Abstract
Ngrol genes (NgrolB, NgrolC, NgORF13, and NgORF14) that are similar in sequence to genes in the left transferred DNA (TL-DNA) of Agrobacterium rhizogenes have been found in the genome of untransformed plants of Nicotiana glauca. It has been suggested that a bacterial infection resulted in transformation of Ngrol genes early in the evolution of the genus Nicotiana. Although the corresponding four rol genes in TL-DNA provoked hairy-root syndrome in plants, present-day N. glauca and plants transformed with Ngrol genes did not exhibit this phenotype. Sequenced complementation analysis revealed that the NgrolB gene did not induce adventitious roots because it contained two point mutations. Single-base site-directed mutagenesis at these two positions restored the capacity for root induction to the NgrolB gene. When the NgrolB, with these two base substitutions, was positioned under the control of the cauliflower mosaic virus 35S promoter (P35S), transgenic tobacco plants exhibited morphological abnormalities that were not observed in P35s-RirolB plants. In contrast, the activity of the NgrolC gene may have been conserved after an ancient infection by bacteria. Discussed is the effect of the horizontal gene transfer of the Ngrol genes and mutations in the NgrolB gene on the phenotype of ancient plants during the evolution of N. glauca.
Various features of living organisms elicit our interest in a better understanding of biodiversity. Many biologists agree that the diversification of creatures originated as a consequence of mutations introduced during their evolution. Horizontal gene transfer is a mutational process for transfer of nucleotides between organisms (1). Horizontal gene transfer is a mechanism for the acquisition of novel capability within a generation and is considered important in the divergence and adaptation of bacterial populations. Horizontal transfer of genes is also observed between other kingdoms. A typical example of such transfer was found in the interaction of Agrobacterium species with plants. Soil microorganisms in the genus Agrobacterium elicit morphological responses in certain plant species by transferring one or two small regions of DNA [transferred DNA (T-DNA)] from its plasmid to the host plant’s genome (2). Agrobacterium rhizogenes is responsible for the formation of adventitious roots known as “hairy roots.” The agropine-type strain of A. rhizogenes has two distinct T-DNA regions, left transferred DNA (TL-DNA) and right transferred DNA, in its Ri plasmid. Four loci involved in root induction have been identified from an analysis of TL-DNA by insertional mutagenesis (3). They are designated rolA, rolB, rolC, and rolD. These genes correspond to ORFs 10, 11, 12, and 15 among the 18 ORFs of TL-DNA (4). Of these, the rolB locus is thought to be the most important for hairy-root induction (3, 5–7). Studies of transgenic plants that carry various combinations of the TL-DNA genes have shown that two other ORFs, ORF13 and ORF14, also play significant roles in the induction of roots on carrot disks and tobacco leaf segments (6, 7). Whole plants regenerated from hairy roots exhibit a characteristic abnormal phenotype known as “hairy-root syndrome” (8). These plants stably transmit TL-DNA genes to their progeny.
Sequences similar to TL-DNA have been found in untransformed plants of the genus Nicotiana (9, 10). A sequence similar to TL-DNA was obtained from the genome of Nicotiana glauca. This region contained four ORFs that corresponded to rolB, rolC, ORF13, and ORF14 (11, 12). These ORFs were designated NgrolB, NgrolC, NgORF13, and NgORF14. Expression of the Ngrol genes has been detected in the genetic tumors of hybrids between N. glauca and Nicotiana langsdorffii (12–14). The reading frames of TL-DNA in A. rhizogenes are referred to herein as Rirol loci, to distinguish them from the corresponding plant genes. Previous reports have suggested that an ancient infection of Nicotiana by an A. rhizogenes-like ancestor resulted in the horizontal transfer of T-DNA genes to their genomes (10, 11, 15). It was proposed that the horizontal transfer of these Ngrol genes might have affected the phenotype of N. glauca (11, 15–17), but little is known about the role of these genes. We are therefore interested in the effects of these Ngrol genes on plants. We considered the following questions: (i) whether the function of Ngrol genes has been conserved in plants after the ancient integration event, and (ii) why N. glauca plants do not exhibit characteristics of hairy-root syndrome. The differences in function between the genes of plants and bacteria are analyzed, and the history of Ngrol genes in the evolution of N. glauca is discussed.
Materials and Methods
Nucleic Acid Analysis.
Ngrol genes were subcloned from clone λNg31 (kindly provided by M. P. Gordon and E. W. Nester, University of Washington), which included the TL-DNA-similar region from the genome of N. glauca (10). A plasmid, pNBC-1314, was constructed by subcloning the SpeI-XhoI fragment of 7,718 bp of λNg31 at the unique XbaI site into the T-DNA region of the binary vector pMM454-Km (kindly provided by M. Sekine, Nara Institute of Science and Technology, Japan) (Fig. 1A). Similarly, pNB was constructed by subcloning the SpeI-SalI fragment of 2,972 bp in pMM454-Km. Rirol genes were subcloned from plasmid pLJ1, which contained the TL-DNA region of pRiHRI of A. rhizogenes strain HRI (2). Construction of plasmids pNC1314, pRBC1314, pRB, and pRC1314 was described previously (6, 18). A binary vector pHTS6.1 was also used for the establishment of transformants (19).
