Abstract
The phylogeny of human T cell lymphotropic virus type II (HTLV-II) was investigated by using strains isolated from Amerindian and Pygmy tribes, in which the virus is maintained primarily through mother-to-child transmission via breast-feeding, and strains from intravenous drug users (IDUs), in which spread is mainly blood-borne via needle sharing. Molecular clock analysis showed that HTLV-II has two different evolutionary rates with the molecular clock for the virus in IDUs ticking 150–350 times faster than the one in endemically infected tribes: 2.7 × 10−4 compared with 1.71/7.31 × 10−7 nucleotide substitutions per site per year in the long terminal repeat region. This dramatic acceleration of the evolutionary rate seems to be related with the mode of transmission. Mathematical models showed the correlation of these two molecular clocks with an endemic spread of HTLV-II in infected tribes compared with the epidemic spread in IDUs. We also noted a sharp increase in the population size of the virus among IDUs during the last decades probably caused by the worldwide increase in intravenous drug use.
Human T lymphotropic virus type I (HTLV-I) and type II (HTLV-II) are the first two human retroviruses discovered (1, 2). In contrast to HIV, HTLV-I and HTLV-II are characterized by their low pathogenicity and high genomic stability (3). HTLV-I has been associated with adult T cell leukemia (4) and tropical spastic paraparesis/HTLV-associated myelopathy (5, 6). The role of HTLV-II in human disease remains to be clearly established, even though there is increasing evidence that the infection may be associated with certain neurological diseases (7–9).
HTLV-II has been isolated in blood donors and polytransfused patients, but it is mainly present among intravenous drug users (IDUs), usually co-infected by HIV, in urban areas throughout the world (10–12). Infection has also been shown to be endemic in a number of Amerindian and Pygmy tribes (13–19). Among endemically HTLV-II-infected tribes, the virus is transmitted via sexual contact from husband to wife (rarely the reverse) (20) and from mother to child, via breast-feeding (21). Because the mainly unidirectional sexual transmission cannot perpetuate infection, HTLV-II is maintained in endemically infected populations mainly by vertical transmission (22). In contrast, blood-to-blood contact via needle sharing is the primary way of HTLV-II transmission in IDUs (11).
There are three main subtypes of HTLV-II. HTLV-IIa and HTLV-IIb (23) are both present in IDUs in Europe (24–26), the United States (17, 27), Vietnam (28), and several native Amerindian tribes (13, 14, 19). HTLV-IId was recently isolated in a Mbuti Efe Pygmy (29). An HTLV-IIb strain has been isolated from Bakola pygmies living in Cameroon (15). It has been suggested that HTLV-II was brought into the Americas some 15,000–35,000 years ago during one or more migrations of HTLV-II-infected Asian populations over the Bering land bridge (13, 18, 33, 34) and that HTLV-II among IDUs originated from Amerindians with at least two separate introductions during this century: one for the IIb subtype and another one for the IIa subtype (35). HTLV-II among IDUs seems to evolve at a constant rate. The rate was roughly estimated to be 10−4/10−5 nucleotide substitutions per site per year in the long terminal repeat (LTR) (35). However, a general molecular clock for all the HTLV-II strains does not exist, implying that non-IDU strains evolve at a different rate (35).
In the present paper, using molecular phylogenies and analyzing the branching structure of phylogenetic trees, we have investigated the population dynamics of HTLV-II in IDUs and endemically infected tribes, and we have specifically addressed the evolutionary rate of HTLV-II in these populations.
Materials and Methods
Compilation of Sequence Data.
All of the HTLV-II LTR sequences available at the start of this study (the beginning of 1998) in the European Molecular Biology Laboratory and GenBank databases were retrieved. The LTR is the most variable region in the HTLV-II genome and the largest dataset available for the fragment corresponding to nucleotides 315–706 of the HTLV-IIa Mo isolate (36). Two alignments were obtained by using this fragment: the first one, called IIend, included 26 IIa and IIb strains isolated from different Amerindian tribes, the Efe2 IId strain isolated from a Mbuti Efe pygmy (29), and the HTLV-IIb strain PYGCAM-1 isolated from Bakola pygmies (15); the second one, called IIidu, included 42 IIa and IIb IDU strains. All of the strains in the two alignments were isolated and sequenced in the last 4 years. An additional alignment, called IIiduT, was obtained with 13 IIa IDU strains plus 1 IIb IDU strain isolated between 1995 and 1997 and the HTLV-IIa Mo strain, which was isolated and sequenced in 1983 (37) and which in all previous phylogenetic analyses has been shown clustering with IIa IDU strains (25, 35). The full list of the sequences and the alignments are available at http://kuleuven.ac.be/aidslab/align.htm.
Analysis of the Phylogenetic Signal.
