Abstract
Although integration of viral DNA into host chromosomes occurs regularly in bacteria and animals, there are few reported cases in plants, and these involve insertion at only one or a few sites. Here, we report that pararetrovirus-like sequences have integrated repeatedly into tobacco chromosomes, attaining a copy number of ≈103. Insertion apparently occurred by illegitimate recombination. From the sequences of 22 independent insertions recovered from a healthy plant, an 8-kilobase genome encoding a previously uncharacterized pararetrovirus that does not contain an integrase function could be assembled. Preferred boundaries of the viral inserts may correspond to recombinogenic gaps in open circular viral DNA. An unusual feature of the integrated viral sequences is a variable tandem repeat cluster, which might reflect defective genomes that preferentially recombine into plant DNA. The recurrent invasion of pararetroviral DNA into tobacco chromosomes demonstrates that viral sequences can contribute significantly to plant genome evolution.
Most plant viruses have single-stranded RNA genomes. Only two classes of plant viruses, caulimoviruses and badnaviruses, contain genomes of double-stranded DNA (1). Because these double-stranded DNA viruses use a virally encoded reverse transcriptase to replicate their genomes, they, together with vertebrate hepadnaviruses, are classified as pararetroviruses to distinguish them from true retroviruses, which have RNA genomes. Retroviruses have not yet been conclusively identified in plants, although recent findings of retrotransposons that encode envelope-like proteins suggest that they might exist (2–4). Pararetrovirus replication in plants proceeds by nuclear transcription of a slightly greater than genome-length RNA with terminal repeats that is generated by the host RNA polymerase II. This is followed by reverse transcription in the cytoplasm of the terminally redundant RNA, which also serves as an mRNA for viral proteins (5). Although retrovirus DNA integrates into host chromosomes by means of a virally encoded integrase (6), pararetroviruses generally lack the gene for this enzyme, and integration is not required for virus replication. However, pararetroviral DNA can in principle integrate into host DNA, as exemplified by mammalian hepatitis B (hepadna)virus, which has been found integrated into host chromosomes in hepatic tissue, where it is associated with liver carcinomas (7). Until recently, there were no data suggesting comparable integration of pararetroviral sequences into plant DNA.
In contrast to bacterial and animal viruses, plant viral sequences are generally thought to integrate rarely, if at all, into host genomes. One well characterized example concerns a single insertion of sequences related to a geminivirus, which has a single-stranded circular DNA genome, into tobacco nuclear DNA (8–10). Although the geminivirus case has been considered exceptional, several recent reports prompt a reconsideration of the possibility that plant pararetrovirus DNA might integrate more commonly into host chromosomes. First, petunia vein-clearing virus, which is the sole known member of one group of plant pararetroviruses, atypically contains core sequences for an integrase function (11). Second, DNA of banana streak (badna)virus (BSV), which does not encode an obvious integrase, has been found integrated at several sites in the genome of banana plants (12, 13). The possibility that pararetrovirus DNA might insert regularly into host genomes, either by means of a viral integrase or by illegitimate recombination, has considerable implications for plant genome evolution. Similar to vertebrate endogenous retroviruses (6), integrated pararetroviral DNA could act as an insertional mutagen; could contribute strong constitutive promoters to neighboring plant genes; or could accumulate to generate a new repetitive sequence family.
In this paper, we report data demonstrating that sequences from a heretofore undescribed pararetrovirus have integrated repeatedly into tobacco nuclear DNA, forming a dispersed repetitive sequence family. Unusual characteristics of the integrated viral sequences suggest that they are derived from defective virus genomes that have an increased probability of recombining into host sequences. Based on these atypical features and the boundaries of viral sequence inserts, a possible mechanism for virus DNA integration is discussed.
Materials and Methods
λ cloning and Nucleotide Sequence Analysis.