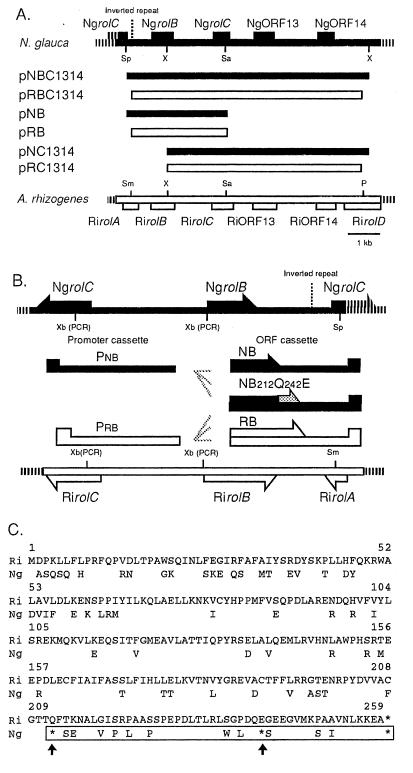
Figure 1.
(A) Schematic representation of constructs. A restriction map of the NgrolB-NgORF14 region (Top, solid line) is compared with the RirolB-RiORF14 region (Bottom, open box). The various fragments and chimeric constructs are indicated by solid bars (Ngrol genes) and open boxes (Rirol genes). P, PvuI; Sa, SalI; Sm, SmaI; Sp, SpeI; X, XhoI. (B) Promoter and ORF cassettes. Pnb, NgrolB promoter cassette; Prb, RirolB promoter cassette; NB, cassette harboring the coding and 3′ untranslated region of NgrolB; NB212Q242E, NB cassette under the modifications of site-directed mutagenesis; RB, cassette harboring the coding and untranslated region of RirolB; Xb (PCR), new restriction sites for XbaI that were created by PCR. (C) Comparison of the amino acid sequences deduced from the RirolB and NgrolB genes (4, 11). Amino acids encoded by NgrolB that are the same as those encoded by RirolB are not shown. Asterisks indicate termination codons. Arrows indicate the termination codons that were changed by site-directed mutagenesis. The restored region in the amino acid sequence encoded by NgrolB212Q242E is indicated by a box.
ORF and promoter cassettes of rolB homologues were constructed for the promoter-swap experiment. Each cassette was fused and subcloned into the modified binary vector pMM454-KmRi13, which included a ClaI-HindIII 3,229-bp fragment of pLJ1 at the ClaI/HindIII site in the T-DNA region of pMM454-Km beforehand. A fragment containing the 5′ region of the RirolB ORF was amplified by PCR from pLJ1 with the following primers: 5′-GCTCTAGAATGGACCCCAAATTGCTA-3′ and 5′-TCGGATCCCCTCGAA-3′. The ORF cassette of RirolB (RB) was constructed by subcloning this PCR-amplified fragment into the RirolB sequence. A fragment that included part of the NgrolB-coding region was generated by PCR from λNg31 with the following primers: 5′-GCTCTAGAATGGCTTCCCAATCGCAATTC-3′ and 5′-GTCCCTTTCTCGAGCCAAATC-3′. The ORF cassette of NgrolB (NB) was constructed by introducing this fragment into the NgrolB sequence. The promoter cassette Pnb, with the promoter sequence of the NgrolB gene, was amplified from λNg31 with the following primers; 5′-CATTCTAGAGTGAGTTGGA-3′ and 5′-ATCTCTAGATTCTCACCCGGCTCG-3′. By using the same primers, the promoter cassette Prb, with the promoter sequence of the RirolB gene, was generated from pLJ1.
The chimeric genes cauliflower mosaic virus 35S promoter (P35S)-NgrolB and P35S-RirolB included the NgrolB and RirolB coding regions, respectively, under the control of P35S (20). The NB and RB cassettes were used to construct these chimeric genes. These cassettes were also positioned under the chimeric promoter for high-level expression (21). A chimeric gene P35s-glucuronidase (GUS), which included the GUS gene ORF under the control of P35s, was used as the negative control.
The following oligonucleotide primers were synthesized for site-directed mutagenesis of NgrolB: 5′-GGCACTACACAGTTCAGCG-3′ and 5′-CTTGACCAGGAAAGCGAGG-3′. Nucleotide substitutions were made (Mutan-K kit, Takara Shuzo, Kyoto, Japan) by using the λNg31 sequence as the template. The resulting mutagenized NgrolB sequence was used to construct chimeric genes under the control of the above-mentioned promoters.
