Abstract
Confirmation of gene expression by a second methodology is critical in order to detect false-positive findings associated with microarrays. However, the impact of methodology upon the measurement of gene expression has not been rigorously evaluated. In the current study, we compared differential gene expression between PC3 and PC3-M human prostate cancer cell lines using three separate methods: microarray, quantitative RT/PCR (qRT/PCR), and Northern blotting. The PC3 to PC3-M ratio of gene expression was determined for each of 24 different genes evaluated, by each of the three methods. Comparison of gene expression ratios between Northern and microarray, Northern and qRT/PCR, and microarray and qRT/PCR, gave correlation coefficients (r) of 0.72, 0.39, and 0.63, respectively. In each instance, one to two outlier genes were apparent. Their exclusion from analysis gave r values of 0.79, 0.72, and 0.83, respectively. These findings demonstrate that the assessment of differential gene expression is dependent upon the methodology used in each situation where outcome between different methodologies was compared, the presence of a relatively limited number of outlier genes precludes high overall correlation between the methods. Validation of gene expression by different methods should be performed whenever possible.
Keywords: microarray, quantitative PCR, Northern blot, gene expression
DNA microarrays are a leading platform for monitoring the expression of thousands of genes in parallel. This technology has been applied to many situations, including disease diagnosis, drug discovery, and toxicology.1–5 As microarray technology evolves, evaluation of upwards of 20,000 genes is becoming routine.6,7 With the capability to screen large portions of the human genome, microarrays are typically used for up-front screening. Use as an up-front screening method, as well as involvement of large numbers of genes, are both contributors to the high proportion of false positives associated with the use of microarrays.8 Thus, confirmation of gene expression by a second methodology is critically important. Confirmation is typically performed as a second step after initial gene array analysis. Once differential gene expression is confirmed by a second methodology, significant resources are focused upon further investigating the identified genes of interest. Given the importance of confirming initial gene array findings, surprisingly little attention has been paid to the methodology used to confirm them.
Because of its high sensitivity, wide availability, and requirement of only a small amount of input RNA, reverse transcription, followed by quantitative polymerase chain reaction (qRT/PCR), is widely used to confirm initial microarray findings.9–13 It is important to note, however, that qRT/PCR is the methodology of choice for confirming gene array findings largely because of the convenience- related factors listed above. That is, qRT/PCR has not been shown to be the optimal methodology. When using qRT/PCR to evaluate microarray findings, general rates of confirmation vary as a function of the particular system under investigation, but a rate of ∼70% can be considered representative.14,15 Under such circumstances, qRT/PCR is considered the gold standard measure, and thus differences are attributed to factors related to microarray-based methodology.
Northern blot analysis is the gold standard to which newer technologies should be compared. This is because factors inherent to Northern blotting serve to negate the impact of gene-specific effects upon measurement outcome. Specifically, the impact of primary base sequence, as well as its effect upon secondary and tertiary gene structure, are relatively less important for Northern blot analysis compared to either microarray or qRT/PCR. The later two methods are dependent upon one or more enzymatic reactions. These enzymatic reactions, as a group, are differentially affected by template base sequence and structure. It is therefore surprising that a rigorous comparison between Northern blot–, microarray-, and qRT/ PCR-based methods for measuring differential gene expression has not been performed. We thus endeavored to make such a comparison in this study.
MATERIALS AND METHODS
Cell Culture
The origin and culture conditions for PC3 and PC3-M human prostate cancer cell lines were previously described.16 Cells were maintained at 37°C in a humidified atmosphere of 5% carbon dioxide, with biweekly media changes. Cell growth was monitored daily, and was continually non-confluent and exponential. Cells were routinely monitored for mycoplasma.
Isolation of RNA
Total cellular RNA was isolated and evaluated as previously described.17 Briefly, total RNA was isolated from whole cells with Trizol reagent (Invitrogen, Carlsbad, CA), according to manufacturer’s instructions. RNA was treated with RNase-free DNase (Qiagen, Valencia, CA), per manufacturer’s instructions. The quality and quantity of RNA were evaluated by measuring OD 260/280. In addition, RNA quality was evaluated by separation on an agarose gel, followed by visual inspection, as previously described.18
Measurement of Gene Expression by Microarrays
The methods for the manufacture of gene arrays, their hybridization, and subsequent scanning, have previously been described in detail.17 Briefly, arrays were custom-printed with a MagnaSpotter robot (BioAutomation Corp., Dallas, TX), by a dedicated core facility located at the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE (by DLK). They were constructed from a set of sense oligonucleotide (60-mer) probes designed for each human target gene by Compugen, Inc. (Rockville, MD), and manufactured by Sigma-Genosys, Inc. (The Woodlands. TX). In addition to 12,107 different genes, individual arrays contained 28 replications of GAPDH, and negative controls. All arrays were from a single print batch. Array quality was evaluated upon initial synthesis, as well as over time. This was done by hybridizing with previously characterized stock RNA, and comparing resultant gene expression profiles using array median centering and correlation coefficient mapping programmed in MATLAB (The Mathworks, Inc., Natick, MA). Probe was constructed by using 50 μg total RNA to synthesize cDNA using StrataScript reverse transcriptase (Stratagene), in the presence of aminoallyl-dUTP (Promega), and then conjugating with either Cy3 or Cy5 monofunctional NHS-ester (Amersham Pharmacia). Resultant probes from PC3 and PC3-M cells were hybridized together in a two-channel fashion on the same microarray chip. After hybridization and washing, array slides were scanned on a ScanArray 4000 confocal laser system (Perkin-Elmer), and the QuantArray software package (Perkin-Elmer) was used to subtract background fluorescence. The process of probe synthesis and gene array profiling was repeated at a separate time, and the resultant mean level of gene expression used for further analysis.