Two genomic DNA λ libraries were prepared from Nicotiana tabacum cv. petite havana SR1 DNA digested partially with Sau3AI or completely with EcoRI, respectively, using the λ FIX II (V-clones) or λ ZAP II (E-clones) from Stratagene as described (14). Using a “partial fill-in” strategy, the λ FIX II system is specifically designed to prevent the formation of cloning artifacts. For screening, a cloned 1.0-kilobase (kb) PCR fragment derived from the reverse transcriptase (RT) region of clone V1 was used. The resulting λ clones were subcloned, and nucleotide sequence analysis was performed with a Li-Cor DNA Sequencer Long Read IR 4200 system (Li-Cor, Omaha, NE) using a ThermoSequenase cycle sequencing kit (Amersham Pharmacia) and infrared-labeled oligonucleotides (MWG Biotech, Ebersberg, Germany). Database searches were performed by using the blast algorithm (15).
DNA Blot Analysis.
Plant genomic DNA used for blot analysis was isolated from leaves of symptomless adult tobacco plants using a Plant DNA Isolation Kit (Roche, Vienna, Austria). DNA gel electrophoresis and transfer to nitrocellulose were done following standard procedures. For DNA slot blots, the MilliBlot-D system (Millipore) was used according to the manufacturer’s instructions. A hybridization probe labeled with 32P was synthesized from an isolated 0.8-kb EcoRI-XbaI fragment of clone V6 (Fig. 1) by using a Multiprime DNA labeling system (Amersham Pharmacia). Hybridization and washing were performed according to Thomashow et al. (16) under moderately stringent conditions [3× standard saline citrate (SSC) at 64°C; 1× SSC = 0.15 M NaCl/0.015 M sodium citrate].
Figure 1.
Structure of integrated TPVL sequences. Twenty-two independent clones from two tobacco genomic λ libraries, made with Sau3AI (V) or EcoRI (E), were fully or partially sequenced. White bars represent TPLV sequences; black bars indicate plant DNA. The extent of the putative TPV genome, beginning with nucleotide 1 (tRNA binding site) at the left and ending variably around nucleotide 8,000 at the right, is bound by two vertical lines. The order of TPV ORFs and relevant restriction enzyme sites are shown at the top. Repeated regions are shaded, including the block of tandem repeats in the putative TPV leader region at the end of ORF4; the NTS9 tandem repeat in flanking plant DNA in clone V1; and short duplications of TPLV sequences at the right of V3, V6, and E21. In-frame deletions are indicated by paired vertical lines within the white bars and are spaced according to the size (3, 6, 12, or 15 bp). Frameshifts are denoted by arrowheads; stop codons by diamonds. Narrow lines connecting white bars represent gaps in the sequence; spaces between unconnected white bars represent unsequenced regions. Gray bars above junctions in V1, V5, V7, V8, and V13 indicate sequenced PCR fragments synthesized from tobacco DNA. Asterisks at the left and right of V14 indicate a short triplication of TPLV sequence. The × in E21 and V3 indicates a short region in inverse orientation in the two clones relative to the shaded sequence, which is duplicated in E21. The inverted duplication of TPVL sequences in V4 is represented by white arrowheads. T7 and T3 signify the end of clones; the extent of other clones was not determined. Because this represents a linear projection of the circular map, ends of clones—depending on their position—can appear to be located internally. Vertical arrows at the top point out the position of junctions between TPLV sequences and tobacco DNA. Abbreviations: CP, coat protein; MP, movement protein; POL, polyprotein; TAV, transactivation protein. Three short ORFs of unknown function are between 7 and 8 kb.
Northern Blot Analysis.
Preparation of poly(A)+ RNA from tobacco plants and callus tissue and Northern blot analysis were done as described (17). The 32P-labeled DNA probe was synthesized from an isolated DNA fragment extending 5.5 kb from the T7 end of clone V6 (Fig. 1).
Virus Detection.
For the attempted preparation of virion DNA from tobacco, the procedure described by Covey et al. (18) was performed following exactly the authors’ recommendations. Two-dimensional gel electrophoresis (18) was used to search for circular forms of virus DNA in tobacco DNA prepared by using the CTAB (N-cetyl-N,N,N-trimethyl-ammonium bromide) procedure (19).
Results
Deduced DNA Sequence of a New Tobacco Pararetrovirus.
