Abstract
Understanding nuclear receptor signaling in vivo would be facilitated by an efficient methodology to determine where a nuclear receptor is active. Herein, we present a feedback-inducible expression system in transgenic mice to detect activated nuclear receptor effector proteins by using an inducible reporter gene. With this approach, reporter gene induction is not limited to a particular tissue, and, thus, this approach provides the opportunity for whole-animal screens. Furthermore, the effector and reporter genes are combined to generate a single strain of transgenic mice, which enables direct and rapid analysis of the offspring. The system was applied to localize sites where the retinoic acid receptor ligand-binding domain is activated in vivo. The results identify previously discovered sources of retinoids in the embryo and indicate the existence of previously undiscovered regions of retinoic acid receptor signaling in vivo. Notably, the feedback-inducible nuclear-receptor-driven assay, combined with an independent in vitro assay, provides evidence for a site of retinoid synthesis in the isthmic mesenchyme. These data illustrate the potential of feedback-inducible nuclear-receptor-driven analyses for assessing in vivo activation patterns of nuclear receptors and for analyzing pharmacological properties of natural and synthetic ligands of potential therapeutic value.
Our understanding of the biological functions of nuclear receptors has been hampered by difficulties in precisely localizing the cells and tissues in which a given nuclear receptor is active. To enable such analyses, we have recently used an assay in transgenic mice, which is based on the detection of a reporter gene induced by an effector protein, expressed under the control of an efficient transgenic promoter (1). The effector is a fusion protein consisting of the DNA-binding domain of the yeast transcription factor GAL4, linked in frame with the ligand-binding domain of the studied receptor. The reporter gene is a bacterial lacZ gene preceded by a promoter containing multiple binding sites for GAL4. Thus, activated effector protein will induce expression of the reporter gene in vivo. The assay has been used to study the activities of retinoic acid receptor (RAR) and the retinoid X receptor in vivo (1).
Although useful for identifying sites of nuclear receptor activation, the strategy has limitations; in our previous experiments, the effector transgene was expressed under the control of a tissue-specific promoter, thus limiting the sites and stages where activated effector protein can be detected. A ubiquitous transgenic promoter driving the expression of the GAL4 effector protein would allow reporter gene detection in a broader range of tissues. The potential problem of phenotypic side effects, however, suggests that it would be beneficial to specify the expression to tissues in which the receptor is naturally signaling. Moreover, it is advantageous to combine the study of different effector and reporter genes in a single line of transgenic mice, because separate transgenic effector and reporter mouse strains require intercrossing before analysis of the offspring.
To attain these goals, we have developed a type of inducible assay where a single effector-reporter gene cassette is used to generate transgenic mice. The effector and reporter genes are both driven by GAL4-binding sites (UASs), resulting in autoregulated gene expression in cells in which the GAL4 effector fusion protein is active. This strategy provides a means whereby transgenic expression is specified to tissues in which the GAL4 nuclear receptor derivative is active. We used a fusion between the yeast GAL4 DNA-binding domain and the ligand-binding domain of the RAR to test the strategy. This principle for expression and detection of nuclear receptor activation [referred to as feedback-inducible nuclear-receptor-driven (FIND) expression] is shown to function efficiently in transfected cells in culture as well as in transgenic mice where localized expression of β-galactosidase and green fluorescent protein (GFP) indicated presumptive sites of retinoid signaling in vivo.
Methods
Plasmid Constructions.
To generate UAS-hsp-lacZ (hsp, heat shock protein), four UASs were subcloned from MH100-tk-luc (ref. 2; tk, thymidine kinase) into pKS-hsp-lacZ (3). The UAS sequences are followed by the hsp68 promoter, linked to the lacZ gene. UAS-hsp-gRAR was generated by PCR of the GAL4 DNA-binding domain (amino acids 1–84) from pCMV-gRAR (ref. 1; CMV, cytomegalovirus). The PCR fragment was cloned into UAS-hsp-lacZ, replacing the lacZ coding region and poly(A) signal sequence. The remaining GAL4 DNA-binding domain (amino acids 75–147) and the RAR ligand-binding domain (amino acids 155–461) were derived from pCMV-gRAR, containing the human RARα gene. The effector and reporter constructs were cloned in tandem into the same vector, UAS-hsp-gRAR/lacZ, generating a plasmid containing the UAS-hsp-gRAR poly(A) sequence, followed by the UAS-hsp-lacZ poly(A) sequence. UAS-hsp-g was constructed by deletion of the RAR ligand-binding domain in UAS-hsp-gRAR. UAS-tk-lacZ was published previously under the name lacZr (1).