One way to visualize the phylogenetic content of a particular dataset of aligned sequences is to perform a likelihood-mapping analysis investigating a number of random quartets of sequences (38). For each quartet, there are three unrooted tree topologies, and their likelihoods can be simultaneously represented inside an equilateral triangle as a dot. In such an analysis, the corners of the triangle represent the three possible fully resolved tree topologies for the quartet, whereas the center of the triangle represents a completely unresolved tree topology, or star-like evolution, and the sides reflect network-like evolution. For n sequences (n!/4!) quartets are possible. When n is large, a random sample of 10,000 quartets is sufficient to obtain a comprehensive picture of the kind of phylogenetic signal present in a particular alignment. The likelihood mapping analysis, with 10,000 random quartets, was performed on the IIend and IIidu alignments, using Tamura–Nei with γ-distributed rates across sites (TNg) implemented in the program puzzle (38) as the best nucleotide substitution model (39).
Phylogenetic Analysis and Molecular Clocks.
Nucleotide distance matrices for the IIend and the IIidu alignment were estimated under the TNg model with puzzle (38, 39). For each dataset, transition/transversion bias, pyrimidine transition/purine transition bias, and the α shape parameter for the γ-distribution of the rates across sites were also empirically estimated with the maximum likelihood method. The molecular clock hypothesis was tested on the two datasets and on a general alignment including both IIidu and IIend strains with the likelihood ratio test implemented in puzzle. Because the clock hypothesis was accepted for the IIend and the IIidu alignment (see Results), we calculated for each of the two datasets new phylogenetic trees with the unweighted pair group method with arithmetic mean (UPGMA) and the Kitch method implemented in phylip (40), using the same model of nucleotide substitutions as above for the subsequent analysis of the branching pattern (see below). Bootstrap support for the internal branches of each tree was obtained by using 1,000 replicates.
Analysis of Branching Patterns.
Using the end-epi program (41), we analyzed branching patterns of the phylogenetic trees. We calculated the “relative cladogenesis statistic” Pk, which is the probability that a particular lineage, existing at time t, will have k tips or daughter lineages (compared with the total number of tips) by time 0 (the present) (42), for the IIend and the IIidu trees with clock-like branch lengths. Lineages-through-time plots (LTTP) were obtained by plotting the semi-logarithmic number of lineages in the tree against the time at which they appear, or in this case the genetic distance from the root, which represents the relative time when the molecular clock assumption holds. Depending on the shape of the LTTP, it is possible to say whether the population is more likely to have grown exponentially (negative curvature) or to have maintained more or less a constant size (positive curvature). For an exponentially growing population, a further y-axis transformation, called epidemic transform, can be applied to determine whether the rate of the exponential growth is increasing (positive curvature), decreasing (negative curvature), or constant (linear graph). For a non-exponentially growing population, a y-axis transformation, called endemic transform, can be applied to speculate if the population is growing linearly (negative curvature), declining (positive curvature), or maintaining a constant size (linear graph) (42).
Evolutionary Rate Estimations for HTLV-II.
Phylogenetic trees were obtained for the IIiduT alignment by using the TNg model with the neighbor–joining (NJ) and Fitch methods. One thousand bootstrap replicates were performed to support the internal branches of the trees. The evolutionary rate was estimated on the trees dividing the difference in branch lengths from different viral strains to their common ancestor, by the difference in their isolation times. Slightly different rates were obtained by comparing each IIa IDU strain of recent isolation with Mo strain (the earliest taxon), and the average was calculated.
A possible range for the evolutionary rate of the virus infecting endemic tribes was calculated by using the Kitch and the UPGMA trees with clock-like branches obtained from the IIend alignment (TNg model) and by assuming that the introduction of HTLV-II into the Americas occurred 15,000–35,000 years ago (13, 18, 33, 34).
Split Decomposition Method.
The split decomposition method is a transformation-based approach. Evolutionary data is transformed or, more precisely, “canonically decomposed” into a sum of “weakly compatible splits” and then represented by a so-called splits graph. For ideal data, this is a tree; whereas less ideal data will give rise to a tree-like network that can be interpreted as possible evidence for different and conflicting phylogenies. The program splitstree2.3f (Daniel H. Huson, Bielefeld, Germany) was used to generate splits graphs for the IIend and the IIidu dataset, respectively, using the LogDet method (43) for computing distances.
Results
Analysis of Phylogenetic Information.
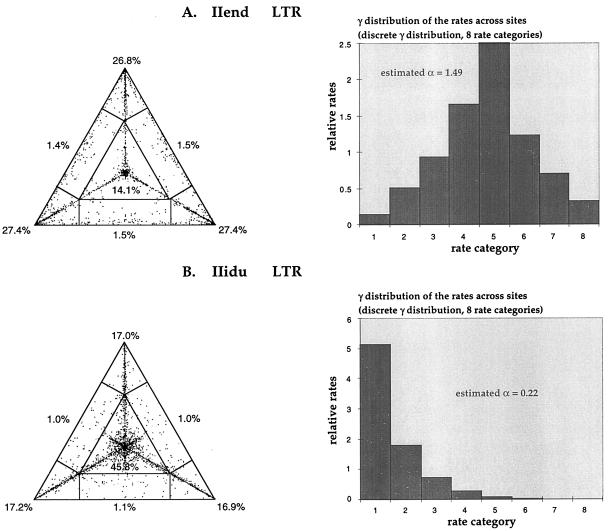
The evaluation of 10,000 random quartets with the likelihood-mapping method gave different answers regarding the phylogenetic signal present in the LTR of HTLV-II endemically infected populations (IIend alignment) compared with HTLV-II-infected IDUs (IIidu alignment). As shown in Fig. 1A, most of the random quartets for the IIend alignment are equally distributed in the three corners of the triangle, representing well-resolved phylogeny, whereas just 14.1% of the quartets are in the center of the triangle, representing star-like phylogeny. The estimated shape parameter α = 1.49 ± 1.27 of the γ-distribution resulted in a bell-shape distribution of the rates along the sites (Fig. 1A), meaning that most sites in the LTR of HTLV-II present in endemically infected populations have intermediate substitution rates and that just a few of them have much lower or much higher rates. In contrast, 45.8% of quartets fall in the center of the triangle when the IIidu alignment is considered, and the estimated shape parameter α = 0.22 ± 0.08 of the γ-distribution resulted in an L-shape distribution of the rates along the sites (Fig. 1B), meaning that most sites in the LTR of HTLV-II isolated in IDUs have very low rates; they are virtually invariable, whereas a few sites exist with very high rates (substitutional “hot spots”). In the latter case, the standard error in estimating α was quite low, strengthening our confidence in the results.
