Abstract
Alzheimer's disease (AD) is associated with regional reductions in fluorodeoxyglucose positron emission tomography (FDG PET) measurements of the cerebral metabolic rate for glucose, which may begin long before the onset of histopathological or clinical features, especially in carriers of a common AD susceptibility gene. Molecular evaluation of cells from metabolically affected brain regions could provide new information about the pathogenesis of AD and new targets at which to aim disease-slowing and prevention therapies. Data from a genome-wide transcriptomic study were used to compare the expression of 80 metabolically relevant nuclear genes from laser-capture microdissected non-tangle-bearing neurons from autopsy brains of AD cases and normal controls in posterior cingulate cortex, which is metabolically affected in the earliest stages; other brain regions metabolically affected in PET studies of AD or normal aging; and visual cortex, which is relatively spared. Compared with controls, AD cases had significantly lower expression of 70% of the nuclear genes encoding subunits of the mitochondrial electron transport chain in posterior cingulate cortex, 65% of those in the middle temporal gyrus, 61% of those in hippocampal CA1, 23% of those in entorhinal cortex, 16% of those in visual cortex, and 5% of those in the superior frontal gyrus. Western blots confirmed underexpression of those complex I–V subunits assessed at the protein level. Cerebral metabolic rate for glucose abnormalities in FDG PET studies of AD may be associated with reduced neuronal expression of nuclear genes encoding subunits of the mitochondrial electron transport chain.
Keywords: gene expression, Affymetrix microarrays, laser capture micro-dissection
Alzheimer's disease (AD) is associated with characteristic and progressive reductions in regional positron emission tomography (PET) measurements of the cerebral metabolic rate for glucose (CMRgl). These CMRgl reductions have been reported in the posterior cingulate, parietal, and temporal cortex, and in the frontal cortex and whole brain in more severely affected patients (1–5). Other studies have reported CMRgl reductions in anatomically well characterized hippocampal and entorhinal cortical regions of interest (6–10). The posterior cingulate cortex (PCC) and the neighboring precuneus are metabolically affected in the earliest clinical and preclinical stages of AD (4, 11), and the primary visual cortex is relatively spared (4, 11). In an ongoing series of studies, we have detected CMRgl reductions in cognitively normal carriers of the apolipoprotein E (APOE) ε4 allele (11–15), a common late-onset AD susceptibility gene (16–18). CMRgl reductions in AD-affected areas were correlated with APOE ε4 gene dose (i.e., three levels of genetic risk for AD) and were progressive in late-middle-aged persons (19). These reductions were also apparent in young adult APOE ε4 heterozygotes (13), more than four decades before the anticipated median onset of dementia, years before the expected onset of the major histopathological features of AD (neurofibrillary tangles and amyloid plaques), and, indeed, in anticipating the initial regional appearance of fibrillar amyloid deposition. (20, 21).
AD-related CMRgl reductions could reflect reductions in the density or activity of terminal neuronal fields or peri-synaptic glial cells (22, 23), a metabolic dysfunction in neurons or glial cells not related to neuronal activity (24, 25), or a combination of these factors. These changes do not appear to be solely attributable to the combined effects of atrophy and partial-volume averaging (26). In a postmortem histochemistry study, we previously found that AD cases had lower cytochrome c oxidase activity than controls in the PCC, and that this reduction was significantly greater than that in primary motor cortex (27), another region that is relatively spared. Based on this observation, we proposed that AD might be related to an impairment in neuronal metabolism.
Molecular evaluation of the cells from metabolically affected brain regions could provide new information about the pathogenesis of AD and new targets at which to aim disease-slowing and prevention therapies. Data from a genome-wide transcriptomic study were used to compare, in each of our sampled brain regions, the expression of metabolically relevant nuclear genes from laser-capture microdissected non-tangle-bearing neurons of expired cases with clinically characterized and histopathologically verified AD and expired controls who did meet clinical criteria for dementia and histopathological criteria for AD.
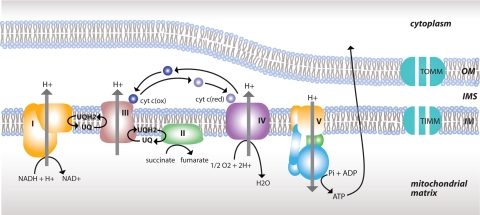
In particular, the data were used to compare cases and controls in the expression of 80 nuclear genes encoding mitochondrial electron transport chain (ETC) subunits along with translocases of the inner and outer mitochondrial membranes (TIMMs and TOMMs, respectively), in six brain regions. These nuclear genes included 39 complex I genes coding for NADH dehydrogenase, all 4 nuclear-encoded complex II genes coding for succinate dehydrogenase, 9 complex III genes coding for ubiquinol-cytochrome c reductase, 13 complex IV genes coding for cytochrome c oxidase, and 15 complex V genes coding for ATP synthase, as well as 11 TIMMs and 6 TOMMs, which regulate the transport of nuclear-encoded electron transport subunits into the mitochondria. These ETC complexes and translocases are illustrated in Fig. 1. The six brain regions included the PCC, which is associated with unusually early CMRgl reductions in AD (4, 28); the middle temporal gyrus (MTG); the hippocampal field CA1 (HIP) and entorhinal cortex (EC), which are also metabolically affected in AD (1, 20); the superior frontal gyrus (SFG), which is associated with preferential CMRgl reductions in normal aging (29–33); and the primary visual cortex (VC), which is relatively spared from CMRgl reductions in both aging and AD (4, 11).
Fig. 1.