UAS-hsp-EGFP, encoding the enhanced GFP (EGFP) gene (4), was generated by exchanging the lacZ gene in UAS-hsp-lacZ with the EGFP gene of pEGFP-N1 including the simian virus 40 poly(A) signal (CLONTECH). UAS-tk-gRAR/IRES-LacZ (IRES, internal ribosome entry site) was generated by replacing the βgeo-coding sequence in IRES-βgeo (5) with a lacZ gene, inserted downstream of the IRES, derived from the encephalomyocarditis virus 5′ UTR. The IRES-lacZ fragment was cloned into UAS-tk-lacZ to generate a plasmid containing four UASs upstream of a tk minimal promoter and IRES-lacZ. Finally, the coding regions of GAL4-RAR from CMV-gRAR were cloned downstream of the tk promoter. Primer sequences and further cloning details can be obtained on request.
Transgenic Animals.
Transgenic embryos were generated as described (1). Briefly, UAS-hsp-g or UAS-hsp-gRAR and UAS-hsp-lacZ or UAS-hsp-EGFP DNA were coinjected into the pronucleus of fertilized C57BL/CBA mouse eggs, and transient transgenic embryos were identified by genotyping of amnion-derived DNA by PCR.
Transfections.
Human choriocarcinoma JEG-3 cells were transfected in triplicates in 24-well plates by the calcium phosphate method (2). At 6–8 h after transfection, 0.1 μM of TTNPB ligand {(E)-4-[2-(5,5,8,8-tetramethyl-5,6,7,8 tetrahydro-2-naphtalenyl)-1-propenyl] benzoic acid} or ethanol was added to the cells. At 36 h, cell extracts were assayed for β-galactosidase and luciferase activity in a microplate luminometer/photometer reader (Lucy-1, Anthos, Salzburg, Austria). All β-galactosidase values were normalized to reference luciferase activities.
Explant Cultures and Retinoid Detection.
Defined regions of the spinal cord, midbrain/hindbrain boundary, and nasal processes were excised from embryonic day (E)11.5 and E12.5 wild-type (NMRI) embryos. Tissues were applied to JEG-3 cells cultured in 24-well plates and transfected with CMV-gRAR and UAS-tk-luc plasmids 6 h before explanting. Cells were cultured overnight in the absence or presence of the RAR-selective antagonist Ro 41-5253 at 10 μM (6). Mean luciferase values of triplicate samples were analyzed and normalized to reference β-galactosidase activities.
Histochemical Analyses.
For examination of EGFP expression and for indirect immunocytochemistry, JEG-3 cells were fixed in 4% (vol/vol) paraformaldehyde. Cells were incubated overnight with the monoclonal antibody RK5C1 (Santa Cruz Biotechnology) against the GAL4 DNA-binding domain (dilution 1:20) or directly observed under the microscope (Nikon Eclipse TE 300) with an FITC filter block for analysis of EGFP expression.
Embryos were analyzed by whole-mount X-Gal (5-bromo-4-chloro-3-indoyl β-d-galactoside) staining, at E11.5 and E12.5, as described (1), followed by paraffin embedding and microtome sectioning at 5 μm. After fixation in 4% (vol/vol) paraformaldehyde, embryos carrying the EGFP reporter gene were observed directly under a microscope (Nikon SMZ-2T) with a blue filter block. Paraffin sections were observed under the microscope (Nikon Eclipse, E1000M) by differential interference contrast. Images were collected with a Spot2 camera (Diagnostic Instruments, Sterling Heights, MI).
Results
FIND Expression Vectors.