Figure 1.
(A) Likelihood mapping and γ-distribution of rates among sites for the 26 HTLV-II strains, aligned in the LTR region, isolated from Amerindian and Pygmy tribes. Dots in the corners represent fully resolved phylogeny, whereas dots in the center represent phylogenetic noise possibly caused by star-like evolution. To approximate a γ-distribution of nucleotide substitution rates across sites, we used a discrete γ-model with eight rate categories: the horizontal axis represents the rate category, and the vertical axis represents relative rates. The shape of the curve is determined by the parameter α, estimated (together with the relative rates) for each group of aligned sequences with the maximum likelihood method: the curve is L-shaped with α < 1, bell-shaped with α > 1. (B) Likelihood mapping and discrete γ-distribution of rates among sites for the 42 HTLV-II strains, aligned in the LTR region, isolated from IDUs.
Different Molecular Clocks in Different Populations.
Table 1 shows that although the clock hypothesis, evaluated using the likelihood ratio test (see Materials and Methods), has to be rejected when considering the IDU strains (IIidu alignment) and the Pygmy/Amerindian strains (IIend alignment) together; it cannot be rejected when they are analyzed separately. Thus, HTLV-II evolves at a constant rate within IDUs or within endemically infected tribes, irrespective of the subtypes, but at different rate when comparing the two populations.
Table 1.
Maximum likelihood ratio test for molecular clock hypothesis
| Strains used | No. of strains, n | Transition/transversion bias | Pyrimidine transition/purine transition bias | ln likelihood without the clock (ML) | ln likelihood with the clock (MLK) | 2 (lnML − lnMLK) | Significance, P value | Rejection of the molecular clock |
|---|---|---|---|---|---|---|---|---|
| IIend+IIidu | 68 | 2.38 | 1.39 | −1540.86 | −1595.98 | 110.22 | 0.0005 | Yes |
| IIend | 26 | 3.85 | 1.50 | −1059.00 | −1076.13 | 34.26 | 0.08 | No |
| IIidu | 42 | 1.38 | 2.07 | −1158.12 | −1180.09 | 43.93 | 0.31 | No |
IIend includes the HTLV-IIa, HTLV-IIb, and HTLV-IId strains, isolated from Amerindian and Pygmy tribes as described in the text. IIidu includes the HTLV-IIa and HTLV-IIb strains, isolated from United States, European, and Vietnamese IDUs. The P value is calculated according to a χ2 test with n − 2 degrees of freedom. The clock is not rejected when the difference between the likelihoods with and without the clock is not significant (P > 0.05).
Patterns of HTLV-II Cladogenesis.