Quantitative Reverse Transcription and Polymerase Chain Reaction (qRT/PCR)
The reactions for qRT/PCR, the primer and probe sequences, and the method of PCR assay development, validation, and analysis have previously been described in detail.17 Briefly, 0.2 μg RNA were reverse transcribed with TaqMan reverse transcriptase (Applied Biosystems) using random hexamers. PCR reactions were performed with a TaqMan Universal PCR kit (Applied Biosystems) on an Applied Biosystems 7500 Real Time Quantitative PCR System workstation. Each reaction employed an upstream and a downstream, primer, a central probe, and all were gene specific. Reactions used cDNA from the equivalent of 40 ng RNA, were run in a reaction volume of 20 μL, run in replicates of two, run in a 96-well format (thus permitting parallel analysis of all genes from both cell lines), and always included RT-minus negative controls, GAPDH (for positive control and normalization purposes), and identical stock RNA as an inter-reaction quality control. Reactions were repeated in a similar fashion at a separate time, also in replicates of two, and the resultant mean threshold cycles were used for further analysis. The threshold cycle (Ct) for individual reactions was identified through Applied Biosystems 7500 Real Time PCR System software. Relative gene expressions were presented with the 2−ΔΔCt method.19
Additional important characteristics of qPCR reactions included the following: (a) The use of exon-spanning primers. Even though RNA was DNase treated, use of exon-spanning primers further optimized quality control. (b) For each individual qPCR reaction, products were gel separated, excised, and sequenced to confirm identity. (c) Finally, as all assays were developed and validated by us, we required that each primer/probe combination employed in a given qPCR reaction generate a sigmoidal curve of reaction product versus cycle number. Further, through the use of differing amounts of stock RNA that contained the target gene being amplified, we demonstrated that the qPCR reaction was able to accurately measure changes in the amount of target gene.
Northern Blotting
Northern blot hybridizations were performed as previously described.18 Briefly, 20 μg total RNA were size fractioned on a 1% denaturing formaldehyde-agarose gel, and then transferred to a nylon membrane (Ambion, Austin, TX). Gene-specific probes for Northern blots ranged in length from 114 to 361 base pairs, and encompassed the gene sequences amplified with qPCR reactions. The length of qPCR products ranged from 60 to 90 base pairs; (data not shown). See Table 1. Probes were manufactured by performing conventional RT/PCR using gene-specific primers, as previously described.20 Briefly, 1 μg RNA was reverse transcribed using an Amersham Pharmacia cDNA synthesis kit (Piscataway, NJ) per manufacturer’s instructions. PCR was performed on a PCR-200 DNA Engine (MJ Research, Inc., Waltham, MA) thermocycler in a reaction volume of 50 μL, consisting of 10 mM Tris-HCl, 50 mM potassium chloride, 1.5 mM magnesium chloride (pH 8.3), 0.2 mM deoxynucleoside triphosphates, 50 pmol of each up-and downstream primer, and 2.5 units of TaqDNA polymerase (Invitrogen). Primer sequences for each of the 24 different genes are shown in the lower half of Table 1. Amplification was performed for 30 sec at 94°C, 1 min at 55°C, for 33 cycles, and an extension step was carried out for 2 min at 72°C. RT-minus negative controls were always run. Amplification products were separated on a 2% agarose gel, and resultant bands were excised and sequenced to confirm identity. Probe for GAPDH was purchased from Ambion.
TABLE 1.