In a study of plant DNA sequences that flank transgene inserts in tobacco (N. tabacum cv. petit havana SR1), we cloned a 1.8-kb DNA fragment that showed sequence homology to the reverse transcriptase gene of several pararetroviruses. In this clone, the homology to viral DNA terminated abruptly at a junction with nonviral sequences that were presumably derived from tobacco, suggesting covalent linkage of plant and viral DNA. Because pararetroviral sequences are not generally believed to integrate into plant nuclear DNA, we searched for additional pararetrovirus–plant DNA junctions using part of the 1.8-kb fragment to probe two different λ genomic libraries prepared from healthy tobacco plants. From hundreds of positive clones, 22 were chosen for further analysis. All 22 clones contained fragments of tobacco pararetroviral-like DNA (Fig. 1). From these fragments, an 8-kb virus genome could be assembled based on the order of open-reading frames in cassava vein mosaic virus (CsVMV) (20), the pararetrovirus exhibiting the highest sequence similarity to the putative tobacco pararetrovirus (TPV). Fourteen junctions with plant DNA were isolated. Sequence analysis of PCR fragments synthesized across a number of these junctions confirmed that they were indeed present in tobacco nuclear DNA (Fig. 1).
Attempts to isolate free TPV from tobacco plants using standard procedures for caulimovirus isolation and DNA extraction (18) failed to recover observable amounts of virions or the 8-kb viral genomic DNA. Because the viral DNA-containing clones were isolated from symptomless tobacco plants that did not contain detectable quantities of the corresponding virus, they are referred to as tobacco pararetrovirus-like (TPVL) sequences.
ORFs and Sequence Comparisons.
A consensus DNA sequence of the putative TPV genome was deduced from overlapping portions of the integrated TPVL fragments. The first nucleotide of the tRNA binding site, which serves as a primer for reverse transcriptase (RT), was designated as nucleotide 1, in accordance with other pararetroviral genomes (20). The nucleotide sequence similarity of the 22 integrated TPVL sequences ranged from 91 to 98%, suggesting that they resulted from relatively recent insertion events. These viral sequences were translationally defective, as indicated by the presence of numerous frameshifts and stop codons, as well as several rearrangements of viral sequences (see below). Some of the observed sequence variation among the integrated TPVL sequences resulted from in-frame deletions that comprised 3, 6, 12, or 15 bp, suggesting that multiple strains of the putative TPV were involved.
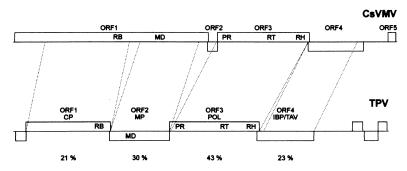
The level of amino acid identity between TPV and CsVMV ranges from 43% for the RT gene encoded by ORF3 to 21% for ORF1, which encodes a putative coat protein (Fig. 2). The arrangement of ORFs in the tobacco virus is similar to CsVMV, particularly with respect to the order of coat protein/movement protein/polyprotein genes, which is different in all other plant pararetroviruses (20). Relative to CsVMV, the movement protein of TPV is in a frameshift, and the short ORF2 of CsVMV is missing entirely in TPV (Fig. 2). Both CsVMV and TPV have low G + C contents (25 and 28%, respectively) compared with the genomes of caulimoviruses (34–40%) and badnaviruses (34–44%) (20).
Figure 2.
Comparison of the genomic organization of cassava vein mosaic virus (CsVMV) (20) and the putative tobacco pararetrovirus (TPV). The putative TPV genome, which was assembled from cloned TPLV sequences, did not contain a recognizable ORF5. Three short TPV ORFs of unknown significance in the putative leader after ORF4 are unlabeled. Such short ORFs are present in the leader regions of other pararetroviruses (21). The percent identity on the amino acid level between TPV and CsVMV is shown below the TPV ORFs. Abbreviations: CP, coat protein; RB, RNA binding site; MD, movement domain; MP; movement protein; POL, polyprotein; PR, proteinase; RH, RNase H; IBP, inclusion body protein; TAV, transactivation protein.