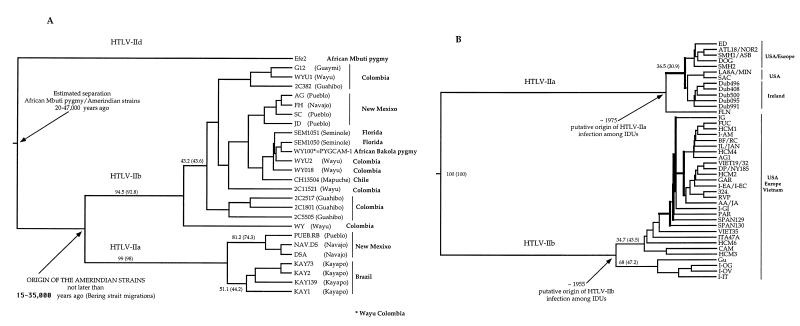
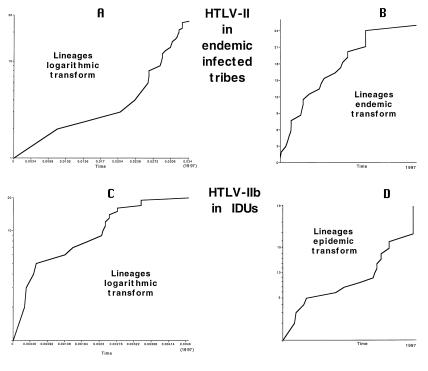
Fig. 2A shows the UPGMA phylogenetic trees for the 26 HTLV-II LTR strains isolated from Pygmy and Amerindian tribes (IIend alignment). Relative statistic cladogenesis (see Materials and Methods) showed that none of the 26 IIend strains produced significantly more daughters than expected under a null model of a uniform rate of cladogenesis. The distribution of lineage-splitting events through time for the IIend strains is shown in the LTTP in Fig. 3A (see Materials and Methods). The entire LTTP curve has positive curvature (Fig. 3A), indicating that the virus did not expand exponentially (41, 42). As a consequence, the so-called endemic transform was applied to the number-of-lineages axis to determine whether the population had stayed constant or had been increasing or declining through time (42). The endemic transform (Fig. 3B) shows a line with negative curvature, indicating a steady growth of the number of virus strains over time (42). The hypothesis of a constant population size was rejected by using the uniform conditional test on the curve of the endemic transform (44). The UPGMA tree of the 42 HTLV-II IDU strains in the LTR is shown in Fig. 2B. Of the 13 and 29 lineages of the IIa and IIb subtypes, respectively, 12 within the IIa and 20 within the IIb produced significantly more daughters than expected (see the clades indicated with the bold line in Fig. 2B) under a null model of uniform rate of cladogenesis (P < 0.05 for the IIa and P < 0.01 for the IIb). Because the analysis of the branching structure with the LTTP is applicable only if there are no major differences in the rate of cladogenesis between branches (42), we analyzed only the LTTP of these 12 IIa and of the 20 IIb strains, respectively, of the phylogenetic tree in Fig. 2B, all of which share a uniform high rate of cladogenesis. The LTTP for the 20 HTLV-IIb strains is reported in Fig. 3C (IIa strains gave similar results, data not shown). The curve clearly has a negative curvature, indicating a population that may have been growing exponentially (41, 42). The so-called epidemic transform was applied to the number-of-lineages axis to determine whether the exponent had stayed constant or had been increasing or decreasing through time (42). In Fig. 3D, the epidemic transform is given. Significant departure of the curve from linearity was tested by using the Wilcoxon-signed rank test (44). A one-tailed test has Z = 1.62 and P > 0.05; thus the line does not depart significantly from linearity, indicating a more or less constant exponential growth of the virus strains among IDUs.
Figure 2.
(A) UPGMA phylogenetic tree of the 26 HTLV-II strains, in the LTR, isolated from Amerindian and Pygmy tribes. The bootstrap statistical analysis was applied by using 1,000 bootstrap samples. Percentage bootstraps for the UPGMA tree are reported (in brackets bootstrap values for the Kitch tree). The dates on the tree indicate the time suggested for the origin of the Amerindian strains (given in bold) and the estimated divergence time between the African Efe2 strain and the other HTLV-II strains. (B) UPGMA phylogenetic tree of the 42 HTLV-II strains, in the LTR, isolated from IDUs. The dates on the tree represent the estimated origin of the HTLV-IIa and HTLV-IIb epidemic in IDUs. Clades indicated with the bold line are those showing a uniform higher rate of cladogenesis with respect to the other strains in the tree (see Results).
Figure 3.
(A) LTTP of the 26 HTLV-II strains, in the LTR, isolated from Amerindian and Pygmy tribes. The time at which each lineage division occurs on a constant-rate (UPGMA) phylogenetic tree is plotted with the time axis scaled as the number of nucleotide substitutions from the root of the tree to the tips. (B) Endemic transform of the same data. This transformation of the vertical axis determines whether the population has been constant (straight line) or linearly growing (downward curvature, as here) or declining (upward curvature) through time. (C) LTTP of the 20 HTLV-IIb strains, in the LTR, isolated from IDUs with higher than expected rate of cladogenesis (branches indicated in bold in Fig. 2). (D) Epidemic transform of the same data. This transformation of the vertical axis determines whether the rate of population growth has been constant (straight line), increasing (upward curvature), or decreasing (downward curvature) through time.
HTLV-II cladogenesis and branching pattern structure for the IIend and IIidu alignments, respectively, were also analyzed on trees obtained with the Kitch method with similar results (data not shown).
HTLV-II Evolutionary Rates.
Neighbor–joining (NJ) and Fitch trees were obtained by using the alignment IIiduT, joining the LTR Mo sequence isolated in 1983 and other HTLV-IIa IDU strains recently isolated. In both trees evaluated with 1,000 bootstrap replicates, Mo was on the most external branch with 64.7% and 63.7% support in NJ and Fitch, respectively (data not shown). Differences in branch lengths between Mo and the most recent taxa were then used for the calculation of the evolutionary rate for HTLV-IIa in IDUs, resulting in 2.7 × 10−4 nucleotide substitutions per site per year (nt/site/yr). The rate calculated on the Fitch tree was slightly higher. Because IIa and IIb IDU strains evolve following the same molecular clock, the rate can be considered an estimate for the LTR evolutionary rate of both HTLV-IIa and -IIb among IDUs. By using this rate on the tree in Fig. 2B, it was possible to estimate the beginning of the HTLV-IIa epidemic among IDUs to be ≈1955 and the beginning of the HTLV-IIb epidemic to be ≈1975. The 20 more recent HTLV-IIb strains, used in the LTTP of Fig. 3C, originated around 1979.