Genes Evaluated by Northern Blot, qRT/PCR, and Microarray
| Gene Name | PCR Forward Primer* | PCR Reverse Primer* | PCR Product Length* |
|---|---|---|---|
| Vimentin | TTCTCAGCATCACGATGAC | TTGGTTGGATACTTGCTGG | 315 |
| laminin receptor 1 (LAMR1) | GTTTGATGTGGTGGATGCTG | CGCTCCAGTCTTCAGTAGGG | 291 |
| Macrophage migration inhibitory factor (MIF) | AGAACCGCTCCTACAGCAAG | GAGTTGTTCCAGCCCACATT | 123 |
| Eukaryotic translation initiation factor 3, subunit 3 (EIF3S3) | GTCGCCAGCAGGAGAATAT | AGAGTTGCCCTGGTGTGAC | 283 |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 (NDUFB8) | TACAACAGGAACCGTGTGGA | CTGGTTCTTTGGAGGGATCA | 202 |
| Ubiquinol-cytochrome c reductase hinge protein (UQCRH) | ATGCGAGCAGTTGGAGAAAT | CTACCAGCCTCAAGCCAAAC | 310 |
| Thioredoxin-like (TXNL) | CTCCAGTCCAGGCAACAAAT | GCTTGGCAAAAGTGGTGACT | 269 |
| Proteasome (prosome, macropain) subunit, beta type, 4 (PSMB4) | GCTCGAAGATGAACCCTTTG | ACCTTTTTCGGTGACAGTGG | 290 |
| Ribosomal protein S20 (RPS20) | AAGGGCTGAGGATTTTTGGT | GGAAACGATCCCACGTCTTA | 341 |
| Syntaxin binding protein 2 (STXBP2) | GAGCCCACCTATCAGCTGTC | GCTTCTTGTCCAGTGCCTTC | 361 |
| Ribosomal protein, large, P0 (RPlP0) | TCGACAATGGCAGCATCTAC | CGACTCTTCCTTGGCTTCAA | 335 |
| Small nuclear ribonucleoprotein polypeptide F (SNRPF) | TGTCTGGACATCTGGGTGAA | TCCCCCACAAAAGATGCTAT | 114 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 (ATP5A1) | CGTGCGTCTAACTGAGTTGC | AGCTTCAAATCCAGCCAAGA | 268 |
| S100 calcium-binding protein A13 (S100A13) | TAATGGCAGCAGAACCACTG | TTGAGCTCCGAGTCCTGATT | 217 |
| Cytochrome c oxidase subunit VIIb (COX7B) | AGCTGTATTGCCGCAGTTCT | TTTGGGGTAACTCTGCCAAC | 259 |
| B-cell associated protein (REA) | GATCGCCACATCACAGAATC | AGCAGATCCACTTCCTCTGG | 156 |
| proteasome (prosome, macropain) subunit, alpha type, 3 (PSMA3) | CAAGCTGCAAAGACGGAAAT | CCCAGCTGAGTTCTAGTTCAAAA | 139 |
| sapiens ring-box 1 (RBX1) | CAAGAAGCGCTTTGAAGTGA | TGGACACACCTGTCGTGTTT | 232 |
| Arsenate resistance protein ARS2 (ARS2) | GATCTTGGGCTATGGAGCTG | TTCCCTGTGGAAGTGCTTTT | 337 |
| Ribosomal protein L13a (RPl13A) | CTGGACCGTCTCAAGGTGTT | TATTGGGCTCAGACCAGGAG | 326 |
| Ribosomal protein S16 (RPS16) | GGCAATGGTCTCATCAAGGT | TCTCCTTCTTGGAAGCCTCA | 241 |
| 2,4-Dienoyl CoA reductase 1, mitochondrial (DECR1) | AACTCGCAAATCTTGCTGCT | TTCTATGGTGTCCCACTGCTC | 153 |
| Signal sequence receptor, delta (translocon-associated protein DELTA) (SSR4) | ACGTCGGTGGAAAACAATTC | GATGGTCCACGCTGACTGTA | 210 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | CCAGGTGGTCTCCTCTGACT | AGGGGTCTACATGGCAACTG | 303 |
| Gene Name | qPCR Forward Primer** | qPCR Reverse Primer** | qPCR Probe** |
| Vimentin | GGACACTTCTGATTAAGACGGTTGA | GTGTGCAATTTTTATTCAAGGTCATCGT | cAggTTATcAAcgAAAcTT |
| Laminin receptor 1 (LAMR1) | TGCCCTCTGTGCCTATTCAG | GGAGCTGCAGACCAGTCTTC | cAAttccctActGAAGActG |
| Macrophage migration inhibitory factor (MIF) | GCGCCTGCGCATCAG | CCGCGTTCATGTCGTAATAGTTGAT | cccGGAcAGGGtctAc |
| Eukaryotic translation initiation factor 3, subunit 3 (EIF3S3) | GATGGACTCGCTGCTCATTG | GGGCAGTGAACTCCTTGATGTT | cAGGccAGAtAAAcAc |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 (NDUFB8) | GGGACGTGTACCCTGTCTAC | CCTCGTTCCAGGTACAGATTATTGT | ctttGGtcccAcAGGctG |
| Ubiquinol-cytochrome c reductase hinge protein (UQCRH) | AGGAGCTCTTTGACTTCTTGCAT | GGTGCAACTTAAGTCCACACATTTAT | ccAttGcGtGGcccAc |
| Thioredoxin-like (TXNL) | TCCAGTCCAGGCAACAAATATGAAT | TGTCTTTTGTACCTTAGTGGCTTTCT | ccAAc TAcTcgTTTgAAgTc |