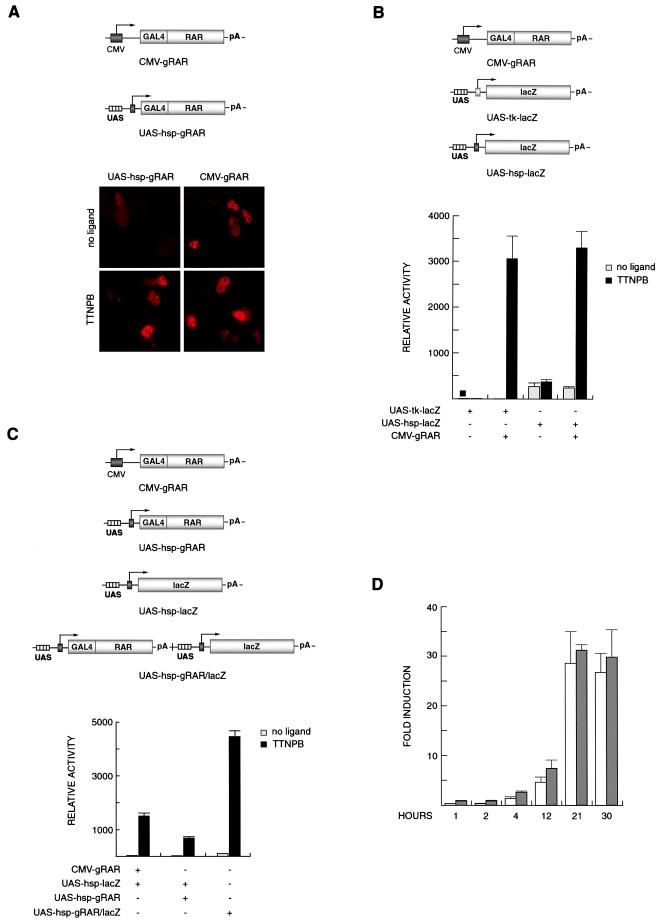
CMV-gRAR (Fig. 1A) is a previously described expression vector encoding the GAL4-RAR linked to the CMV enhancer and promoter (1). In contrast to traditional expression vectors (such as CMV-gRAR), expression from the construct UAS-hsp-gRAR (Fig. 1A) is autoregulated and driven by four UASs upstream of a minimal promoter from the hsp68 gene. This promoter has been used in other studies in reporter gene constructs both in vitro and in transgenic mice (3, 7). We first tested the efficacy of autoregulated expression of GAL4-RAR protein in transfected human JEG-3 choriocarcinoma cells by immunocytochemistry by using antibodies against the GAL4 DNA-binding domain (Fig. 1A). As predicted, strong immunoreactivity could be detected in cells transfected with UAS-hsp-gRAR after incubation with the synthetic RAR ligand, TTNPB. Only a faint signal was detected in TTNPB-treated cells transfected with a construct containing UASs, the hsp promoter, and a GAL4 DNA-binding domain but lacking the ligand-binding domain of RAR (UAS-hsp-g; see also Fig. 5A; data not shown). A quantitative estimate of the number of immunoreactive cells indicated that feedback-induced expression of GAL4-RAR was at least as efficient as the ligand-independent constitutive expression from CMV-gRAR (Fig. 1A and data not shown).
Figure 1.
A FIND expression system. (A) Nuclear staining of JEG-3 cells transfected with indicated plasmids and analyzed for effector protein expression (GAL4-RAR) by indirect immunocytochemistry with an antibody against the GAL4 DNA-binding domain. The CMV-driven expression of GAL4-RAR is constitutively high, whereas basal expression from UAS-hsp-gRAR is low but is induced in the presence of TTNPB. (Magnification ×40.) (B) Comparison of induction with two reporter constructs driven by the minimal tk or hsp promoter. Basal levels, in the absence of TTNPB, are higher with UAS-hsp-lacZ compared with UAS-tk-lacZ, whereas the maximal activation is equal. (C) Reporter activity is up-regulated with equally efficiency by CMV-gRAR and by UAS-hsp-gRAR in the presence of TTNPB. High induction is also observed when the gRAR and lacZ genes are present on a combined effector-reporter vector (UAS-hsp-gRAR/lacZ). (D) Time-course study of autoregulated reporter induction. The induction profiles are similar when UAS-hsp-lacZ is cotransfected with CMV-gRAR (white bars) or UAS-hsp-gRAR (gray bars), reaching maximal values at 21 h after ligand addition. β-Galactosidase activities are shown as means (±SD) of triplicate values.
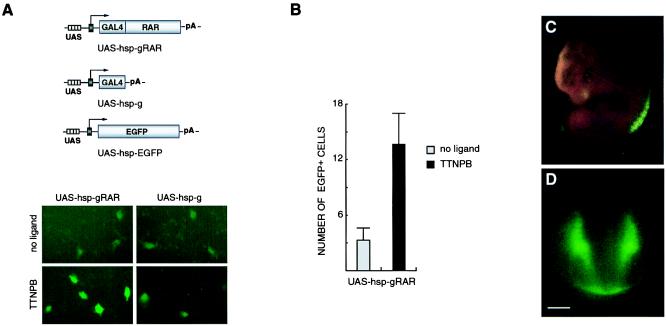
Figure 5.