The evolutionary rate of HTLV-II in the LTR among Amerindian and Pygmy infected tribes can be estimated, considering the tree in Fig. 2A. HTLV-II in Amerindians is present in both paleo-Indian and Na-dene populations, but so far it has not been found in Aleut, suggesting that the virus must have been carried along with the earliest human migrations over the Bering strait 15,000–35,000 years ago (13, 18, 33, 34, 45). The divergence in the Amerindian population therefore has been accumulated during 15,000–35,000 years or even more because the divergence of HTLV-IIa and HTLV-IIb might have occurred in Asia when the two populations responsible for the two earliest Amerindian migration waves separated from each other. We used this time interval to calibrate the molecular clock in Amerindians, resulting in an evolutionary rate of 1.71 × 10−6 to 7.31 × 10−7 nt/site/yr. Calculating back toward the separation of HTLV-IId from IIa and IIb, the origin of the HTLV-II tree was putatively placed between 20,000 and 47,000 years ago (Fig. 3A). Since the origin of the virus, the infected population grew slowly as can be deduced from the shape of the curvature given in Fig. 3A. By using the estimated evolutionary rates, the separation between the Bakola pygmy strain PYGCAM-1 and that of the Seminole Amerindian SEM1050 must have occurred only ≈400 years ago, and the separation with the Wayu strain WY100, which has a sequence identical to that of PYGCAM-1, occurred even more recently.
SplitsTrees of HTLV-II Among IDUs and Pygmy/Amerindian Tribes.
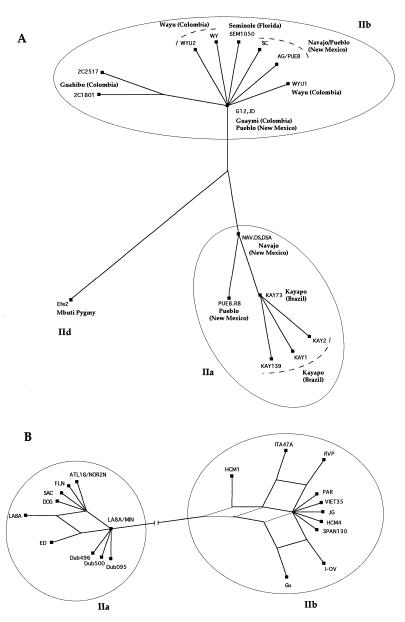
The split decomposition trees, using the IIend alignment and the IIidu alignment are given in Fig. 4. Because of the computational limits of the method, only 22 IIidu and 18 IIend strains were used, representing all of the major clades of the trees in Fig. 2. The IIend alignment shows a classical tree-like behavior (Fig. 4A). For HTLV-IIa, the earliest internal node is occupied by Na-dene populations (NAV.DS and DSA), suggesting transmission of HTLV-IIa from Na-dene (Navajo) to Paleo-Indian (Kayapo). For HTLV-IIb, two Paleo-Indian strains (Pueblo, strain JD and Guaymi, strain G12) occupy the earliest internal node, suggesting that the origin of HTLV-IIb is among Paleo-Indians. These data support the introduction of HTLV-II in two separate waves, one with Paleo-Indian migrations resulting in HTLV-IIb and the second with Na-dene migrations resulting in HTLV-IIa. In contrast, the IIidu SplitsTree shows a complex network reflecting different conflicting phylogenies (Fig. 4B), and it could be attributed to the fact that each virus strain among IDUs could be transmitted several times to a number of different individuals by needle sharing.
Figure 4.
(A) SplitsTree obtained with the split-decomposition method, of 18 HTLV-II strains, in the LTR, isolated from Amerindian and Pygmy tribes. (B) SplitsTree of 20 HTLV-II strains, in the LTR, isolated from IDUs.
Discussion
Different Evolutionary Rates of HTLV-II in Different Populations.
The main goal of the present study is to demonstrate that HTLV-II has two rather different population dynamics in two different infected groups, leading to dramatic differences in its evolutionary rate. In a previous paper, we estimated the evolutionary rate in the LTR of HTLV-II among IDUs to be ≈10−4/10−5 nt/site/yr, assuming a time interval of between 25 and 100 years for the introduction of intravenous drug usage in the United States (35). The results of the present study show that two different molecular clocks exist for HTLV-II: the one for the virus infecting IDUs ticking 150–350 times faster than the one ticking for the virus present in endemically infected Amerindian and African pygmy tribes. RNA virus evolutionary rates generally range from ≈10−2 to 10−4 nt/site/yr (46). The LTR of HTLV-II among IDUs evolves at a rate, 2.7 × 10−4, still compatible with the rates reported for retroviruses. Among endemically infected tribes the estimated rate, 1.71 × 10−6/7.31 × 10−7, is much lower, justifying the classification of HTLV-II as a slowly evolving retrovirus. This and previous phylogenetic analyses performed on different gene regions including the LTR, env, and pX gene regions show that this difference cannot be due to different genomic features of the virus infecting the two populations because both contain closely related IIa and IIb strains (13, 24–29, 35). Different HTLV-II subtypes evolve at the same rate or at a different rate depending only on the populations (IDUs or Amerindian/African pygmy tribes) from which they are isolated. In contrast, in endemically infected tribes, all the sites of the HTLV-II LTR seem to evolve more or less at the same rate, suggesting neutral evolution. This latter statement has to be considered cautiously because the error in estimating the α parameter of the γ-distribution of rates across sites for the IIend dataset is quite high. Both the likelihood-mapping method and the split decomposition method reveal different evolution dynamics in the two populations. Among endemically infected tribes, HTLV-II evolves after a tree-like process, whereas among IDUs, there is much more phylogenetic noise, implying a star-like evolution. Indeed, branching pattern analysis of the phylogenetic trees is in agreement with the idea of a virus linearly increasing its population through time among endemically infected tribes (Fig. 3A) vs. a virus exponentially growing among IDUs (Fig. 3C). Although LTTP seem quite powerful in describing population dynamics of viruses (47), the analysis is limited by the fact that only a tiny proportion of the contemporary lineages can be sampled.