| Proteasome (prosome, macropain) subunit, beta type, 4 (PSMB4) | CTACAGAGACCAACTGGGATATTGC | GCAACTTCAGTTCTGGATAATGCA | cAcAtGAtcAGtGGctttG |
| Ribosomal protein S20 (RPS20) | GTGAAAGGACCAGTTCGAATGC | ACGTCTTAGAACC TTCACCACAAG | ccAAGActttGAGAAtcAc |
| Syntaxin binding protein 2 (STXBP2) | GCCACCGAGGGCAAGT | GCCTTCAGGTCATCCAGGAA | ctcAttGGctcctcAcAcAt |
| Ribosomal protein, large, P0 (RPLP0) | GGATTACACC TTCCCACTTGCT | GCCACAAAGGCAGATGGATCA | AAGGccttGAccttttc |
| Small nuclear ribonucleoprotein polypeptide F (SNRPF) | ATGGAGCTTTGTCTGGACATCTG | TTCCTCTTCTTCCACACCTCTGATA | AcAttAttAcAccttAttAAAActtc |
| ATP synthase, H+ transporting, mitochondrial F1 COMPLEX, ALPHA SUBUNIT, ISOFORM 1 (ATP5A1) | CCAGCACCAAGCCTTGTTG | AGCTTTGCATCTGATTGTTCTGAGA | cAtcAGccctGAtAGtGc |
| S100 calcium-binding protein A13 (S100A13) | CCAGCAGTTGCCCCATCT | CACATCCAAGCTCTTCATCTTCTCA | AAGGAtGtGGGctctctt |
| Cytochrome c oxidase subunit VIIb (COX7B) | TGGAGCCACTTTCTGTATTGTTACA | CAGCTGGGATGATTACTGATTCCT | cttGtGttGctAcAtAtGtc |
| B-cell associated protein (REA) | CTTGTGCTGAACCTACAGGATGAA | TCTTACCCTTGATGAGGCTGTCA | cttcccctGGtGAAAct |
| Proteasome (prosome, macropain) subunit, alpha type, 3 (PSMA3) | GATCCATTAGGGTTACCAGACAGG | ACTTGTTGTGGCTGCTTGGA | cAtAGAtGAcAtGcctAtAAAA |
| Sapiens ring-box 1 (RBX1) | CATTGGACAACAGAGAGTGGGAATT | CAAAACAATTAAGCTTGATGGAAGAAGTCT | ctAGtGcccAtActtttG |
| Arsenate resistance protein ARS2 (ARS2) | AGGCCAGGGAGGTTATCCT | AGGTCCCGATATTCCACAATGG | tcGcAAcAGGAtGGttc |
| Ribosomal protein L13a (RPL13A) | CAAGATCCACTACCGGAAGAAGAAA | ACGTTCTTCTCGGCCTGTTT | ccGtAGcctcAtGAGctG |
| Ribosomal protein S16 (RPS16) | GCCCTGGTGGCCTATTACC | GGATGTCTTTGATCTCCTTC TTGGAA | cctcAtccAcAtAtttc |
| 2,4-dienoyl CoA reductase 1, mitochondrial (DECR1) | TGACGGTGGAGAGGAAGTACTTATT | TGTCTTCCTGATGAGTTCTTCTATGGT | tccttGGtGAcctttctc |
| Signal sequence receptor, delta (translocon-associated protein DELTA) (SSR4) | CCCGCCTCTGTTTACAGTCA | GCAGCCAGCACCTCAGT | AAGtGccccGAtGGtc |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | CGACCACTTTGTCAAGCTCATTTC | CCACCACCCTGTTGCTGTAG | cTggTATgAcAAcgAATTT |
Probes were labeled with [α32P] dCTP (Amersham Biosciences) using Ready-To-Go DNA labeling Beads kit (Amersham Biosciences), per manufacturer’s instructions. Blots were hybridized overnight at 42°C in UltraHyb Hybridization Buffer (Ambion). After washing, membranes were exposed to Hyperfilm MP (Amersham Biosciences) at −80°C for 0.5–144 h, as needed. Individual bands were quantified using AlphaEaseFC software (Alpha Innotech Corporation, San Leandro, CA), as previously described.21 All bands were within the linear range of detection for each exposure. Membranes were stripped with 0.5% SDS at 95°C, stripping confirmed by re-exposure, and membranes were re-probed with different gene-specific probes. A given blot was sequentially probed for multiple genes according the following rules: It was probed for only one gene at a time. It was first probed for genes with the lowest abundance, then for genes of increasingly greater signal intensity, and finally each blot was probed for GAPDH. All genes on a given blot were of different molecular weights, thus further ensuring no overlap of signal. Gene expression was normalized to that of GAPDH. Northern blots for each gene were repeated at a separate time, using a separate batch of RNA. The resultant mean gene expression level was used in all subsequent analysis.