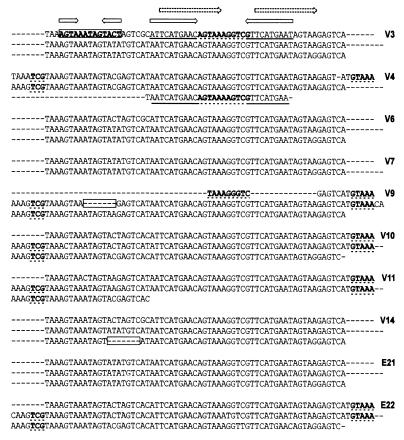
An unusual feature of the TPV is the variable putative leader region immediately following the last complete ORF (ORF4) (Fig. 1). Atypically, this region contains a short cluster of tandem repeats that vary in monomer length and copy number among the different TPVL sequence clones (Fig. 3). The tandemly repeated region could comprise two (V7) or three (V3, V6, E21) complete copies of a 63-bp monomer, or one (V4) to approximately two (V9, V10, V11, E22) copies of a somewhat longer version (76 bp) that was extended at both ends relative to the 63-bp unit (Fig. 3). Partial (V4, V9) and internally deleted (V9, V14) copies were also present in some clones (Fig. 3). The region beyond this tandem repeat contained several short ORFs, which do not encode any known peptides, and continued to be variable among the integrated viral sequences, with a number of clones containing deletions immediately after the tandem repeats (V3, V7, V9, V10, V14, E22) (Fig. 1).
Figure 3.
Variations in the tandem repeat in the putative TPV leader region beyond ORF4. The TPLV sequence clones indicated to the right contained this tandem repeat block (Fig. 1). The 63-bp monomer comprises internal inverted and direct repeats (arrows, top). Length heterogeneity of the 63-bp monomer involves specific sequences (bold) that could form RNA stem-loop structures. Extensions of the 63-bp monomeric unit creating a 76-bp unit involve sequences present in the large loop of a possible RNA hairpin (bold, dotted underline). Partial copies in V4 and V9 consist of sequences in the stem and loop of a possible large hairpin (bold; dotted and heavy underline). Internal deletions (boxed regions; V9, V14) involve sequences in a second putative small stem-loop region (bold and boxed). In V11, the third monomer copy is partial because of the end of clone.
TPVL Sequence Junctions with Tobacco DNA.
One complicated and thirteen simple junctions between TPVL sequences and plant DNA were recovered and sequenced. The simple fusions consisted of TPVL sequences leading directly into plant DNA. The complicated junction comprised a 466-bp plant DNA fragment inserted into TPVL sequences that were contiguous apart from a 14-bp duplication at the insertion site (V10). The TPVL sequences at the junctions with tobacco DNA were not randomly distributed across the putative TPV genome (Fig. 1, top arrows). Of the 14 junctions, 7 involved sequences close to the beginning (V11, V14) or end (V3, V6, 2 × V9, E22) of the TPV genome. Six junctions involved sequences present in or close to TPV ORF1 [V1, V5, V7, V8, V10 (double arrow because of complex junction), E20].
There was no significant homology between TPVL sequences and tobacco DNA at the integration sites. The plant DNA at three of the junctions was related to previously identified sequences of various types. Copies of the NTS9 tandem repeat, a highly repetitive sequence that is found specifically in the Nicotiana sylvestris fraction of the tobacco genome (22), were found adjacent to viral sequences in clone V1. Plant DNA homologous to the 3′ noncoding region of an N. sylvestris Lhcb gene (GenBank accession no. AB012638) was integrated as a 466-bp filler comprising the complex junction in clone V10. Plant DNA to the left of TPV DNA in clone V8 showed 76% amino acid similarity to the reverse transcriptase region of a gypsy-like retrotransposon of pineapple (23).
Virus–Virus Junctions and Rearrangements.
Junctions and rearrangements comprising TPVL sequences were present in several clones. These included an inverted repeat (V4); an internal duplication (V7); a short triplication (V14); and a duplication/inversion (E21 vs. V3). Most of these rearrangements of TPVL sequences and virus–virus junctions were located in the same two regions as the junctions with plant DNA, namely at the beginning and end of the putative TPV genome and in ORF1.
TLV Transcription and Fluorescence in Situ Hybridization Analysis.
Faint TPVL sequence transcripts were detected in poly(A)+ RNA isolated from tobacco leaves. No increased levels of RNA were observed in regenerating callus tissue derived from stems and roots (data not shown). We were unable to detect TPVL sequences on tobacco metaphase chromosomes by fluorescence in situ hybridization analysis, possibly because of the relatively small size and dispersed nature of the inserts. Integrated geminivirus (8, 10) and BSV sequences (12) that have been detected by fluorescence in situ hybridization have consisted of large, complex inserts 50–150 kb in size.