On the basis of these observations, the evolutionary rate of HTLV-II appears to depend on the way the virus is spread and thus on the mode of transmission. Mother-to-child transmission via breast-feeding in endemically infected tribes or transmission via needle sharing among IDUs might contribute to differences in evolutionary rate of related HTLV-II strains. HTLV-II-infected cells in vivo are subjected to clonal expansion (48). Hence, the reverse transcription step is less necessary for the virus to maintain its population in the host during its lifetime, whereas it might be considered necessary for the infection of a new host. Consequently, the higher the transmission rate, the higher the probability of mutations and thus the evolutionary rate of the virus. Other mechanisms might be also involved. Many HTLV-II seropositive IDUs are co-infected with HIV. Therefore, the presence of HIV and of a weaker immune system could also affect the HTLV-II evolutionary rate. However, no effects of HIV on the expression and replication cycle of HTLV-II have been reported so far (48–50). In conclusion, the dramatic increase in the evolutionary rate within IDUs does not seem to be related to co-infection with HIV but rather to the increased rate of transmission of HTLV-II.
Considering the implications of the evolutionary rate of retroviruses for virulence, pathogenesis, and the ability to develop effective antiviral drugs and vaccine, the possibility of a dramatic increase in the rate as a result of different modes of transmission should be considered with attention as a potential source of development of new and more aggressive viral variants.
Early History of HTLV-II in Endemically Infected Tribes.
HTLV-II was introduced into the Americas some 15,000–35,000 years ago, which is 500–2,000 human generations ago. In such a long period, we can assume that any Amerindian strain, a descendent of an ancient virus inherited via breast-feeding, is a result of at least 500–2,000 transmissions or even more considering also the reported sexual transmission of this virus in Amerindian tribes (22). Following this reasoning, it is clear that the HTLV-IIb PYGCAM-1 strain isolated from a Bakola Pygmy in Cameroon and identical to the Colombian Wayu WY100 strain is unlikely to be the descendent of an ancient virus separated from the other Amerindian HTLV-IIb strains before the Bering straits migrations because we would assume that this strain has been vertically transmitted for at least 500–2,000 human generations without any single mutation in the LTR. We estimated that PYCAM-1 diverged no more than 400 years ago. Thus, the infection by this strain more than likely resulted from the increased human mobility and decreased isolation of Bakola pygmies during the last centuries (45). In contrast, by using the Bering strait migration time to date the origin of Amerindian strains, the highly divergent HTLV-IId Efe2 strain, isolated from a Mbuti pygmy, diverged from the other HTLV-II strains at least 20–47,000 years ago, which is in agreement with the view that there has been a long separation between African Mbuti pygmies and non-African human population following the exodus out of Africa of modern humans (45).
Recent History of HTLV-II in IDUs.
In the last few decades, HTLV-II among IDUs has spread exponentially because of needle sharing. In just 1 year, a single IDU could potentially transmit his or her strain to many other IDUs. According to our calculations, HTLV-IIb was introduced in IDUs during the 1950s and HTLV-IIa during the 1970s. In particular, HTLV-IIb seems to have sharply increased its population at the end of the 1970s. The increase in morphine usage after the Second World War, as a painkiller, and the increase of intravenous drug usage, especially among young people during the last 20 years, could be responsible for these epidemic waves of HTLV-II in humans.
Acknowledgments
We are grateful to Prof. Esteban Domingo and Dr. Korbinian Strimmer for helpful discussions. This study was supported in part by the HTLV European Research Network (HERN) and by Grant 3009894N of the Belgian Foundation for Scientific Research. M.S. is the recipient of a Training Mobility Research Marie Curie Fellowship from the European Commission. W.W.H. is supported by the Japanese Foundation for AIDS Prevention.