Feedback-induced EGFP reporter gene expression in vitro (A and B) and in vivo (C and D). (A) GFP expression is up-regulated by TTNPB treatment in cells cotransfected with UAS-hsp-EGFP and UAS-hsp-gRAR but not when the EGFP reporter plasmid is coexpressed with UAS-hsp-g, which lacks the RAR ligand-binding domain. (Magnification ×20.) (B) The number of EGFP-positive cells transfected with UAS-hsp-EGFP and UAS-hsp-gRAR is increased 4-fold in the presence of TTNPB. All cells with detectable fluorescence, irrespective of signal intensity, were rated as positive. (C) EGFP expression in a E11.5 transgenic embryo is detected at the forelimb and hindlimb levels of the spinal cord and in the dorsal aspect of the eye. (D) Transection showing fluorescence in the dorsolateral half and the ventral margin of the spinal cord. (Bar = 100 μm.)
We next investigated whether feedback induced GAL4-RAR-promoted induction of a GAL4-dependent reporter gene in transfected cells. Two different reporter constructs were analyzed. In previous transgenic mouse experiments, we used a reporter containing four UASs upstream of a minimal tk promoter from the herpes simplex virus, followed by a bacterial lacZ gene (ref. 1; UAS-tk-lacZ; Fig. 1B). However, we have noticed that the inducibility of this reporter transgene is highly sensitive to sites of chromosomal integration (data not shown). Published data suggest that the activity of the hsp minimal promoter used in the UAS-hsp-gRAR plasmid construct is relatively insensitive to the site of integration (see, e.g., refs. 2 and 3). Therefore, a reporter construct was designed containing four UASs upstream of the hsp promoter, followed by the lacZ gene (UAS-hsp-lacZ; Fig. 1B). Coexpression of CMV-gRAR with either reporter plasmid resulted in strong β-galactosidase induction after cells had been incubated in the presence of TTNPB (Fig. 1B), although the UAS-hsp-lacZ reporter construct gave rise to a higher basal activity in the absence of ligand. Notably, the ligand-induced maximal activation levels were similar in cells transfected with either of the two lacZ reporter constructs.
We next tested the induction of UAS-hsp-lacZ by feedback-regulated GAL4-RAR. Fig. 1C shows that TTNPB induced the UAS-hsp-lacZ reporter both when GAL4-RAR was expressed from an individual feedback-inducible cotransfected plasmid (UAS-hsp-gRAR) and when the effector and reporter genes were present on a single plasmid construct (UAS-hsp-gRAR/lacZ; Fig. 1C), the latter being consistently more effective.
Because the expression of GAL4-RAR effector protein from UAS-hsp-gRAR depends on a ligand-induced autoregulatory mechanism, we wished to evaluate the time course of reporter gene activation after addition of ligand, as compared with when the GAL4-RAR effector protein is constitutively expressed. No delay was evident (as indicated in Fig. 1D) where reporter gene activation at different time points was assessed in cells transfected with the autoregulated effector and reporter vectors (gray bars) or with CMV-gRAR and UAS-hsp-lacZ (white bars). In conclusion, the FIND expression vectors are remarkably efficient when used in a combined effector/reporter system.
Using an alternative strategy, we tested a bicistronic effector/reporter construct. A UAS-tk-gRAR sequence was linked to a downstream lacZ gene inserted after the IRES from the encephalomyocarditis virus (5). Although lacZ expression was induced by addition of TTNPB, the maximal activation with the bicistronic construct was less than 1/10 of that observed when CMV-gRAR and the UAS-tk-lacZ plasmids were coexpressed.
Hence, FIND expression of GAL4-RAR is effective and can drive reporter gene expression either from a combined effector/reporter gene construct or from a bicistronic expression cassette. Because the combination of two individual UAS-dependent genes proved to be more potent compared with the bicistronic approach, we continued to characterize the more effective, feedback-inducible expression system in transgenic mice.
In Vivo Analysis of RAR Signaling with the FIND Expression System to Drive lacZ and EGFP Reporter Genes.