Species Distribution of TPVL-Related Sequences.
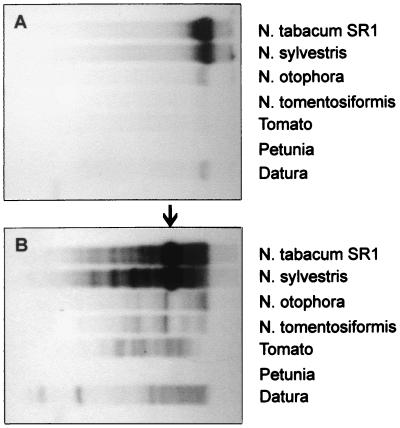
DNA blot analysis of uncut tobacco DNA demonstrated that TPVL-specific probes hybridized to high molecular weight DNA (Fig. 4A), as would be expected for integrated viral sequences. When DNA was cleaved with XbaI, several stronger bands overlaid on a number of weaker bands were observed (Fig. 4B). The predominant strong band at ≈3.5 kb presumably reflects an internal XbaI fragment cut from full-length TPV inserts (Fig. 1). The background smear of weaker bands probably represents less than full-length inserts or sequence polymorphism at the XbaI site. The diploid progenitors of allotetraploid tobacco (N. sylvestris and either Nicotiana tomentosiformis or Nicotiana otophora) also were tested (Fig. 4 A and B). Strong signals were observed with DNA isolated from N. sylvestris. Considerably weaker signals were obtained with N. tomentosiformis and N. otophora. Two other solanaceous plants, Datura and tomato, produced positive signals; a third, petunia, was negative. No hybridization of TPVL probes was seen with DNA isolated from a selection of nonsolanaceous plants, including Arabidopsis and pea (data not shown). The copy number in tobacco was estimated by slot blot analysis to be ≈103 copies/diploid genome (data not shown).
Figure 4.
Southern blot analysis using a TPVL sequence probe on different plant DNA preparations. Approximately 5 μg of DNA were loaded in each lane. (A) Uncut DNA. (B) DNA digested with XbaI. The position of two XbaI sites in the putative TPV genome is shown above the ORFs in Fig. 1. The probe consisted of the XbaI-EcoRI fragment containing part of ORF3 (Fig. 1). The hybridizing fragments in A are >20 kb; the major band in B (arrow) is ≈3.5 kb.
Discussion
Sequences derived from a previously unidentified tobacco pararetrovirus (TPV) have been found by molecular cloning and nucleotide sequence analysis to have integrated repeatedly—apparently by illegitimate recombination—into tobacco nuclear DNA. The integrated tobacco pararetroviral-like (TPVL) sequences are characterized by frame shifts and stop codons as well as rearrangements and complex junctions with plant DNA, indicating that they are not functional copies. Although integrated BSV sequences have recently been detected at a few sites in the banana genome (12, 13), our observations extend these results by showing that viral sequences can accumulate to form a family of moderately repetitive, dispersed repeats in a plant genome. The findings raise a number of questions about how and why these pararetroviral sequences have inserted regularly into tobacco DNA, and the extent to which similar viral sequences have invaded other plant genomes.
The frequency of TPVL sequence integration is extraordinarily high, having resulted in ≈103 copies per tobacco diploid genome. Insertion into the host chromosomes is not a normal part of pararetrovirus replication and is not thought to occur regularly on a random basis. Indeed, other integrated pararetroviral sequences, including BSV in banana (12, 13) and hepatitis B (hepadna)virus in human liver (7), are present at only low copy numbers in the respective host chromosomes. Therefore, to attain the copy number observed with TPVL sequences, it is likely that something unusual occurred with the TPV genome to facilitate or promote integration. Moreover, as with geminivirus-related sequences in tobacco (10), integration of TPVL sequences must have taken place in cells that contribute to the germ line. Although we cannot yet account for the frequent insertion of TPV DNA into tobacco chromosomes or describe the mechanism of integration, possible clues can be found in the sequence of plant–virus and virus–virus junctions, as well as unusual and variable features in different TPVL sequence clones. On the basis of this information, we propose that failed gap repair in defective versions of open circular TPV DNA led to enhanced recombination between viral and plant sequences.