Statistics
The gene expression intensities measured by microarray, Northern blot, and qRT/PCR were normalized by the values of GAPDH, as measured by that particular technology. Thus, if MA(X) was the original microarray measure of expression for gene X and MA(GAPDH) that for GAPDH, then MA(X)/MA(GAPDH) was the normalized microarray measure. Similarly, we had NB(X)/ NB(GAPDH) and qPCR(X)/qPCR(GAPDH) as normalized measures of expression of gene X in Northern blot and in qPCR technology, respectively. For comparison of expressions we used plots of all 24 genes, with one technology on the horizontal and other on the vertical axis. In such bivariate graphs, the Pearson and Spearman correlations were computed to estimate the strength of the linear association. As no major difference between results for these two correlation measures was observed, Pearson correlation results were presented.
RESULTS
Measurement of Gene Expression by Northern Blot, Microarray, and Quantitative Reverse Transcription/ PCR (qRT/PCR)
Total RNA was isolated from PC3 and PC3-M human prostate cancer cells. RNA was aliquoted and stored as stock. The quantity of stock RNA was evaluated by optical density. After confirming that the OD260/OD280 ratio was greater than 1.8, quality was further confirmed by agarose electrophoresis (Figure 1). In this manner we created a large stock of identical RNA from each cell line that was used for downstream gene expression analysis. This process was repeated at a separate time, thus allowing us to conduct replicate evaluations of gene expression (i.e., N = 2).
FIGURE 1.
Evaluation of stock RNA by agarose gel electrophoresis. PC3 and PC3-M total RNA were isolated and separated by agarose gel electrophoresis as described in Materials and Methods. Ethidium bromide–stained gels were visualized under ultraviolet light, and photographed. RNA from each cell line was isolated at two separate times, experiment #1 and experiment #2, as indicated.
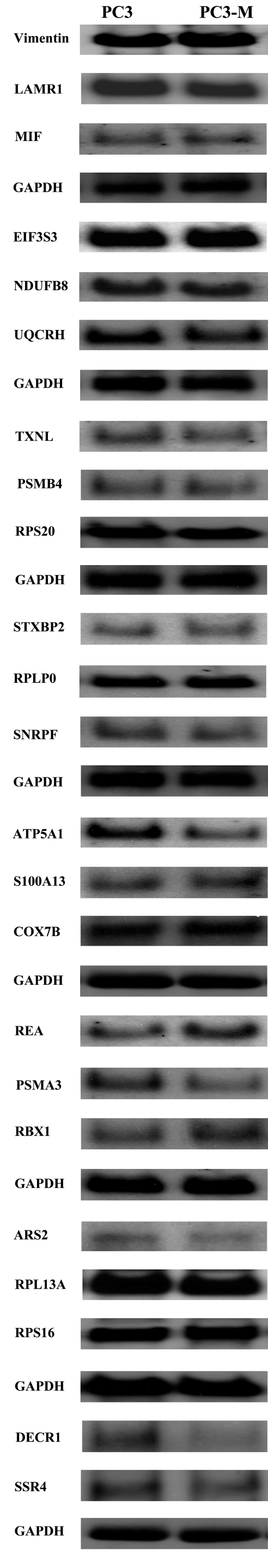
We had previously described the differential gene expression between PC3 and PC3-M cells on a 12,000-gene microarray chip using this stock RNA.17 In that same study, 32 genes were evaluated by qRT/PCR using this stock RNA. Genes selected for evaluation by qRT/PCR had exons within 500 bp of the poly-A tail. This permitted use of exon-spanning primers, and optimized overlap with 3′-enriched oligonucleodides used to construct microarrays. Further, these 32 genes contained genes that were expressed at high, intermediate, and low levels, as measured by microarray. For the current study, this prior analysis was expanded by evaluating the expression of these 32 genes by Northern blot hybridization. Of these 32 genes, a non-trivial signal was detected by Northern blot for 24 genes. Representative blots are depicted in Figure 2. For each of these 24 genes, a single prominent band was detected at the molecular weight expected for the mature transcript. For 8 genes, no discernable band was apparent despite extended exposure times, and despite multiple attempts at optimization of hybridization conditions (data not shown). The 24 genes that gave a detectable signal on Northern blot thus served as the focus of the current investigation. For each of the three different methods of analysis, two separate measures were taken, corresponding to the two separate preparations of RNA (N = 2). From this, a mean value was determined, and used for downstream analysis. For each of the three methods, the level of gene expression was normalized to that of GAPDH. Next, for each gene, and for each of the three methods, the ratio of expression in PC3 to PC3-M cells was determined (Table 2).
FIGURE 2.