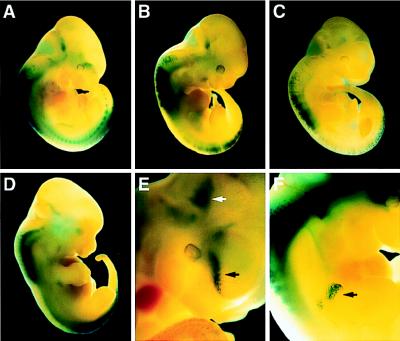
UAS-hsp-gRAR, combined with UAS-hsp-lacZ, was used to generate transgenic embryos, and β-galactosidase expression was analyzed at E11.5 and E12.5 by whole-mount X-Gal staining. In 12 (n = 13) embryos, a characteristic staining was detected, most prominently in the developing spinal cord, at the forelimb and hindlimb levels (Fig. 2A–D). (Three transgenic embryos had very low lacZ expression, although it was consistently confined to these regions of the spinal cord.) Thus, 70–90% of the transgenic embryos showed a clear and reproducible pattern of reporter gene expression. Importantly, these findings are highly consistent with earlier studies of embryonic RAR signaling showing that the brachial and lumbar segments of the spinal cord are “hot spots” for retinoid synthesis (1, 3, 8–15). Additional sites of expression were the ventral forebrain and the proximal forelimb buds, also in keeping with previous reports (refs. 9, 16, and 17; Fig. 2 E and F, black arrows).
Figure 2.
In vivo detection of RAR signaling. Transgenic embryos, at E11.5 (A–C, E, and F) and E12.5 (D), analyzed for X-Gal-staining patterns. (A–D) Staining is observed at the limb levels of the developing cord. (E) Blue-stained cells are detected in the developing forebrain (black arrow) and in a region at the midbrain/hindbrain junction (white arrow). Higher magnification of embryo shown in B. (F) The black arrow indicates staining of the proximal forelimb. (Magnifications: A–C, ×3.5; D, ×3; E and F, ×10.)
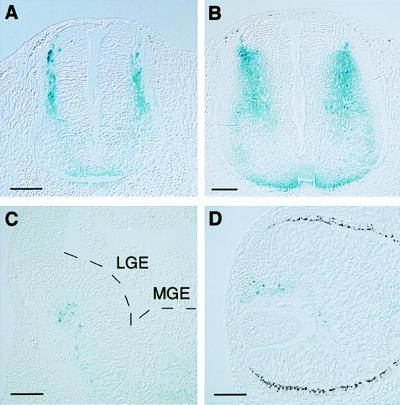
Examination of spinal cord transections in five different embryos revealed X-Gal-positive cells localized predominantly in the dorsal half, in agreement with published results (refs. 1, 3, 9, 10, and 12; Fig. 3 A and B). X-Gal staining in the ventral forebrain was mainly confined to the lateral ganglionic eminence (Fig. 3C). This staining is consistent with an analysis showing that the lateral ganglionic eminence is a localized source of retinoids (17). Also the presumptive neural retina contained cells positive for β-galactosidase (Fig. 3D), in accordance with other studies (9, 10, 18).
Figure 3.
Horizontal (A and B) and coronal (C and D) sections of transgenic embryos showing blue-stained regions in the E11.5 (A) and E12.5 (B) spinal cord, the forebrain region (C), and the eye (D). (A and B) β-Galactosidase-positive cells are located in the dorsolateral half of the spinal cord. Fainter staining is also noted ventrally. (C) Blue-stained cells are mainly confined to the subventricular zone of the lateral ganglionic eminence (LGE) in the forebrain. MGE, medial ganglionic eminence. (D) In the eye, X-Gal-positive cells are present dorsally in the inner layer of the optic cup. (Bars in A–C and D = 100 and 50 μm, respectively.)