The P35S-NgrolC gene was constructed as previously described in the construction of P35S-RirolC (22). A fragment consisting of the NgrolC gene containing the restriction site HpaI was generated by PCR from the N. glauca genome with the following two primers: 5′-CCTACTTTGTTAACATGGC-3′ and 5′-GTCGGGCAGTCGACGTA-3′. The PCR-generated fragment of the 5′ region of NgrolC was digested with HpaI/SalI and subcloned into pHTS6.1 (19), which previously harbored a partial sequence of NgrolC.
Northern analysis was carried out with some tissues of plants by using antisense RNA probes as described (13).
Inoculations of Leaf Segments to Induce Hairy Roots.
Agrobacterium tumefaciens EHA101 (23) that harbored constructs derived from pMM454-Km was tested for its ability to induce the formation of roots on tobacco-leaf segments. Inoculation was performed as described previously (6).
Molecular Evolutionary Analysis.
Molecular evolutionary analysis of the NgrolB and RirolB genes was performed with the oden computer program (Natural Institute of Genetics, Mishima, Japan). Numbers of nucleotide substitutions per site (Kn) were estimated by the method of Tajima and Nei (24). Numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per site were estimated by the method of Nei and Gojobori (25).
Results
Comparison of Function Between Rirol and Ngrol Genes.
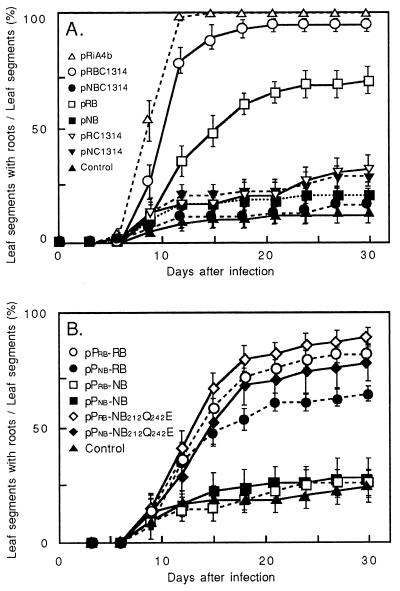
The function of Ngrol genes was analyzed by transforming leaf segments of N. tabacum with A. tumefaciens that harbored either pRBC1314 or pNBC1314 (Fig. 1A). Almost all of the leaf segments inoculated with pRBC1314 developed vigorous roots that had the features of hairy roots (Fig. 2A). No extensive development of roots, however, appeared on the segments infected with A. tumefaciens that harbored pNBC1314. Phenotypes of regenerated plants that had been transformed with these genes were then studied. Whole plants resulting from introduction of the region, including the four Rirol genes, displayed some abnormal phenotypic features, such as internodes of reduced length and wrinkled leaves (data not shown). Roots grew rapidly with a highly branched pattern and reduced geotropism. These features are characteristic of hairy-root syndrome (8). In contrast, transgenic plants that harbored Ngrol genes did not exhibit any of these traits of hairy-root syndrome.
Figure 2.
Root Induction by Ngrol or Rirol genes. The frequency of root production was determined by calculating the number of segments that formed roots as a percentage of the total number of segments in each flask. Binary vector pMM454-Km was used as the control. (A) Leaf segments were infected with A. tumefaciens harboring constructs of Ngrol and Rirol genes. Sixty leaf segments in ten flasks were used for each construct. Bars indicate SE (n = 10). (B) Leaf segments were inoculated with the constructs of rolB homologues. Fifty-four leaf segments in nine flasks were examined for each construct. Bars indicate SE (n = 9).
Functions of NgrolB and RirolB Genes in Root Induction.
Deletion analysis of the RirolB-RiORF14 region revealed that the RirolB locus is indispensable for adventitious root induction (6). To compare the functions of the RirolB and NgrolB genes in root induction, constructs pRB and pNB were introduced into tobacco-leaf segments (Fig. 1A). When pRB was introduced into N. tabacum, significant induction of hairy roots was observed on leaf segments (Fig. 2A; refs. 5, 6). pNB, however, was unable to induce more root formation than the control (pMM454-Km). pNC1314 and pRC1314 were unable to induce roots as vigorously as did pNB.
Exchange of Promoter Regions Between NgrolB and RirolB Genes.