Abbreviations
- HTLV-I or -II
human T cell lymphotropic virus type I or II
- IDU
intravenous drug users
- IDUIIend
HTLV-II strains isolated from endemically infected populations
- IIidu
HTLV-II strains isolated from intravenous drug users
- LTR
long terminal repeat
- LTTP
lineages-through-time plots
- UPGMA
unweighted pair group method with arithmetic
- TNg
Tamura–Nei model of nucleotide substitution with γ-distributed rates across sites
References
- 1.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J A, Gallo R C. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalyanaraman V S, Sarngadharan M G, Robert-Guroff M, Miyoshi I, Golde D, Gallo R C. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 3.Gessain A, Gallo R C, Franchini G. J Virol. 1992;66:2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida M, Miyoshi V, Hinuma Y. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calendar A, de Thé G. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 6.Osame M, Matsumoto M, Usuku K, Izumo S, Ijichi N, Amitani H, Tara M, Igata A. Ann Neurol. 1987;21:117–122. doi: 10.1002/ana.410210203. [DOI] [PubMed] [Google Scholar]
- 7.Hall W W, Takayuki K, Ijichi S, Takahashi H, Zhu S W. Semin Virol. 1994;5:165–178. [Google Scholar]
- 8.Hall W W, Ishak R, Zhu S W, Novoa P, Eiraku N, Takahashi H, Ferreira C, Azevedo V, Ishak M O, Ferreira C, et al. J Acquired Immune Defic Syndr. 1996;13:S204–S214. doi: 10.1097/00042560-199600001-00031. [DOI] [PubMed] [Google Scholar]
- 9.Murphy E L, Friday J, Smith J W, Engstrom J, Sacher R A, Miller K, Gibble J, Stevens J, Thomson R, Hansma D, et al. Neurology. 1997;48:315–320. doi: 10.1212/wnl.48.2.315. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich G D, Glaser S B, La Vigne K, Quan D, Mildvan D, Sninsky J J, Kwo S, Papsidero L, Poiesz B J. Blood. 1989;74:1658–1664. [PubMed] [Google Scholar]
- 11.Lee H, Swanson P, Shorty V S, Zack J A, Rosenblatt J D, Chen I S Y. Science. 1989;244:471–475. doi: 10.1126/science.2655084. [DOI] [PubMed] [Google Scholar]
- 12.Zella D, Mori L, Sala M, Ferrante P, Casoli C, Magnani G, Achilli G, Cattaneo E, Lori F, Bertazzoni U. Lancet. 1990;335:575–576. doi: 10.1016/0140-6736(90)92140-d. [DOI] [PubMed] [Google Scholar]
- 13.Biggar R, Taylor M E, Neel J V, Hjelle B, Levine P H, Black F, Shaw G M, Sharp P M, Hahn B. Virology. 1996;216:165–173. doi: 10.1006/viro.1996.0043. [DOI] [PubMed] [Google Scholar]
- 14.Duenas-Barajas E, Bernal J E, Vaught D R, Briceno I, Duran C, Yanagihara R I, Gajdusek D C. AIDS Res Hum Retroviruses. 1993;8:1851–1855. doi: 10.1089/aid.1992.8.1851. [DOI] [PubMed] [Google Scholar]
- 15.Gessain A, Mauclère P, Froment A, Biglione M, Hesran J Y L, Tekaia F, Millan J, de Thé G. Proc Natl Acad Sci USA. 1995;92:4041–4045. doi: 10.1073/pnas.92.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goubau P, Desmyter J, Ghesquiere J, Kasereka B. Nature (London) 1992;359:201. doi: 10.1038/359201a0. [DOI] [PubMed] [Google Scholar]
- 17.Hjelle B, Zhu S W, Takahashi H, Ijichi S, Hall W W. J Infect Dis. 1993;168:737–740. doi: 10.1093/infdis/168.3.737. [DOI] [PubMed] [Google Scholar]
- 18.Lairmore M D, Jacobson S, Gracia F, De L, Castillo B K, Larreategui M, Roberts B D, Levine P H, Blattner W A, Kaplan J E. Proc Natl Acad Sci USA. 1990;87:8840–8844. doi: 10.1073/pnas.87.22.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Switzer W M, Owen S M, Pieniazek D, Nerurkar V, Duenas-Barajas E, Heneine W, Lal R B. Virus Genes. 1995;10:153–162. doi: 10.1007/BF01702596. [DOI] [PubMed] [Google Scholar]
- 20.Hjelle B, Cyrus S, Swenso S G. Ann Intern Med. 1992;116:90–91. doi: 10.7326/0003-4819-116-1-90. [DOI] [PubMed] [Google Scholar]
- 21.Van-Dyke R B, Heneine W, Perrin M E, Rudolph D, Starszak E, Woods T, Switzer W M, Kaplan J E. J Pediatr. 1995;127:924–928. doi: 10.1016/s0022-3476(95)70029-3. [DOI] [PubMed] [Google Scholar]
- 22.Black F. Hum Biol. 1997;69:467–482. [PubMed] [Google Scholar]
- 23.Hall W W, Takahashi H, Liu C, Kaplan M H, Scheewind O, Ijichi S, Nagashima K, Gallo R C. J Virol. 1992;66:2456–2463. doi: 10.1128/jvi.66.4.2456-2463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salemi M, Cattaneo E, Casoli C, Bertazzoni U. J Acquired Immune Defic Syndr. 1995;8:516–520. doi: 10.1097/00042560-199504120-00013. [DOI] [PubMed] [Google Scholar]