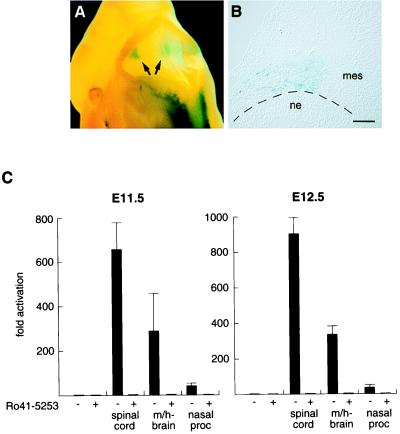
Interestingly, a recurrent staining pattern was detected in the ventral midbrain/hindbrain region, a structure that had not been reported previously to contain retinoids at this stage. This pattern was observed at both E11.5 and E12.5 in 7 of 13 embryos (Fig. 2E, white arrow; Fig. 4A, arrows). Careful examination of sections from this region revealed that the lacZ expression was confined to mesenchymal cells adjacent to the neuroepithelium of the isthmic-pontine area (Fig. 4B and data not shown). To confirm that the staining signal in this region reflected the presence of retinoids and to estimate the levels of RAR-activation, an explant experiment was performed (17). Defined regions from E11.5 and E12.5 wild-type embryos were excised and cultured overnight, together with JEG-3 cells transfected with CMV-gRAR and UAS-tk-luc expression vectors in the absence or presence of the RAR-selective antagonist Ro 41-5253 (6). As depicted in Fig. 4C, potent activation of GAL4-RAR was induced by midbrain/hindbrain explants of both E11.5 and E12.5 embryos as reflected by a 300-fold increase of luciferase activity, indicating that these tissues contain substantial amounts of retinoids. Importantly, GAL4-RAR activation was totally obliterated by the presence of Ro 41-5253, verifying that this activation is a RAR-specific signaling event. Only spinal cord explants gave rise to a more prominent activation of an approximately 600-fold (E11.5) to 900-fold (E12.5) induction, as expected. In contrast, explant tissues of the medial and lateral nasal processes contained very low amounts of activating retinoids, resulting in less than a 40-fold induction. Another tissue of very low retinoid content is the cortex, as previously shown (17). In conclusion, these experiments strongly support that cephalic mesenchyme of the ventral midbrain/hindbrain region is a source of retinoid synthesis and implicate functions for retinoid signaling in the development of midbrain/hindbrain structures.
Figure 4.
Retinoid signaling in the ventral midbrain/hindbrain region. (A) Dorsal view of a X-Gal-stained transgenic E11.5 embryo expressing UAS-hsp-gRAR and UAS-hsp-lacZ; lacZ staining in the midbrain/hindbrain region is evident (arrows). (B) Coronal section of the midbrain/hindbrain region (left side). β-Galactosidase-positive cells are located in cephalic mesenchyme, adjacent to the isthmic/pontine neuroepithelium. (Bar = 100 μm.) (C) Explant experiments of E11.5 (Left) and E12.5 (Right) wild-type embryonic tissues to detect retinoid production. JEG-3 cells transfected with CMV-gRAR and UAS-tk-luc effector and reporter plasmids were incubated overnight either alone or with indicated explants in the absence (−) or presence (+) of the RAR antagonist Ro 41-5253. Luciferase induction is shown as mean fold activation (±SD) of triplicate values. ne, neuroepithelium; mes, mesenchyme; m/h-brain, midbrain/hindbrain region; nasal proc, medial and lateral nasal processes.
In control experiments, a transgenic construct containing only the GAL4 DNA-binding domain (UAS-hsp-g; see Fig. 5A) replaced the GAL4-RAR fusion gene. LacZ expression was detected in one embryo (n = 4), albeit in a pattern entirely unrelated to that consistently observed in GAL4-RAR expressing embryos (data not shown). The remaining GAL4 transgenic embryos did not show any blue staining.
Although the β-galactosidase assay allows a convenient histological staining methodology, detection of GFP has the advantage of enabling analysis in living cells and embryos. Therefore, a similar feedback-inducible reporter system was generated by using EGFP (4) as a reporter gene. JEG-3 cells transfected with the EGFP reporter gene alone or in combination with UAS-hsp-gRAR or UAS-hsp-g were analyzed for TTNPB-induced fluorescence (Fig. 5). Cells containing the EGFP reporter gene and GAL4-RAR displayed strong fluorescence on incubation with ligand (Fig. 5A). Only a few faintly fluorescent cells were detectable when the ligand-binding domain of RAR was excluded in the transfection. A quantification of fluorescent cells per visual field also showed that the number of EGFP-positive cells was increased after incubation in the presence of TTNPB (Fig. 5B). Finally, transgenic embryos containing an EGFP reporter gene reproduced the pattern generated with the lacZ reporter vector (Fig. 5 C and D). The highest intensity of fluorescence was detected at the limb levels of the spinal cord, and in one of the embryos (n = 2), fluorescence was detected in the eye, consistent with previous data (compare Figs. 3D and 5C). Furthermore, in transections of the spinal cord, fluorescent cells were localized in a pattern virtually identical to that observed in lacZ transgenic embryos (Figs. 3A and 5D). In summary, these data show that the FIND system can be applied successfully for studying in vivo signaling of nuclear receptors by detection of induced lacZ or EGFP reporter genes.