Nagata et al. (14) performed fluorometric and histochemical analyses of transgenic plants that harbored a GUS-reporter gene under the control of the promoters of the NgrolB and RirolB genes. They showed that the pattern of expression of the NgrolB gene was the same as that of the RirolB gene in terms of tissue specificity, but that the promoter of NgrolB was 2- to 3-fold less active than that of RirolB. They suggested that the low level of activity of Ngrol promoters might be one of the reasons that N. glauca plants do not display symptoms of hairy-root syndrome. The effect of promoter activity of these two genes on root induction was examined by exchanging the NgrolB and RirolB promoters. The construct pPnb-RB fuses the promoter region of NgrolB with the RirolB-coding region in pMM454-KmRi13 (Fig. 1B). The positive control, pPrb-RB, contains the RirolB gene under the control of its own promoter. Both pPnb-RB and pPrb-RB were able to elicit root formation (Fig. 2B). Root induction was not notably enhanced after inoculation with pPrb-NB or pPnb-NB. Therefore, the exchange of promoters did not significantly affect the functions of the coding regions of RirolB and NgrolB genes. Slight depression of the frequency of root induction was, however, detected after transformation with pPnb-RB when compared with pPrb-RB. A difference in the activity of cis-acting sequences between RirolB and NgrolB as shown by Nagata et al. (14) may cause this depression of root induction.
Transcription of Ngrol Genes in N. glauca.
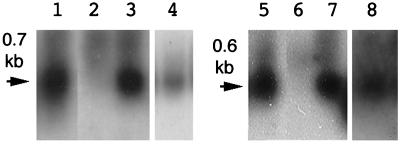
An explanation for the normal phenotype of N. glauca presented by Sinkar et al. (26) is that the Ngrol genes in N. glauca might not be transcribed in plant tissues, allowing plants to display the normal phenotype. Northern analysis was performed on some tissues of N. glauca. Transcripts of the NgrolB and NgrolC genes were detected in the stem tissue of N. glauca as well as the genetic tumor of hybrids of N. glauca and N. langsdorffii (Fig. 3). Callus tissues derived from N. glauca also contained the transcripts of these genes. In contrast, no signal was observed in the extract from leaf tissues. Recently, some reports showed that trolC, torf13–1, and torf13–2 homologues of TL-DNA genes in the genome of N. tabacum are also transcribed in regulated patterns in plant tissues (16, 27).
Figure 3.
RNA gel blot analysis of NgrolB (lanes 1–4) and NgrolC (lanes 5–8) in plant tissues. Lanes 1 and 5, genetic tumor of the hybrids of N. glauca and N. langsdorffii. Lanes 2 and 6, leaf tissue of N. glauca. Lanes 3 and 7, stem tissue of N. glauca. Lanes 4 and 8, callus tissue of N. glauca cultured in MS medium supplemented with 1-mg liter-1 kinetin. Poly(A)+RNA (3 μg for lanes 1–3 and 5–7, and 10 μg for lanes 4 and 8) was subjected to electrophoresis. The probes for analysis of expression of the NgrolB and NgrolC were a 1.1-kbp PstI-HindIII fragment and a 0.8-kbp HincII-EcoRI fragment of pLJ1, respectively (13).
Molecular Evolutionary Analysis.
A comparison of the sequence of NgrolB with that of RirolB revealed a difference between these genes in the lengths of their coding regions (Fig. 1C; ref. 11). rolB homologues of strains of A. rhizogenes other than the agropine-type strain also have coding regions longer than that of the NgrolB gene (28–30). Although the ORF of NgrolB starts at the corresponding base of RirolB, differences in bases at sites 633 and 723 bp from the initiation codon generate early-termination codons. The first termination codon reduces the ORF of NgrolB by 144 bp from that of RirolB. We then compared the features of nucleotide-sequence differences between NgrolB and RirolB genes. Numbers of nucleotide substitutions per site (Kn) were calculated for four different regions: the 5′ noncoding region, the coding region of 633 bp, the next region of 144 bp, and the 3′ noncoding region (Table 1). This 144-bp region is a 3′ untranslated region of NgrolB gene that corresponds to a 3′ coding sequence in the RirolB gene. This region has a small Kn value that may indicate the presence of functional constraints against nucleotide changes. We can see that the Kn value in the 144-bp region is smaller than those in the 5′ and 3′ noncoding regions, but is much the same as that in the ORF region (Table 1). Frequencies of synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions and Ka/Ks ratios were also calculated. A Ka/Ks ratio <1 may result from the elimination of most nonsynonymous changes through natural selection. When the coding region and the 144-bp region were examined, Ks exceeded Ka for both the regions. These results revealed that the 144-bp region following the ORF of NgrolB might have been conserved as a coding sequence for amino acids. Two single-base substitutions can change these two termination codons to amino acids. The resulting 48-aa extension is similar to the corresponding sequence of the RirolB protein. These results indicate that two point mutations of the NgrolB gene occurred during the evolution of N. glauca.
Table 1.
