group of Arg1699 in the BRCT-N domain (Fig. 3B). However, as the C-terminal residues are not ordered in all the p-ACC1 peptides (Fig. 1B), it is likely that these interactions are not essential for the binding of the peptide.
Roughly 650 Å2 of the surface area of the p-ACC1 peptide shown in Fig. 3A is buried upon interaction with the BRCT domains. Consistent with the structural analysis described above, the pS-P-T-F motif in the center of the p-ACC1 peptide, especially the pSer1263 and Phe1266 residues, make the largest contribution to the surface area burial in the complex (Fig. 3C). About 43% of the total buried surface area is from the pSer1263 and Phe1266 residues. In the other 7 complexes in the crystallographic asymmetric unit, the N- and C-terminal segments of the peptide (outside the pS-P-T-F motif) have different conformations or are disordered (Fig. 1B), and the buried surface area of these peptides are somewhat smaller.
Conformational differences to other BRCT/phospho-peptide complexes
The overall structure of the BRCT domains and the binding mode of the central portion of the phospho-peptides, essentially the pS-P-(T/V)-F motif, are similar among the structures of the p-ACC1, p-BACH1 and p-CtIP complexes (Fig. 4A) (17-19). The rms distance between the Cα atoms of the BRCT domains in the p-ACC1 complex and the equivalent atoms in the other two complexes is 0.9 Å. In all three structures, the pSer and the Phe residues are involved in the most interactions with the BRCT domains, providing the anchoring points for the binding of the peptides (Fig. 3C).
Figure 4.
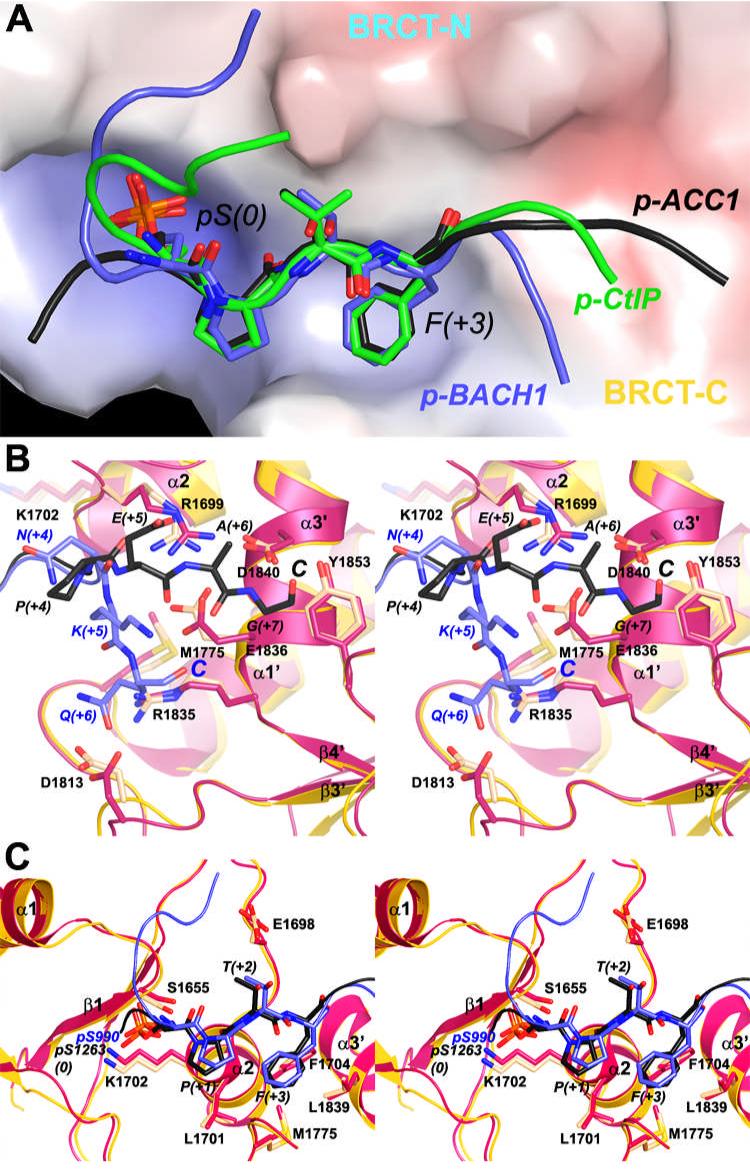
Conformational differences to other BRCT/phospho-peptide complexes. (A). The overall binding modes of the p-ACC1, p-BACH1 and p-CtIP peptides to the BRCA1 BRCT domains. The molecular surface of the BRCT domains is colored by the electrostatic potential. (B). Different binding modes of the C-terminal regions of the p-ACC1 and p-BACH1 peptides. The BRCT domains from the BRCT/p-ACC1 complex are colored in yellow (main-chain) and wheat (side-chain), and those from the BRCT/p-BACH1 complex are colored in red. The p-ACC1 (black) and p-BACH1 (light blue) peptides are drawn as stick and ribbon models. (C). Different binding modes in the central motif of the p-ACC1 and p-BACH1 peptides.