Northern blot analysis of gene expression. Eight Northern blots were prepared from 20-μg aliquots of total RNA from PC3 and PC3-M cells. . Each blot was probed for the indicated genes, as described in Materials and Methods. Depicted are results are from a single experiment. Similar results were obtained in a replicate Northern blot analysis, conducted at a separate time, and using RNA prepared at a separate time.
TABLE 2.
Ratio of PC3/PC3-M Gene Expression as Measured by Microarray, qRT/ PCR, or Northern Blot Analysis
| Gene | Microarray | qRT/PCR | Northern Blot |
|---|---|---|---|
| Vimentin | 1.1 | 1.3 | 1.0 |
| LAMR1 | 1.4 | 1.0 | 0.9 |
| MIF | 1.0 | 0.9 | 1.0 |
| EIF3S3 | 1.0 | 1.3 | 1.0 |
| NDUFB8 | 0.9 | 0.8 | 1.1 |
| UQCRH | 1.0 | 0.8 | 1.4 |
| TXNL | 1.9 | 1.9 | 1.6 |
| PSMB4 | 1.5 | 1.3 | 1.1 |
| RPS20 | 0.8 | 0.8 | 0.9 |
| STXBP2 | 0.9 | 2.7 | 0.9 |
| RPlP0 | 1.0 | 1.4 | 1.0 |
| SNRPF | 1.1 | 0.9 | 1.0 |
| ATP5A1 | 1.5 | 1.4 | 1.7 |
| S100a13 | 1.0 | 1.3 | 0.9 |
| COX7B | 1.3 | 0.9 | 1.1 |
| REA | 0.6 | 0.5 | 0.7 |
| PSMA3 | 2.1 | 1.3 | 2.4 |
| RBX1 | 1.3 | 1.2 | 0.8 |
| ARS2 | 1.1 | 1.7 | 1.2 |
| RPl13A | 1.3 | 0.8 | 1.1 |
| RPS16 | 0.9 | 0.9 | 0.9 |
| DECR1 | 3.5 | 3.0 | 1.8 |
| SSR4 | 1.4 | 1.4 | 1.3 |
| GAPDH | 1.0 | 1.0 | 1.0 |
Comparison of Differential Gene Expression as Measured by Northern Blot, Microarray, and Quantitative Reverse Transcription/PCR (qRT/PCR)
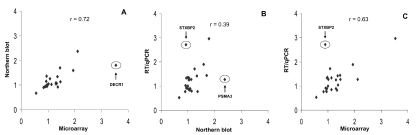
A primary goal of microarray analysis is to detect differences in gene expression between two populations of cells. Differences in expression are typically expressed as a ratio between the two cell types of interest. Therefore, current investigations focused upon evaluating method-dependent differences in the PC3 to PC3-M gene expression ratio. The ratio of gene expression for each of the 24 different genes was determined by Northern blot, qRT/ PCR, and microarray. A comparison of gene expression ratios between Northern blot and microarray gives a correlation coefficient (r) of 0.72 (Figure 3A). As can be seen, the DECR1 gene is an outlier. Exclusion of this gene from analysis leads to an improvement in r to 0.79. A comparison of gene expression ratios between Northern blot and qRT/PCR gives an r of 0.39 (Figure 3B). Here too, a limited number of genes appear to be outliers; in particular, STXBP2 and PSMA3. Exclusion of these two genes from analysis leads to a substantial improvement in r to 0.72. A comparison of gene expression ratios between microarray and qRT/PCR gives an r of 0.63 (Figure 3C). Exclusion of the outlier gene STXBP2 gives r = 0.83.
FIGURE 3.
Measurement of gene expression by different methods. The ratio of PC3/PC3-M gene expression was measured by Northern blot, qRT/PCR, and microarray for each of the 24 genes listed in Table 1, as described in Materials and Methods. Gene expression was normalized to that of GAPDH. Depicted values are the mean of two different measurements, performed at separate times. The first measurement used RNA from experiment #1, while the second used RNA from experiment #2, as described in Figure 1. For each plot, the resultant correlation coefficients are depicted, as are outlier genes. A: Correlation of Northern blot and microarray; B: Correlation of qRT/PCR and Northern blot; C: Correlation of qRT/PCR and microarray.
DISCUSSION
Microarray expression analysis has revolutionized many facets of biology, and will continue to be applied widely. However, significant questions remain with regard to the generation, analysis, and, in particular, the interpretation of microarray data.8 Although the validation of microarray expression results obtained for specific genes using independent techniques is still considered a central component of any microarray experiment, the genes selected for validation are usually identified from the microarray data. Their selection is based upon the implicit assumption that there is a good correlation between the microarray data and actual mRNA levels in the cells under investigation. Subsequent confirmation of microarray data is in turn based upon the implicit assumption that qRT/PCR provides an accurate measure of actual mRNA levels. These are fundamental issues that have not been adequately addressed by any single study wherein the performance characteristics of Northern blot, microarrays, and qRT/ PCR are all rigorously evaluated. By evaluating the ability of each of these three methods to detect differential gene expression between two cell populations in parallel, by using high-quality identical stock RNA for each method, by evaluating a relatively large number of genes, and by incorporating replicate measurements, this represents the first such study.