Even though integration of TPVL sequences did not take place at a specific nucleotide(s), junctions with tobacco DNA and with other TPVL sequences were clustered at two general sites in the putative TPV genome, suggesting that these regions are particularly recombinogenic. About 43% (6/14) of the TPVL sequence junctions with tobacco DNA were concentrated in 11% of the putative TPV genome in ORF1, and 50% were clustered in 11% of the genome comprising the beginning and end of the putative TPV DNA. Three of four virus–virus junctions or duplications/rearrangements were also concentrated in these regions. These findings are strikingly similar to those made with integrated hepatitis B (hepadna)virus sequences, in which ≈40% of the junctions with host DNA and a number of virus–virus junctions were clustered in a region representing 7% of the viral genome (7). The two preferred integration sites of the TPV genome might correspond to the location of gaps in the open circular form of pararetroviral DNA, based on the cauliflower mosaic virus genome as a general model for pararetroviral replication (1). These gaps are normally repaired in a presumably sequential manner in the nucleus to produce supercoiled DNA (24), which is the substrate for transcription by host RNA polymerase II. Delayed repair of these gaps could conceivably extend the lifetime of open circular DNA and increase opportunities for recombination between free ends of TPV DNA at the gaps and plant sequences. In particular, the presence of one cluster of TPVL sequence junctions and rearrangements close to the beginning or end of the putative TPV genome would be consistent with the involvement of gap 1, which is present at the tRNA binding site beginning at nucleotide 1. To explain the second cluster of junctions, a second gap would be postulated in ORF1, which is not incompatible with the position of gap 2 in ORFII of cauliflower mosaic virus open circular DNA (1).
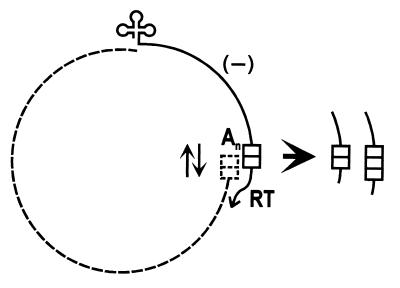
Despite suggestive data indicating the involvement of gaps in viral DNA integration, it is unclear which features of the TPVL sequences could have contributed to delayed gap repair and accumulation of open circular DNA. One unusual and conspicuous characteristic of many TPVL sequence clones is the putative leader region that contains the variable tandem repeat cluster. Given the general absence of such repeats in viral genomes, it is reasonable to assume that the infectious TPV genome contained a single copy of the 63-bp sequence; duplications in this region would be associated with defective virus genomes that were unable to complete one or more steps of the multiplication cycle. Changes in the copy number of the 63-bp monomer could have been generated by misalignment during the template switch by reverse transcriptase if the tandem repeat were present in the terminal repeats of the template RNA (Fig. 5). Alterations in monomer length might have been produced by aberrant reverse transcription through the shorter inverted repeats/direct repeat within the 63-bp monomer, as indicated by the involvement of sequences primarily in stem-loop regions of an RNA secondary structure (Fig. 3). Secondary structures formed in this region could possibly prevent repair of one or more gaps, leaving them free to recombine with plant DNA. Further identification of other integrated pararetrovirus sequences as well as characterization of the natural TPV genome are required to determine the validity of this proposal.
Figure 5.
Hypothetical way to increase the copy number of the tandem repeat during the reverse transcriptase step of TPV genome replication. Based on the position of the RNA polymerase II promoter/leader after ORF4 in the closely related CsVMV (25), it is assumed that the tandem repeat containing two copies of the 63-bp monomer (two blocks) in the putative TPV leader will be present in the terminal repeats of the slightly greater than genome-length TPV RNA (dotted line). Using tRNA (cloverleaf) as a primer at nucleotide 1 of TPV DNA, reverse transcriptase (RT) synthesizes minus strand DNA (solid line) and degrades the RNA template until it reaches the terminal redundancy, where the DNA hybridizes to the complementary RNA sequence before RT switches strands. Because of the presence of the repeat in this region, there could be misalignment in the hybrid (oppositely pointing arrows), leading to the addition of one copy to produce three copies of the monomer. No misalignment maintains two copies. An signifies the poly(A) tail on the RNA.