A comparison of Kn, Ks and Ka values in NgrolB and RirolB genes
| 5′UTR* | ORF region† | 144-bp portion‡ | 3′ UTR§ | |
|---|---|---|---|---|
| Kn | 0.339 ± 0.021 | 0.197 ± 0.020 | 0.187 ± 0.042¶ | 0.362 ± 0.044 |
| Ks | 0.341 ± 0.058 | 0.267 ± 0.100 | ||
| Ka | 0.153 ± 0.019 | 0.128 ± 0.038 | ||
| Ka/Ks | 0.449 | 0.479 |
5′ untranslated region between rolB and rolC homologues (1,207 bp).
ORF region of NgrolB and RirolB gene (633 bp).
3′ untranslated region of NgrolB gene that corresponds to 3′ coding sequence of RirolB gene (144 bp).
3′ untranslated region from the site of RirolB termination codon to the site of the end of the left arm of the cellular T-DNA (318 bp; ref. 11)
Kn is significantly higher in the 5′ and 3′ UTRs than in the 144-bp portion, but is not significantly different between the ORF and 144 regions (P < 0.01; t-test).
Reversion of the Capacity for Root Induction by Site-Directed Mutagenesis.
Two single base substitutions were made at sites 633 and 723 nt from the initiation codon of NgrolB by site-directed mutagenesis. These substitutions changed the two termination codons (TAG and TAA) to a Gln (CAG) and a Glu (GAA), respectively. The putative reading frame of the resulting ORF, designated NgrolB212Q242E (259 aa), closely matches that of RirolB. Tobacco-leaf segments were transformed with A. tumefaciens that carried the NgrolB212Q242E construct under the control of the NgrolB promoter in pMM454-KmRi13 (pPnb-NB212Q242E; Fig. 1B). More intense and earlier root formation was repeatedly observed after transformation with pPnb-NgrolB212Q242E than with pPnb-NB (Fig. 2B). The ORFs of RirolB and NgrolB212Q242E were equally effective in evoking root induction. The chimeric gene NgrolB212Q242E under the control of the promoter of RirolB (Prb-NB212Q242E) was also able to elicit induction of roots. Thus, the restored region of 48 aa seems to be crucial for root induction of the NgrolB gene.
Transformation with P35S-NgrolB212Q242E.
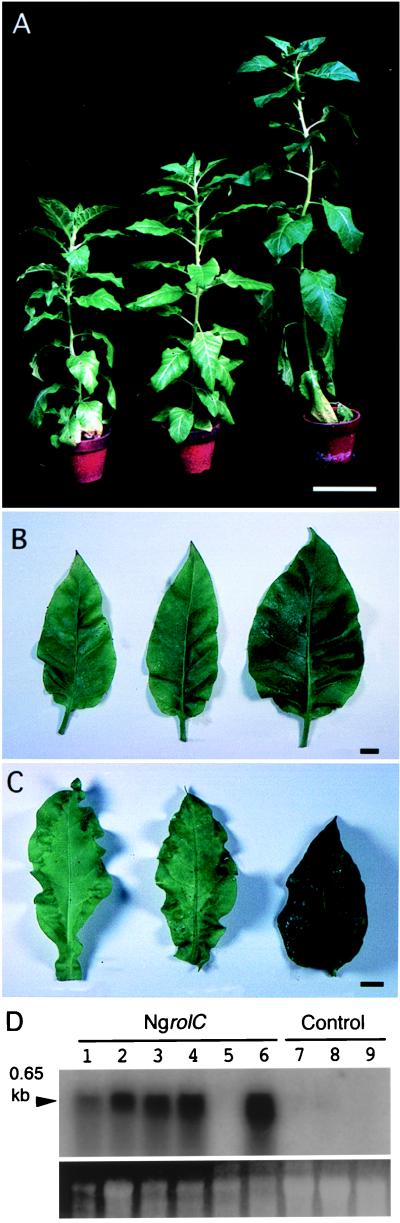
We further studied the function of the rolB homologues under the control of P35S. We used P35S because the growth-regulating function of these genes should become more clearly observable with high-level expression. More than six independent transformants were regenerated for each gene. Leaf disks inoculated with P35S-NgrolB212Q242E exhibited reduced morphogenic capacity. Transformed plants grew slowly, the apex was distorted, the internodes were short, and the leaves exhibited wrinkled and epinastic growth (Fig. 4 A and B). Inflorescence had small numbers of irregularly shaped flowers with large sepals, wavy petals, and short pistils (Fig. 4C). Spontaneous self-pollination occurred normally, but production of seeds was reduced. These traits of P35S-NgrolB212Q242E plants were also observed with another promoter for high-level expression (21). Transcripts of NgrolB212Q242E were detectable in leaves of primary transformants (T0) and their progeny plants (T1) that exhibited anomalies (Fig. 4D). The level of transcription was weak in progeny of another line, evidencing mild phenotype with slightly distorted stems and wrinkled leaves (Fig. 4D, lane 6). By contrast, regenerated plants transformed with P35S-NgrolB had a normal phenotype, as did their progeny (Fig. 4E). Established P35S-NgrolB transgenic lines that exhibited stable normal morphology expressed the NgrolB gene at varying levels. The size of the transcripts of P35S-NgrolB was larger than that under the control of its own promoter. Enlargement of the transcript size was also reported when the RirolB gene was controlled by P35S (31). Overexpression of the RirolB gene was also examined in tobacco plants of six independent lines. As shown by Schmulling et al. (31), P35S-RirolB transformed plants had flat, round, and highly necrotic leaves (Fig. 4F). In contrast, P35S-NgrolB212Q242E transgenic leaves were wrinkled, with epinastic growth, and were not necrotic. Roder et al. (32) reported that transformation with the RirolB gene under the control of a tetracycline-dependent promoter resulted in extremely stunted plants with necrotic and wrinkled leaves that did not develop a floral meristem. Although delay of growth and wrinkling of leaves were observed into the P35S-NgrolB212Q242E plants, these plants developed inflorescence. We must not forget that, in addition to the generation of termination codons, additional mutations may have taken place in the NgrolB sequence during the evolution of N. glauca. Much still remains to be done to understand the morphogenic activity of the rolB homologues, NgrolB212Q242E and the ancient NgrolB gene.