However, there are also recognizable and significant structural differences among the three complexes (Fig. 4A). The largest difference is seen in the conformation of the N- and C-terminal ends of the peptides. The p-ACC1 peptide is bound in a mostly extended form, whereas the p-BACH1 and p-CtIP peptides assume roughly an “S” shape (Fig. 4A). The N-terminal three residues of the p-BACH1 peptide (at the -5, -4, and -3 positions) have significant interactions with the BRCT domains and contribute to the large surface area burial of this peptide in the complex (Fig. 3C) (18, 19). At the C-terminal end, the p-ACC1 peptide interacts with a distinct region of the BRCT domains as compared to the p-BACH1 or p-CtIP peptides (Fig. 4A). The side-chain guanidinium group of Arg1699 is hydrogen-bonded to the main-chain carbonyl oxygens of both Phe1266 (at the +3 position) and Pro1267 (+4 position) in the extended conformation of the p-ACC1 peptide (Figs. 3B, 4B). In comparison, the p-BACH1 peptide changes direction by roughly 70° at the +4 residue, and the side-chain guanidinium group of Arg1699 assumes a different conformation in that complex, which also breaks the bi-dentate ion-pair interactions with the Glu1836 side chain observed in the p-ACC1 peptide complex (Fig. 4B). Clear electron density for this bi-dentate interaction was observed in our structure for the complex with the best electron density for the p-ACC1 peptide. Conformational changes in the side chains of both residues, as well as a small change in the position of the α3’ helix, allowed for the formation of this bi-dentate ion pair (Fig. 4B). Like p-BACH1, the p-CtIP peptide is also bound in an S conformation (Fig. 4A), and the bi-dentate interaction is not observed in that complex. Despite their different interactions with the BRCT domains, the C-terminal residues of the phospho-peptides contribute roughly the same surface area burial to the complex (Fig. 3C).
The Arg1699-Glu1836 bi-dentate ion pair is observed in the structure of the MDC1 BRCT domains in complex with the H2AX peptide, which is bound in an extended conformation as well (20). This Arg side chain is also crucial for recognizing the C-terminus of the phospho-peptide in that complex, by forming bi-dentate interactions with the carboxylate group. Our structural observations suggest that the Arg1699-Glu1836 bi-dentate ion pair could also be formed in a complex with phospho-peptide having an extended C-terminal segment.
There are also differences in the binding modes for the central portions of the peptides in the three complexes, and an overlay of this region in the structures of the p-ACC1 and p-BACH1 complexes is shown in Fig. 4C. The phosphate group on the pSer residue and the Lys1702 residue that interacts with it show variations of about 0.8 Å among the structures, and this shift in the position of the phosphate group may also be coupled with conformational changes in the β1-α1 loop in the BRCT-N domain (Fig. 4C). In comparison, the Phe residue at the +3 position has smaller conformational variation in the three structures (Fig. 4C), suggesting that the hydrophobic pocket for this residue is more rigid.
The affinity of the BRCA1 BRCT domains for the p-ACC1 peptide
We used isothermal titration calorimetry (ITC) to determine the binding affinity between the p-ACC1 peptide and the BRCA1 BRCT domains. A Kdvalue of 5.2 μM was obtained from the ITC data, and the stoichiometry of the complex is 1.04±0.02 (Fig. 5). The apparent ΔH of the interaction is -9.0±0.2 kcal/mol. The affinity of the BRCT domains for the p-ACC1 peptide is comparable to that for the p-CtIP peptide, with a Kd of 3.7 μM (17), but is 6-fold weaker than that for the p-BACH1 peptide, with a Kd of 0.7∼0.9 μM (17-19). The higher affinity for the p-BACH1 peptide is likely due to the additional interactions of its N-terminal residues with the BRCT domains (Fig. 3C).