The absence of sequence homology between target plant DNA and TPVL sequences and frequent rearrangements of TPVL sequences at the insertion site are consistent with integration by illegitimate recombination. Insertion into tobacco DNA appeared random, as indicated by the different types of plant DNA that flanked TPVL sequences, including a highly repetitive tandem element, a moderately repetitive retrotransposon, and a low copy gene. Interestingly, TPVL sequences are much more abundant in the N. sylvestris genome than in N. tomentosiformis or N. otophora, suggesting that most copies in tobacco are present in the N. sylvestris fraction of the genome. Consistent with this, two of the identifiable flanking plant DNA sequences, the NTS9 tandem repeat (22) and the Lhcb gene (GenBank accession no. AB012638), are indeed specific to the N. sylvestris genome. The species-specific accumulation of TPLV sequences indicates that integrated viral DNA can contribute to genome divergence between two closely related plant species.
The putative TPV inferred from integrated TPVL sequences would be the first pararetrovirus described for tobacco. It is more related to CsVMV than to the only other known pararetrovirus of a solanaceous species, petunia vein-clearing virus (11). Similarities extend to the unique features of CsVMV, including a low GC content and modified order of coat protein/movement protein/polyprotein compared with all other plant pararetroviruses. Therefore, it can probably be placed in the same group as CsVMV, which itself has previously been classified as the sole member of a new pararetrovirus group separate from caulimoviruses and badnaviruses (20). Similar sequences in the high molecular weight DNA of other solanceous plants suggest that additional integrated pararetroviruses remain to be discovered.
We obtained no evidence that infectious episomal genomes could result from homologous recombination of TPVL sequences out of the tobacco genome, as has been reported for integrated BSV sequences in banana (12, 13). All TPVL clones that were sequenced contained frameshifts and stop codons, which would not allow reconstitution of an infectious virus. We were also unable to obtain evidence for virus replication or a productive infection in intact plants or regenerating callus, where circular DNA forms or large quantities of viral RNA were not observed. In contrast to the integrated but potentially infectious BSV sequences in the banana genome, the numerous integrated TPVL sequences might provide a novel type of homology-dependent resistance to the putative TPV (26), which would explain the absence of detectable virus and symptoms of infection in tobacco. Some cytosines in the TPVL sequences are methylated (M.F.M., unpublished data). These modified TPVL sequences could possibly trigger homology-mediated methylation of nonintegrated TPV genomes, which would repress transcription and virus replication (27).
In addition to transposable elements, tandem repeats, and microsatellites, integrated pararetroviral-like sequences must now be considered a type of repetitive DNA in higher plants. The discovery of a new class of repeated DNA justifies sequencing nongenic regions to identify additional novel components of plant genomes (28). Not only are such sequences important for understanding plant genome evolution, they can also potentially provide information about interactions between host genomes and parasitic elements and the vagaries of viral genome replication.
Acknowledgments
We are grateful to Dr. B. Charrier and Prof. P. Meyer for performing several independent PCR analyses. We also thank Dr. W. Aufsatz and Prof. T. Hohn for helpful discussions and Dr. B. Morris for comments on the manuscript. This work has been supported by grants to A.M. and M.M. from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (Grant Z21-MED) and the European Union (Contract BIO4-CT96-0253).
Abbreviations
- BSV
banana streak virus
- TPV
tobacco pararetrovirus
- TPVL
tobacco pararetrovirus-like
- kb
kilobase
- RT
reverse transcriptase
- CsVMV
cassava vein mosaic virus
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ238747).