- 25.Salemi M, Vandamme A-M, Guano F, Gradozzi C, Cattaneo E, Casoli C, Bertazzoni U. J Gen Virol. 1996;77:1193–1201. doi: 10.1099/0022-1317-77-6-1193. [DOI] [PubMed] [Google Scholar]
- 26.Vallejo A, Garcia-Saiz A. J Acquired Immune Defic Syndr. 1994;7:517–519. (lett.). [PubMed] [Google Scholar]
- 27.Takahashi H, Zhu S H, Ijichi S, Vahlne A, Suzuki H, Hall W W. AIDS Res Hum Retroviruses. 1993;9:721–732. doi: 10.1089/aid.1993.9.721. [DOI] [PubMed] [Google Scholar]
- 28.Fukushima Y, Takahashi H, Hall W W, Nakasone T, Nakata S, Song P, Duc D D, Hien B, Quang N X, Trinh T N, et al. AIDS Res Hum Retroviruses. 1995;11:637–644. doi: 10.1089/aid.1995.11.637. [DOI] [PubMed] [Google Scholar]
- 29.Vandamme A-M, Salemi M, Van Brussel M, Liu H-F, Van Laethem K, Van Ranst M, Michels L, Desmyter J, Goubau P. J Virol. 1998;72:4327–4340. doi: 10.1128/jvi.72.5.4327-4340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giri A, Markham P, Digilio L, Hurteau G, Gallo R C, Franchini G. J Virol. 1994;68:8392–8395. doi: 10.1128/jvi.68.12.8392-8395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H F, Vandamme A-M, Van Brussel M, Desmyter J, Goubau P. Lancet. 1994;344:265–266. doi: 10.1016/s0140-6736(94)93032-5. [DOI] [PubMed] [Google Scholar]
- 32.Vandamme A-M, Liu H-F, Van Brussel M, De Meurichy W, Desmyter J, Goubau P. J Gen Virol. 1996;77:1089–1099. doi: 10.1099/0022-1317-77-5-1089. [DOI] [PubMed] [Google Scholar]
- 33.Neel J V, Biggar R J, Suzernik R I. Proc Natl Acad Sci USA. 1994;91:10737–10741. doi: 10.1073/pnas.91.22.10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardi D, Switzer W M, Hadlock K G, Kaplan J E, Lal R B, Folks T M. J Virol. 1993;67:4659–4664. doi: 10.1128/jvi.67.8.4659-4664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salemi M, Vandamme A-M, Gradozzi C, Van Laethem K, Cattaneo E, Taylor G, Casoli C, Goubau P, Desmyter J, Bertazzoni U. J Mol Evol. 1998;46:602–611. doi: 10.1007/pl00006340. [DOI] [PubMed] [Google Scholar]
- 36.Shimotohno K, Takahashi Y, Shimizu N, Gojobori T, Golde D W, Chen I S Y, Miwa M, Sugimura T. Proc Natl Acad Sci USA. 1985;82:3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen I S Y, McLaughlin J, Gasson J C, Clark S C, Golde W. Nature (London) 1983;305:502–505. doi: 10.1038/305502a0. [DOI] [PubMed] [Google Scholar]
- 38.Strimmer K, von Haeseler A. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salemi, M., Desmyter, J. & Vandamme, A.-M. (1999) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 40.Felsenstein J. phylip, Phylogenetic Inference Package. University of Washington, Seattle: Department of Genetics; 1993. , Version 3.5c. [Google Scholar]
- 41.Harvey P H, Rambaut A, Nee S. In: Genesis and Maintenance of Biodiversity. Hochberg M E, Clobert J, Barbault R, editors. Oxford: Oxford Univ. Press; 1995. pp. 60–68. [Google Scholar]
- 42.Nee S, Holmes E C, Rambaut A, Harvey P H. Philos Trans R Soc London B. 1995;349:25–31. doi: 10.1098/rstb.1995.0087. [DOI] [PubMed] [Google Scholar]
- 43.Steel M A. Appl Math Lett. 1994;7:19–24. [Google Scholar]
- 44.Cox D R, Lewis P A W. The Statistical Analysis of Series Events. London: Methuen; 1996. [Google Scholar]
- 45.Cavalli-Sforza L L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 46.Domingo E, Holland J J. In: The Evolution Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 161–184. [Google Scholar]
- 47.de Zanotto P M, Gould E A, Gao G F, Harvey P H, Holmes E C. Proc Natl Acad Sci USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimarelli A, Angelin-Duclos C, Gessain A, Casoli C, Bertazzoni U. Virology. 1996;223:362–364. doi: 10.1006/viro.1996.0487. [DOI] [PubMed] [Google Scholar]
- 49.Cimarelli A, Angelin-Duclos C, Gessain A, Cattaneo E, Casoli C, Biglione M, Mauclère P, Bertazzoni U. J Acquired Immune Defic Syndr. 1995;10:198–204. doi: 10.1097/00042560-199510020-00014. [DOI] [PubMed] [Google Scholar]
- 50.Woods T C, Graber J M, Hershow R C, Khabbaz R F, Kaplan J E, Heneine W. AIDS Res Hum Retroviruses. 1995;11:1235–1239. doi: 10.1089/aid.1995.11.1235. [DOI] [PubMed] [Google Scholar]