Figure 5.
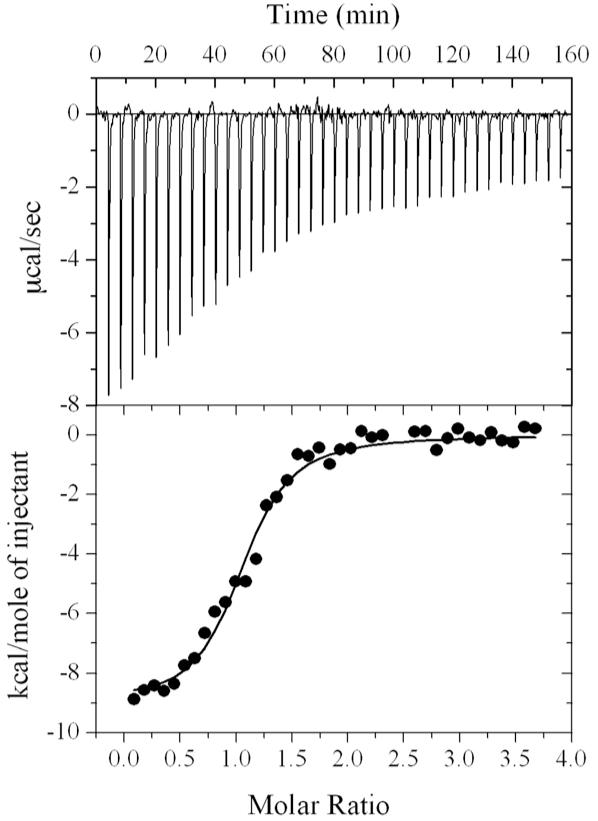
ITC measurement of the affinity between the BRCT domains and the p-ACC1 peptide.
Implications for the function of BRCA1 in fatty acid biosynthesis
The Ser1263 phosphorylation site is distinct from those for AMP-activated protein kinase (AMPK) and cAMP-dependent protein kinase in ACC1 (27, 29, 40). Therefore, a different protein kinase is required for the phosphorylation of this residue. However, the identity of this protein kinase is currently not known, neither is the protein kinase that phosphorylates the Ser residue in BACH1 and CtIP that is recognized by the BRCT domains, with which ACC1 shares the common Ser-Pro-(Thr/Val)-Phe motif. The presence of a Pro residue just after the phosphorylated Ser suggests the possible involvement of a proline-directed protein kinase, for example the MAP kinases or the CDKs. We examined many of these kinases, including p38α, CDK2, CDK6, CDK9 and GSK3β (with the CDKs in complex with the appropriate cyclin), but none of them can phosphorylate a peptide substrate from ACC1, DSPPQSPTFPEA (unpublished data).
BRCA1 is widely considered to function in the nucleus for its roles in DNA damage repair and other processes. BRCA1 also contains two nuclear export sequences, which suggests that this protein can also enter the cytoplasm (41, 42). The heterodimeric BRCA1/BARD1 complex can shuttle between the nucleus and the cytoplasm through the CRM1 export receptor (43). Therefore, BRCA1 is expected to have functions in the cytoplasm as well.
Our studies provide structural evidence for direct interactions between the BRCT domains of BRCA1 and a phospho-peptide from the cytosolic enzyme ACC1 (27). As the BRCA1-ACC1 complex has reduced activity for fatty acid biosynthesis (35), these studies suggest a cytoplasmic component for BRCA1’s function as a tumor suppressor. BRCA1 helps keep fatty acid biosynthesis and lipogenesis in check in normal cells, whereas mutations in the BRCT domains of BRCA1 can abolish the regulation of ACC1 and lead to elevated lipogenesis (28), which is an important requirement for cancer cell growth.
Acknowledgements
We thank J. A. Ladias for the gift of the GST-BRCT plasmid; Amit Reddi for help with the ITC experiment; Yan Zhu and Carol Prives for help with the cell culture experiment; Randy Abramowitz and John Schwanof for setting up the X4A beamline.
This research is supported in part by a grant from the NIH (DK067238) to LT.