References
- 1.Grierson D, Covey S N. Plant Molecular Biology. New York: Chapman & Hall; 1988. [Google Scholar]
- 2.Wright D A, Voytas D F. Genetics. 1998;149:703–715. doi: 10.1093/genetics/149.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laten H M, Majumdar A, Gaucher E A. Proc Natl Acad Sci USA. 1998;95:6897–6902. doi: 10.1073/pnas.95.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A. Trends Plant Sci. 1998;3:371–374. [Google Scholar]
- 5.Kiss-László Z, Hohn T. Trends Microbiol. 1996;4:480–485. doi: 10.1016/s0966-842x(97)82909-5. [DOI] [PubMed] [Google Scholar]
- 6.Patience C, Wilkinson D A, Weiss R A. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 7.Sherker A H, Marion P L. Annu Rev Microbiol. 1991;45:475–508. doi: 10.1146/annurev.mi.45.100191.002355. [DOI] [PubMed] [Google Scholar]
- 8.Kenton A, Khashoggi A, Parokonny A, Bennett M D, Lichtenstein C. Chromosome Res. 1995;3:346–350. doi: 10.1007/BF00710015. [DOI] [PubMed] [Google Scholar]
- 9.Bejarano E R, Khashoggi A, Witty M, Lichtenstein C. Proc Natl Acad Sci USA. 1996;93:759–764. doi: 10.1073/pnas.93.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashby M K, Warry A, Bejarano E R, Khashoggi A, Burrell M, Lichtenstein C. Plant Mol Biol. 1997;35:313–321. doi: 10.1023/a:1005885200550. [DOI] [PubMed] [Google Scholar]
- 11.Richert-Pöggeler K R, Shepard R J. Virology. 1997;236:137–146. doi: 10.1006/viro.1997.8712. [DOI] [PubMed] [Google Scholar]
- 12.Harper G, Osuji J O, Heslop-Harrison J S, Hull R. Virology. 1999;255:207–213. doi: 10.1006/viro.1998.9581. [DOI] [PubMed] [Google Scholar]
- 13.Ndowora T, Dahal G, LaFleur D, Harper G, Hull R, Olszewski N E, Lockhart B. Virology. 1999;255:214–220. doi: 10.1006/viro.1998.9582. [DOI] [PubMed] [Google Scholar]
- 14.Jakowitsch J, Papp I, Moscone E A, van der Winden J, Matzke M, Matzke A J M. Plant J. 1999;17:131–140. doi: 10.1046/j.1365-313x.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:402–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Thomashow M F, Nutter R, Montoya A, Gordon M P, Nester E W. Cell. 1980;19:729–739. doi: 10.1016/s0092-8674(80)80049-3. [DOI] [PubMed] [Google Scholar]
- 17.Mette M F, van der Winden J, Matzke M A, Matzke A J M. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covey S N, Noad R J, Al-Kaff N S, Turner D S. Methods Mol Biol. 1998;81:53–62. doi: 10.1385/0-89603-385-6:53. [DOI] [PubMed] [Google Scholar]
- 19.Taylor B, Powell A. Focus. 1982;4:4–5. [Google Scholar]
- 20.De Kochko, A., Verdaguer, B., Taylor, N., Carcamo, R., Beachy, R. N. & Fauquet, C. Arch. Virol.143, 945–962. [DOI] [PubMed]
- 21.Hohn T, Dominguez D, Schärer-Hernández N, Pooggin M, Schmidt-Puchta W, Hemmings-Mieszczak M, Fütterer J. In: Mechanisms Determining mRNA Stability and Translation in Plants. Bailey-Serres J, Gallie D, editors. Washington, DC: Am. Soc. Plant Physiol.; 1998. pp. 84–95. [Google Scholar]
- 22.Jakowitsch J, Papp I, Matzke M A, Matzke A J M. Chromosome Res. 1998;6:649–651. doi: 10.1023/a:1009265713279. [DOI] [PubMed] [Google Scholar]
- 23.Thomson K G, Thomas J E, Dietzgen R G. Plant Mol Biol. 1998;38:461–465. doi: 10.1023/a:1006083200299. [DOI] [PubMed] [Google Scholar]
- 24.Covey S N, Turner D S. J Gen Virol. 1993;74:1887–1893. doi: 10.1099/0022-1317-74-9-1887. [DOI] [PubMed] [Google Scholar]
- 25.Verdaguer B, de Kochko A, Beachy R, Fauquet C. Plant Mol Biol. 1996;31:1129–1139. doi: 10.1007/BF00040830. [DOI] [PubMed] [Google Scholar]
- 26.Baulcombe D C. Curr Opin Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- 27.Tang W, Leisner S. Biochem Biophys Res Commun. 1998;245:403–406. doi: 10.1006/bbrc.1998.8435. [DOI] [PubMed] [Google Scholar]
- 28.Bennetzen J L. Trends Plant Sci. 1999;15:85–87. [Google Scholar]