Figure 4.
Phenotypes of transgenic plants harboring rolB homologues. (A) A transgenic plant carrying the P35S-NgrolB212Q242E chimeric gene (Left) and a control plant (Right). Bar = 10 cm. (B) Leaves of a NgrolB212Q242E-overexpressing plant. (E) Leaves of a NgrolB-overexpressing plant. (F) Leaves of a RirolB-overexpressing plant. (C) Comparison of floral leaves from transgenic plants. (B, C, E, and F) Bars = 1 cm. (D) Transcripts of the NgrolB212Q242E (lanes 1–6) and NgrolB (lanes 7–9) genes in transgenic plants. Lanes 1–3, T0 transgenic plants; lanes 4–9, T1 plants; lane 10, a control plant. The probe was a 0.8-kbp XbaI-SacI fragment of λNg31. Fifteen micrograms of total RNA per lane was resolved on a formamide gel and blotted onto a nylon membrane.
Transformation with P35S-NgrolC.
Analysis of the NgrolB gene led us further into a consideration of the function of the other Ngrol genes. Comparison of the DNA sequences of NgrolC with RirolC revealed that the reading frames of NgrolC initiate and terminate at the corresponding bases of RirolC. Tobacco-leaf disks were transformed with the P35S-NgrolC chimeric gene. More than 10 T0 regenerants showed a dramatically dwarfed phenotype, probably because of reduced internodal length (Fig. 5A). The P35S-NgrolC transgenic leaves were lanceolate and pale green (Fig. 5B). Their floral organs were slender and small. Transgene expression was detected in transformants that showed the characteristic morphological alterations (Fig. 5D). No transcripts were detectable in leaves from a comparable T0 plant with a normal phenotype (Fig. 5D, lane 5). These characteristics are identical to the phenotype of the P35S-RirolC transgenic plants reported previously (22, 31). We have bred three independent P35S-NgrolC and P35S-RirolC T0 plants that exhibited the same traits of abnormal leaf morphology (Fig. 5C). Their leaves had pale-green inner blades and wrinkled dark-green margins. These phenotypes may be caused by periclinal chimeric regeneration, as reported previously (22, 33).
Figure 5.
Comparison of plants transformed with rolC homologues. (A) Whole plants harboring P35S-RirolC chimeric gene (Left) or P35S-NgrolC gene (Center) and a control plant (Right). Bar = 10 cm. (B) Leaves from transgenic plants harboring P35S-RirolC (Left) or P35S-NgrolC (Center) and from a control plant (Right). (C) Abnormal chimeric morphology of leaves from transgenic plants harboring P35S-RirolC gene (Left) or the P35S-NgrolC gene (Center). A normal leaf of wild-type tobacco is shown as a control (Right). (B and C) Bars = 1 cm. (D) Transcripts of the NgrolC in leaf tissues of transgenic plants. Lanes 1–5, T0 plants infected with A. tumefaciens harboring the P35S-NgrolC gene; lane 6, a T1 plant; lanes 7–9, wild-type and control tobacco plants. The probe was a 1.1-kbp EcoRV-EcoRI fragment of λNg31.
Discussion
Analysis of the DNA sequence supported a model where members of the genus Nicotiana carry a portion of T-DNA from an ancient infection by the A. rhizogenes-like ancestor (10, 11). The acquisition of T-DNA may have altered the phenotype or selective advantage of the infected plants (11, 15–17). This alteration may explain the establishment and persistence of the homologues of rol genes in present-day plants. Alternatively, the T-DNA may have been trapped by chance survival of a regenerated plantlet (15). We are examining the possible role of the Ngrol genes in N. glauca.