Abbreviations used
- BRCA
breast cancer associated
- BRCT
BRCA1 C-terminal
- ACC
acetyl-coenzyme A carboxylase
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- PDB
protein data bank
References
- (1).Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am. J. Hum. Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- (2).King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- (3).Yarden RI, Papa MZ. BRCA1 at the crossroad of multiple cellular pathways: approaches for therapeutic interventions. Mol. Cancer Ther. 2006;5:1396–1404. doi: 10.1158/1535-7163.MCT-05-0471. [DOI] [PubMed] [Google Scholar]
- (4).Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat. Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- (5).Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, Baumbach L, Sutphen R, Pickard-Brzosowicz JL, Nathanson KL, Sali A, Goldgar D, Couch FJ, Radice P, Monteiro AN. Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Glover JNM, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- (8).Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- (9).Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- (10).Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J. Biol. Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- (11).Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat. Res. 2000;460:319–332. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]
- (12).Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- (14).Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- (15).Williams RS, Green R, Glover JNM. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- (16).Williams RS, Lee MS, Hau DD, Glover JNM. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- (17).Varma AK, Brown RS, Birrane G, Ladias JAA. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochem. 2005;44:10941–10946. doi: 10.1021/bi0509651. [DOI] [PubMed] [Google Scholar]
- (18).Shiozki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol. Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- (19).Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechnanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- (20).Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- (21).Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Derbyshire DJ, Basu BP, Serpell LC, Joo WS, Date T, Iwabuchi K, Doherty AJ. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Birrane G, Varma AK, Soni A, Ladias JAA. Crystal structure of the BARD1 BRCT domains. Biochem. 2007;46:7706–7712. doi: 10.1021/bi700323t. [DOI] [PubMed] [Google Scholar]
- (24).Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can’t pSERious? Structure. 2001;9:R33–R38. doi: 10.1016/s0969-2126(01)00580-9. [DOI] [PubMed] [Google Scholar]
- (25).Williams RS, Glover JNM. Structural consequences of a cancer-causing BRCA1-BRCT missense mutation. J. Biol. Chem. 2003;278:2630–2635. doi: 10.1074/jbc.M210019200. [DOI] [PubMed] [Google Scholar]
- (26).Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, Venezia ND. BRCA1 interacts with acetyl-CoA carboxylase through its tandem of BRCT domains. Oncogene. 2002;21:6729–6739. doi: 10.1038/sj.onc.1205915. [DOI] [PubMed] [Google Scholar]
- (27).Ray H, Moreau K, Dizin E, Callebaut I, Venezia ND. ACCA phosphopeptide recognition by the BRCT repeats of BRCA1. J. Mol. Biol. 2006;359:973–982. doi: 10.1016/j.jmb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- (28).Brunet J, Vazquez-Martin A, Colomer R, Grana-Suarez B, Martin-Castillo B, Menendez JA. BRCA1 and acetyl-CoA carboxylase: the metabolic syndrome of breast cancer. Mol. Carcinog. 2008;47:157–163. doi: 10.1002/mc.20364. [DOI] [PubMed] [Google Scholar]
- (29).Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Ann. Rev. Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- (31).Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin. Cancer Res. 1997;3:2115–2120. [PubMed] [Google Scholar]
- (32).Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- (33).Beckers A, Organe S, Timmermans L, Scheys K, Peeters A, Brusselmans K, Verhoeven G, Swinnen JV. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67:8180–8187. doi: 10.1158/0008-5472.CAN-07-0389. [DOI] [PubMed] [Google Scholar]
- (34).Brusselmans K, de Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- (35).Moreau K, Dizin E, Ray H, Luquain C, Lefai E, Foufelle F, Billaud M, Lenoir GM, Venezia ND. BRCA1 affects lipid synthesis through its interaction with acetyl-CoA carboxylase. J. Biol. Chem. 2006;281:3172–3181. doi: 10.1074/jbc.M504652200. [DOI] [PubMed] [Google Scholar]
- (36).Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (37).McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Cryst. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- (38).Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (39).Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- (40).Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metabolism. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- (41).Thompson ME, RobinsonBenion CL, Holt JT. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J. Biol. Chem. 2005;280:21854–21857. doi: 10.1074/jbc.M502676200. [DOI] [PubMed] [Google Scholar]
- (42).Rodriguez JA, Henderson BR. Identification of a functional nuclear export sequence in BRCA1. J. Biol. Chem. 2000;275:38589–38596. doi: 10.1074/jbc.M003851200. [DOI] [PubMed] [Google Scholar]
- (43).Henderson BR. Regulation of BRCA1, BRCA2 and BARD1 intracellular trafficking. Bioessays. 2005;27:884–893. doi: 10.1002/bies.20277. [DOI] [PubMed] [Google Scholar]
- (44).DeLano WL. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]