Of these three methods, Northern blot hybridization represents the standard to which other methods are compared. More importantly, however, with Northern blotting there is no enzymatic manipulation of RNA. This is critical, as it essentially eliminates the artificial changes in differential gene expression introduced by enzymatic manipulation seen when processing RNA from two different cell populations. In contrast, both qRT/PCR and microarray methods require enzymatic manipulation. Enzymatic manipulation is sequence and structure dependent, and can introduce a non-linear bias between two different RNA populations. Further, an inherent limitation of DNA microarrays is that not all of the thousands of genes included on the array are expected to generate a perfectly specific and linear signal. A single hybridization condition is applied to tens of thousands of distinct nucleic acid hybridization reactions, resulting in suboptimal conditions for some reactions. Some degree of non-specific and cross-hybridization for a substantial number of probes is unavoidable. Finally, factors such as the location of the oligonucleotide used for array manufacture on the target mRNA (in particular, distance from the 3′ end), the effect of differential RNA splicing on alternative transcripts, and polymorphisms can all influence the correlation between microarray data and validation techniques.
In the current study, the poorest correlation was observed when Northern blot was compared to qRT/PCR. Given the above considerations, this would in fact be the expected scenario. This is because qRT/PCR is dependent upon three linked enzymatic amplification reactions. In contrast, when Northern blot was compared to microarray, there was a much more robust correlation. While the latter did involve synthesis of cDNA, the two techniques were otherwise similar in that they relied heavily upon sequence-specific hybridization. Thus, this outcome was similarly predictable. In fact, the correlation coefficient of 0.72 observed in the current study for the comparison between Northern blot and microarray compares favorably to findings by other investigators, who report correlation coefficients ranging from 0.78 to 0.85.22–24 It was interesting to find, however, that the correlation coefficient of 0.63 observed between microarray and qRT/PCR was higher than that between Northern blot and qRT/PCR. Our results are in agreement with those of others who have compared outcomes between microarray and qRT/PCR. In studies by other investigators, correlation coefficients ranged from 0.45 to 0.75, with values of 0.68 being representative.25,26 Given that microarray and qRT/PCR both involve a cDNA synthesis step, this step may be responsible, at least in part, for the low correlation seen between Northern blot (no cDNA synthesis step) and qRT/PCR.
It is interesting that with both microarray and qRT/ PCR, outlier genes were evident when compared to the results obtained with Northern blot. Again, this finding is consistent with the factors discussed above related to the introduction of artificial differences. The notion that these factors are relevant is further supported by considering what happens when outlier genes are excluded from analysis. Specifically, there is a modest increase in the correlation coefficient when Northern blot and microarray methods are compared. In contrast, when Northern blot and qRT/PCR methods are compared, there is almost a doubling of the correlation coefficient when outlier genes are excluded. If in fact the multiple enzymatic steps inherently involved with qRT/PCR enhanced the rate of artificial outcome as a function of target sequence (and its associated effects upon the secondary and tertiary structure of target genes), then the presence of outlier genes with qRT/ PCR becomes a natural consequence.
While investigations began with 32 genes for which we had previously developed and validated qRT/PCR assays, only 24 yielded detectable signal on Northern blot. The 8 non-detectable genes represented low and moderate expressing genes, as previously reported by us.17 It was likely that use of mRNA for Northern blot, instead of total RNA, would provide for a readily detectable signal for these low and moderate expressing genes. However, we did not pursue this for two reasons. First, we wanted to avoid any potential bias introduced by the process of mRNA isolation. Second, we still had 24 genes available for analysis, thus permitting a relatively exhaustive evaluation of different genes.
In summary, our findings demonstrate that the assessment of differential gene expression is methodology dependent. In each instance where methodoligical outcome is compared, the presence of a relatively limited number of outlier genes precludes high overall methodological correlation. Our findings call into question the status of qRT/ PCR as the optimal methodology to confirm gene array findings. However, given that sample RNA is typically available only in limited amounts, practical considerations may leave no reasonable alternative when seeking to confirm microarray findings.