We suggest that N. glauca plants do not exhibit the phenotype of hairy-root syndrome because the NgrolB sequence might have been truncated. An A. rhizogenes-like ancestor might have had the NgrolB sequence with more amino acids than that of today in its plasmid. The ancient transformed plant harboring the complete NgrolB gene might have formed hairy roots. Tepfer (8) referred to the observation that hairy roots of tobacco plants regenerated shoots when grown in pots, but roots of normal plants did not regenerate shoots. Progeny of regenerated plants from the ancient hairy roots might have had the Ngrol genes in their genome. Subsequently, nonsense point mutations in the NgrolB gene might have occurred, and the phenotype of hairy-root syndrome might have been lost. The present-day NgrolB gene did not direct hairy-root formation. It is unlikely that a truncated NgrolB gene was transformed to an ancestral plant.
The NgrolC gene elicited biological response in transgenic tobacco plants when controlled by the P35S. This gene was transcribed in some tissues of N. glauca. Nagata et al. (14) have shown that the expression pattern of the promoter of the NgrolC gene exhibited much the same tissue specificity as that of the RirolC promoter. We propose that the functional sequence of the NgrolC gene may have been conserved after the ancient introduction from the bacterium. Transformation of RirolC has altered the resistance reactions of potato to pathogens and the flowering time of carrot (17, 34). Expression of NgrolC may be connected with these phenotypes of N. glauca. We also reported that the functional sequences of NgORF13 and NgORF14 genes have been conserved after the ancient infection event as well as NgrolC has been conserved (18). However, we cannot say for certain that the level of expression of these genes is sufficient for the appearance of their capacities (26). Further analysis of the expression pattern and level of Ngrol genes in tissues of N. glauca should help to clarify the physiological role of these genes.
The natural genetic transformation of rol genes might have generated novel variants that are progenitors of modern species (11, 15–17). The phenotype induced by rol genes could have adaptive significance for the plants (15–17). Phenotypic alterations caused by the expression of Ngrol genes might have contributed to the creation of biodiversity in the genus Nicotiana. Previous studies showed that several plants, including carrot and morning glory, also carry sequences similar to T-DNA (35, 36). These results raise the possibility that horizontal gene transfers, from bacteria to plants, have occurred during the evolution of some plant species. Further analysis of rol-homologues in plants might shed some light on the evolution of these plant species.
Acknowledgments
We thank Dr. M. P. Gordon (University of Washington), Dr. D. Tepfer (Institut National de la Recherche Agronomique, France), Dr. Y. Ohashi (Natural Institute of Agrobiological Resources, Japan), Dr. H. Tsukaya (University of Tokyo, Japan), Dr. M. Sekine (Nara Institute of Science and Technology, Japan), and Dr. H. Kamada (University of Tsukuba, Japan) for providing a clone of cT-DNA, and the plasmids, pLJ1, pE2113-GUS, pHTS6.1, pMM454-Km, and A. tumefaciens R1000 (pRiA4b), respectively. This study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas and a Grant-in-Aid for Scientific Research to K.S. from the Ministry of Education, Science, Sports, and Culture, Japan, and a grant to K.S. from Japan Tobacco, Inc.
Abbreviations
- P35S
cauliflower mosaic virus 35S promoter
- TL-DNA
left transferred DNA
- T-DNA
transferred DNA
- RB
the ORF cassette of RirolB
- NB
the ORF cassette of NgrolB
- pRB
a plasmid harboring the RirolB locus in its T-DNA region
- pNB
a plasmid harboring the NgrolB locus in its T-DNA region
- T0
primary transformants
- T1
progeny plants of T0
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Heinemann J A. Trends Genet. 1991;7:181–185. doi: 10.1016/0168-9525(91)90433-q. [DOI] [PubMed] [Google Scholar]
- 2.Jouanin L. Plasmid. 1984;12:91–102. doi: 10.1016/0147-619x(84)90055-6. [DOI] [PubMed] [Google Scholar]
- 3.White F F, Taylor B H, Huffman G A, Gordon M P, Nester E W. J Bacteriol. 1985;164:33–44. doi: 10.1128/jb.164.1.33-44.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slightom J L, Durand-Tardif M, Jouanin L, Tepfer D. J Biol Chem. 1986;261:108–121. [PubMed] [Google Scholar]