Discussion
Herein, we describe a principle for nuclear receptor expression vectors that depends on a feedback autoregulatory mechanism. Feedback-inducible gene expression is shown to enable effective and rapid detection of nuclear receptor-dependent reporter genes in vivo. Reporter gene expression in transgenic mice indicates sites where ligand-induced activation occurs in vivo. However, it should be emphasized that ligand-independent regulatory mechanisms for nuclear receptor activation, e.g., by phosphorylation (19), have been demonstrated and may also be detected by this strategy. Furthermore, patterns of activation in vivo should be interpreted with caution, because certain receptors might require additional or full-length sequences for activation. Nonetheless, this methodology, which we refer to as the FIND system, should be a useful method to assess nuclear receptor activation in vivo.
The assay is amenable to studies on different issues relating to signaling by nuclear receptors. For example, several nuclear receptors mediate paracrine and/or autocrine signaling, where the ligands are locally synthesized (20, 21). Thus, to understand where a given receptor is functional, it is of critical importance to identify sites and time points of receptor activation. The experiments with the GAL4-RAR effector protein described above provide examples of such an analysis. Furthermore, it has become increasingly clear that certain nuclear receptors control metabolic processes by functioning as “sensors” where dietary or metabolic states are influencing the activity of the receptors (22–24). The assay should provide an opportunity to test predictions concerning such sensor mechanisms by measuring receptor activities under varying dietary and/or other environmental conditions. Moreover, a large number of nuclear receptors, including estrogen receptors, peroxisome proliferator activated receptors, liver X receptor, and pregnane X receptor, are considered highly interesting targets in drug development (22, 24–27). This strategy offers a tool to characterize the pharmacological potency and properties of synthetic nuclear receptor ligands in vivo, as well as to define target tissues of drugs. Finally, the system could be used to analyze the activation potential of various orphan receptors.
The FIND system offers several important advantages. (i) By applying the autoregulatory mechanism, the analysis is not restricted by the specificity of a heterologous transgenic promoter that drives the expression of the effector gene. Thus, nuclear receptor activation can be investigated in any cell type and tissue. (ii) We have noted that high levels of expression of GAL4-RAR can lead to phenotypic side effects (A.M.d.U., L.S., and T.P., unpublished observations). The FIND system directs expression of the GAL4 effector to tissues in which a given nuclear receptor is normally active, thus decreasing the risk of teratogenicity. (iii) Although the minimal promoter used in our previous studies (tk) is highly sensitive to the chromosomal integration site for its inducibility (data not shown), the hsp minimal promoter used here consistently gives a reproducible induction of reporter gene expression in GAL4-RAR transgenic embryos. (iv) Only one line of transgenic mice needs to be generated when analyzing a particular receptor, enabling direct analysis of transient transgenic embryos.
GAL4-RAR was used as effector transgene when analyzing the feedback-inducible system in transgenic mice. Because it is relatively well established how retinoids are distributed in vivo from previous studies, it is possible to correlate this knowledge with the pattern of reporter gene activation observed here. In addition to the spinal cord, other sites where retinoids have been reported in the embryo include the developing eye, ear, limb, heart, and the forebrain (8–10, 16, 17, 28). We detected X-Gal- or EGFP-positive cells in most of these regions [optic cup (n = 4), otic vesicle (n = 1), limb bud (n = 4), forebrain (n = 1)]. Interestingly, the developing midbrain/hindbrain region contained β-galactosidase-expressing cells in a majority of the transgenic embryos, and this area has not been reported to produce retinoids. Expression of a proposed retinoid synthesizing enzyme (ADH4) has, however, been established in this region (29). The midbrain/hindbrain boundary has an important function as an organizer for the patterning and development of structures in the posterior midbrain and anterior hindbrain (30). The presence of retinoids and RAR activation in the ventral part of this region indicates that retinoid signaling might be involved in refining these processes. The data show that the GAL4-RAR signaling is confined to the cephalic mesenchyme adjacent to neuroepithelial cells. Taken together, the experiments clearly illustrate the potential of the FIND system to localize sites in different cell types where retinoids are activating the RARs during embryonic development.
In conclusion, we suggest that the presented assay should provide a valuable tool for studying several aspects of nuclear receptor signaling. As exemplified here with RAR, spatial and temporal relationships defining the distribution of ligands in vivo can be determined for a selected receptor. Furthermore, this assay should provide opportunities for addressing complex issues concerning receptor signaling in vivo and allows the assessment of receptor activation under varying environmental conditions. Finally, the system offers unique possibilities to design endpoint assays whereby synthetic ligands of therapeutic interest can be tested by a rational and efficient experimental design.