ACKNOWLEDGMENTS
Supported by the National Cancer Institute (CA099263, CA37403, and Specialized Program of Research Excellence grant CA90386, to RCB), and a National Center for Research Resources Grant (M01 RR00048), both from the National Institutes of Health, Department of Health and Human Services
REFERENCES
- 1.Schulze A, Downward J. Navigating gene expression using microarrays—a technology review. Nat Cell Biol. 2001;3:E190–195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 3.Debouck C, Goodfellow PN. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48–50. doi: 10.1038/4475. [DOI] [PubMed] [Google Scholar]
- 4.Macoska JA. The progressing clinical utility of DNA microarrays. CA Cancer J Clin. 2002;52:50–59. doi: 10.3322/canjclin.52.1.50. [DOI] [PubMed] [Google Scholar]
- 5.Lobenhofer EK, Bushel PR, Afshari CA, Hamadeh HK. Progress in the application of DNA microarrays. Environ Health Perspect. 2001;109:881–891. doi: 10.1289/ehp.01109881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyal F, Stransky N, Bernard-Pierrot I, Vincent-Salomon A, de Rycke Y, Elvin P, et al. Visualizing chromosomes as transcriptome correlation maps: Evidence of chromosomal domains containing co-expressed genes—a study of 130 invasive ductal breast carcinomas. Cancer Res. 2005;65:1376–1383. doi: 10.1158/0008-5472.CAN-04-2706. [DOI] [PubMed] [Google Scholar]
- 7.Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jovanovic BD, Bergan RC, Kibbe WA. Some aspects of analysis of gene array data. Cancer Treat Res. 2002;113:71–89. doi: 10.1007/978-1-4757-3571-0_5. [DOI] [PubMed] [Google Scholar]
- 9.Ha PK, Benoit NE, Yochem R, Sciubba J, Zahurak M, Sidransky D, et al. A transcriptional progression model for head and neck cancer. Clin Cancer Res. 2003;9:3058–3064. [PubMed] [Google Scholar]
- 10.Hao X, Sun B, Hu L, Lahdesmaki H, Dunmire V, Feng Y, et al. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer. 2004;100:1110–1122. doi: 10.1002/cncr.20095. [DOI] [PubMed] [Google Scholar]
- 11.Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, et al. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931–946. [PubMed] [Google Scholar]
- 12.Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, et al. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinolog y. 2004;145:3913–3924. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- 13.Le Page C, Ouellet V, Madore J, Hudson TJ, Tonin PN, Provencher DM, et al. From gene profiling to diagnostic markers: IL-18 and FGF-2 complement CA125 as serum-based markers in epithelial ovarian cancer. Int J Cancer. 2006;118:1750–1758. doi: 10.1002/ijc.21521. [DOI] [PubMed] [Google Scholar]
- 14.Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi M, Miura K, Iwao H, Yamanaka S. Quantitative assessment of DNA microarrays—comparison with Northern blot analyses. Genomics. 2001;71:34–39. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Chen S, Xu L, Liu Y, Deb DK, Platanias LC, et al. Genistein inhibits p38 map kinase activation, matrix metallo-proteinase type 2, and cell invasion in human prostate epithelial cells. Cancer Res. 2005;65:3470–3478. doi: 10.1158/0008-5472.CAN-04-2807. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Xu L, Chen S, Jovanovic BD, Helenowski IB, Kelly DL, et al. Characterization of a method for profiling gene expression in cells recovered from intact human prostate tissue using RNA linear amplification. Prostate Cancer Prostatic Dis. 2006;9(4):379–391. doi: 10.1038/sj.pcan.4500888. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Jovanovic B, Pins M, Lee C, Bergan RC. Over expression of endoglin in human prostate cancer suppresses cell detachment, migration and invasion. Oncogene. 2002;21:8272–8281. doi: 10.1038/sj.onc.1206117. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz GN, Liu YQ, Tisdale J, Walshe K, Fowler D, Gress R, et al. Growth inhibition of chronic myelogenous leukemia cells by ODN-1, an aptameric inhibitor of p210bcr-abl tyrosine kinase activity. Antisense Nucleic Acid Drug Dev. 1998;8:329–339. doi: 10.1089/oli.1.1998.8.329. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, Chen S, Bergan RC. MAPKAPK2 and HSP27 are downstream effectors of p38 MAP kinase–mediated matrix metallo-proteinase type 2 activation and cell invasion in human prostate cancer. Oncogene. 2006;25:2987–2798. doi: 10.1038/sj.onc.1209337. [DOI] [PubMed] [Google Scholar]
- 22.Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, et al. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agbemafle BM, Oesterreicher TJ, Shaw CA, Henning SJ. Immediate early genes of glucocorticoid action on the developing intestine. Am J Physiol Gastrointest Liver Physiol. 2005;288:G897–906. doi: 10.1152/ajpgi.00454.2004. [DOI] [PubMed] [Google Scholar]
- 24.Kucho K, Tsuchiya Y, Okumoto Y, Harada M, Yamada M, Ishiura M. Construction of unmodified oligonucleotide-based microarrays in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: Screening of the candidates for circadianly expressed genes. Genes Genet Syst. 2004;79:319–329. doi: 10.1266/ggs.79.319. [DOI] [PubMed] [Google Scholar]
- 25.Eom H, Lee CG, Jin E. Gene expression profile analysis in astaxanthin-induced Haematococcus pluvialis using a cDNA microarray. Planta. 2006;223:1231–1242. doi: 10.1007/s00425-005-0171-2. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, et al. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]