- 5.Spena A, Schmulling T, Koncz C, Schell J. EMBO J. 1987;6:3891–3899. doi: 10.1002/j.1460-2075.1987.tb02729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki S, Syono K. Plant Cell Physiol. 1999;40:252–256. [Google Scholar]
- 7.Capone I, Spano L, Cardarelli M, Bellincampi D, Petit A, Costantino P. Plant Mol Biol. 1989;13:43–52. doi: 10.1007/BF00027334. [DOI] [PubMed] [Google Scholar]
- 8.Tepfer D. Cell. 1984;37:959–967. doi: 10.1016/0092-8674(84)90430-6. [DOI] [PubMed] [Google Scholar]
- 9.White F F, Ghidossi G, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1982;79:3193–3197. doi: 10.1073/pnas.79.10.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White F F, Garfinkel D J, Huffman G A, Gordon M P, Nester E W. Nature (London) 1983;301:348–350. [Google Scholar]
- 11.Furner I J, Huffman G A, Amasino R M, Garfinkel D J, Gordon M P, Nester E W. Nature (London) 1986;319:422–427. [Google Scholar]
- 12.Aoki S, Kawaoka A, Sekine M, Ichikawa T, Fujita T, Shinmyo A, Syono K. Mol Gen Genet. 1994;243:706–710. doi: 10.1007/BF00279581. [DOI] [PubMed] [Google Scholar]
- 13.Ichikawa T, Ozeki Y, Syono K. Mol Gen Genet. 1990;220:177–180. doi: 10.1007/BF00260478. [DOI] [PubMed] [Google Scholar]
- 14.Nagata N, Kosono S, Sekine M, Shinmyo A, Syono K. Plant Cell Physiol. 1995;36:1003–1012. doi: 10.1093/oxfordjournals.pcp.a078842. [DOI] [PubMed] [Google Scholar]
- 15.White F F. In: Antibiotic Resistance Genes: Ecology, Transfer and Expression. Levy S B, Novick R P, editors. Plainview, New York: Cold Spring Harbor Lab. Press; 1986. pp. 45–54. [Google Scholar]
- 16.Meyer A D, Ichikawa T, Meins F., Jr Mol Gen Genet. 1995;249:265–273. doi: 10.1007/BF00290526. [DOI] [PubMed] [Google Scholar]
- 17.Limami M A, Sun L, Douat C, Helgeson J, Tepfer D. Plant Physiol. 1998;118:543–550. doi: 10.1104/pp.118.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki S, Syono K. Plant Cell Physiol. 1999;40:222–230. [Google Scholar]
- 19.Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y. Plant Physiol. 1991;97:1414–1421. doi: 10.1104/pp.97.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrzak M, Shillito R D, Hohn T, Potrykus I. Nucleic Acids Res. 1986;14:5857–5868. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsuhara I, Ugaki M, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- 22.Oono Y, Suzuki T, Toki S, Uchimiya H. Plant Physiol. 1993;34:745–752. [Google Scholar]
- 23.Hood E E, Helmer G L, Fraley R T, Chilton M D. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajima F, Nei M. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 25.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 26.Sinkar V P, White F F, Furner I J, Abrahamsen M, Pythoud F, Gordon M P. Plant Physiol. 1988;86:584–590. doi: 10.1104/pp.86.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frundt C, Meyer A D, Ichikawa T, Meins F., Jr Mol Gen Genet. 1998;259:559–568. doi: 10.1007/s004380050849. [DOI] [PubMed] [Google Scholar]
- 28.Hansen G, Larribe M, Vaubert D, Tempe J, Biermann B J, Montoya A L, Chilton M D, Brevet J. Proc Natl Acad Sci USA. 1991;88:7763–7767. doi: 10.1073/pnas.88.17.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyokawa S, Kobayashi K, Kikuhci Y, Kamada H, Harada H. Plant Physiol. 1994;104:801–802. doi: 10.1104/pp.104.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serino G, Clerot D, Brevet J, Costantino P, Cardarelli M. Plant Mol Biol. 1994;26:415–422. doi: 10.1007/BF00039550. [DOI] [PubMed] [Google Scholar]
- 31.Schmulling T, Schell J, Spena A. EMBO J. 1988;7:2621–2629. doi: 10.1002/j.1460-2075.1988.tb03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roder F T, Schmulling T, Gatz C. Mol Gen Genet. 1994;243:32–38. doi: 10.1007/BF00283873. [DOI] [PubMed] [Google Scholar]
- 33.Schmulling T, Schell J. Plant Mol Biol. 1993;21:705–708. doi: 10.1007/BF00014554. [DOI] [PubMed] [Google Scholar]
- 34.Fladung M, Gieffers W. Physiol Mol Plant Pathol. 1993;42:123–132. [Google Scholar]
- 35.Spano L, Pomponi M, Costantino P, Van Slogteren G M S, Tempe J. Plant Mol Biol. 1982;1:291–300. doi: 10.1007/BF00027560. [DOI] [PubMed] [Google Scholar]
- 36.Tepfer D. 2e Colloque sur les Recherches Fruitières. Bordeaux, France: Centre Technique Interprofessionnel des Fruits et Légumes; 1982. pp. 47–59. [Google Scholar]