Acknowledgments
We dedicate this work to the memory of Kazuhiko Umesono. We thank P. Mountford for providing the IRES-βgeo plasmid; M. Klaus for providing Ro 41-5253; U. Lendahl for helpful discussion concerning transgenic methodology and for comments on the manuscript; E. Nilsson and B. Bereczky-Veress at the Karolinska Institute Mouse Camp for excellent technical assistance; and Å. Wallén, R. Zetterström, and R. Pettersson for discussions and comments on the manuscript.
Abbreviations
- RAR
retinoic acid receptor
- FIND
feedback-inducible nuclear-receptor-driven
- tk
thymidine kinase
- hsp
heat shock protein
- CMV
cytomegalovirus
- UAS
GAL4-specific binding site
- IRES
internal ribosome entry site
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- En
embryonic day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Solomin L, Johansson C B, Zetterström R H, Bissonnette R P, Heyman R A, Olson L, Lendahl U, Frisén J, Perlmann T. Nature (London) 1998;395:398–402. doi: 10.1038/26515. [DOI] [PubMed] [Google Scholar]
- 2.Perlmann T, Jansson L. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 3.Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 4.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 5.Mountford P, Zevnik B, Düwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apfel C, Bauer F, Crettaz M, Forni L, Kamber M, Kaufman F, LeMotte P, Pirson W, Klaus M. Proc Natl Acad Sci USA. 1992;89:7129–7133. doi: 10.1073/pnas.89.15.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kothary R, Clapoff S, Darling S, Perry M D, Moran L A, Rossant J. Development (Cambridge, UK) 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds K, Mezey E, Zimmer A. Mech Dev. 1991;36:15–29. doi: 10.1016/0925-4773(91)90068-h. [DOI] [PubMed] [Google Scholar]
- 9.Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Development (Cambridge, UK) 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- 10.Balkan W, Colbert M, Bock C, Linney E. Proc Natl Acad Sci USA. 1992;89:3347–3351. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner M, Jessell T M. Development (Cambridge, UK) 1992;116:55–66. doi: 10.1242/dev.116.1.55. [DOI] [PubMed] [Google Scholar]
- 12.Colbert M C, Linney E, LaMantia A-S. Proc Natl Acad Sci USA. 1993;90:6572–6576. doi: 10.1073/pnas.90.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaffery P, Dräger U C. Proc Natl Acad Sci USA. 1994;91:7194–7197. doi: 10.1073/pnas.91.15.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao D, McCaffery P, Ivins K J, Neve R L, Hogan P, Chin W W, Dräger U C. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 15.Sockanathan S, Jessell T M. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- 16.LaMantia A-S, Colbert M C, Linney E. Neuron. 1993;10:1035–1048. doi: 10.1016/0896-6273(93)90052-s. [DOI] [PubMed] [Google Scholar]
- 17.Toresson H, Mata de Urquiza A, Fagerström C, Perlmann T, Campbell K. Development (Cambridge, UK) 1999;126:1317–1326. doi: 10.1242/dev.126.6.1317. [DOI] [PubMed] [Google Scholar]
- 18.Dräger U C, Wagner E, McCaffery P. J Nutr. 1998;128,Suppl.:4635–4665. doi: 10.1093/jn/128.2.463S. [DOI] [PubMed] [Google Scholar]
- 19.Weigel N L, Zhang Y. J Mol Med. 1998;76:469–479. doi: 10.1007/s001090050241. [DOI] [PubMed] [Google Scholar]
- 20.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 21.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 22.Peet D J, Janowski B A, Mangelsdorf D J. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 23.Kliewer S A, Willson T M. Curr Opin Genet Dev. 1998;8:576–581. doi: 10.1016/s0959-437x(98)80014-2. [DOI] [PubMed] [Google Scholar]
- 24.Blumberg B, Evans R M. Genes Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- 25.Gustafsson J-A. Curr Opin Chem Biol. 1998;2:508–511. doi: 10.1016/s1367-5931(98)80127-0. [DOI] [PubMed] [Google Scholar]
- 26.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterström R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 27.Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter C M, Ong E S, Evans R M. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss J B, Xavier-Neto J, Shapiro M D, Nayeem S M, McCaffery P, Dräger U C, Rosenthal N. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 29.Haselbeck R J, Duester G. Alcohol Clin Exp Res. 1998;22:1607–1613. [PubMed] [Google Scholar]
- 30.Wassef M, Joyner A L. Perspect Dev Neurobiol. 1997;5:3–16. [PubMed] [Google Scholar]