Altered expression of mitochondrial energy metabolism elements. Energy metabolism-relevant elements showing statistically significant underexpression in the PCC are shown. These elements include the five complexes of the ETC and TIMMs and TOMMs. OM, outer mitochondrial membrane; IMS, intermembrane space; IM, inner mitochondrial membrane.
We initially tested the hypothesis that AD would be associated with reduced neuronal expression of metabolically relevant nuclear genes in the PCC and that these AD-related reductions would be significantly greater than those in VC. (Findings from our survey of 55,000 neuronal transcripts have been published in ref. 54.) AD-related reductions in the posterior cingulate neuronal expression of several of the implicated complexes I–IV and ATP synthase (complex V) subunits were subsequently validated at the protein level, suggesting that the reductions in the neuronal expression of metabolically relevant nuclear genes might be associated with the CMRgl reductions found in fluorodeoxyglucose positron emission tomography (FDG PET) studies of AD.
Results
Fold change values and P values for the 80 ETC and translocase genes in each of the sampled regions are posted at www.tgen.org/neurogenomics/pcc, and additional expression results have been reported in ref. 54. The proportion of significantly underexpressed ETC and translocase genes in each of the sampled brain regions (P < 0.01 after correction for multiple comparisons) is shown in Table 1. In comparison with controls, AD cases had significantly lower expression of 70% of the nuclear genes encoding mitochondrial ETC subunits in PCC pyramidal neurons, 65% of those in MTG neurons, 61% in HIP neurons, 23% in EC layer II stellate neurons, 16% in VC neurons, and 5% in SFG neurons. By comparison, AD cases had significantly higher expression of only 4% of the nuclear genes encoding mitochondrial ETC subunits in PCC pyramidal neurons, 13% of those in MTG neurons, 8% of HIP neurons, 1% of EC neurons, 4% of VC neurons, and 0% of SFG neurons (data for these genes are listed on the supplementary data site). Additional factors that displayed statistically significant underexpression in the PCC, and which may influence mitochondrial energy metabolism, include the TIMMs and TOMMs (Table 1; individual P values and fold changes are located on the supplementary data site), which are required for the transportation of proteins, including ETC components, from the cytoplasm to the inner mitochondrial membrane and different compartments in the mitochondria (34–37). Overall, 35% of these translocases demonstrated underexpression in the PCC neurons, 47% in the MTG neurons, 41% in the HIP neurons, 35% in the EC neurons, 6% in the VC neurons, and 24% in the SFG neurons. By comparison, only 6% of the translocases showed overexpression in the PCC and HIP; 12% in the VC; and 0% in the MTG, EC, and SFG.
Table 1.
The proportion of underexpressed metabolism-related genes in each of the sample brain regions
| Brain region | Complex I |
Complex II |
Complex III |
Complex IV |
Complex V |
TIMMs |
TOMMs |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of subunits | Fold | No. of subunits | Fold | No. of subunits | Fold | No. of subunits | Fold | No. of subunits | Fold | No. of subunits | Fold | No. of subunits | Fold | |
| PCC | 28/39 (72) | −3.0 | 2/4 (50) | −2.8 | 8/9 (89) | −3.6 | 7/13 (54) | −2.8 | 11/15 (73) | −3.8 | 3/11 (27) | −3.2 | 3/6 (50) | −2.6 |
| MTG | 27/39 (69) | −2.5 | 2/4 (50) | −2.5 | 6/9 (67) | −2.3 | 8/13 (62) | −2.4 | 9/15 (60) | −3.3 | 3/11 (27) | −2.2 | 5/6 (83) | −2.3 |
| HIP | 25/39 (64) | −2.5 | 2/4 (50) | −2.7 | 5/9 (56) | −2.8 | 7/13 (54) | −2.2 | 10/15 (67) | −2.7 | 3/11 (27) | −2.6 | 4/6 (67) | −2.7 |
| EC | 9/39 (23) | −4.5 | 0/4 (0) | 1/9 (11) | −3.7 | 3/13 (23) | −4.5 | 5/15 (33) | −4.3 | 5/11 (45) | −3.7 | 1/6 (17) | −2.8 | |
| VC | 7/39 (18) | −1.8 | 0/4 (0) | 1/9 (11) | −2.0 | 1/13 (8) | −1.5 | 4/15 (27) | −2.1 | 0/11 (0) | 1/6 (17) | −2.2 | ||
| SFG | 4/39 (10) | −3.9 | 0/4 (0) | 0/9 (0) | 0/13 (0) | 0/15 (0) | 2/11 (18) | −3.6 | 2/6 (33) | −2.4 | ||||
The numerator indicates the number of subunits showing statistically significant (P < 0.01 with multiple testing corrections) underexpression and the denominator indicates the total number of nuclear-encoded subunits for the complex/translocase. This ratio is expressed as a percentage in parentheses. PCC, posterior cingulate cortex; MTG, middle temporal gyrus; HIP, hippocampus; EC, entorhinal cortex; VC, visual cortex; SFG, superior frontal gyrus).
In a post hoc analysis, we compared the proportion of all genes underexpressed in the neurons of AD cases and controls. AD was associated with underexpression of 61% of statistically significant (P < 0.01, corrected for multiple comparisons) genes in the PCC, 43% of those in the MTG, 56% of those in the HIP, 69% of those in the EC, 81% of those in the SFG, and 58% of those in the VC. These findings suggest that although metabolism genes may not be disproportionately affected in AD, the regional pattern of underexpressed metabolic genes (PCC > SFG and VC) more closely reflects the pattern of metabolic reductions observed in PET studies.
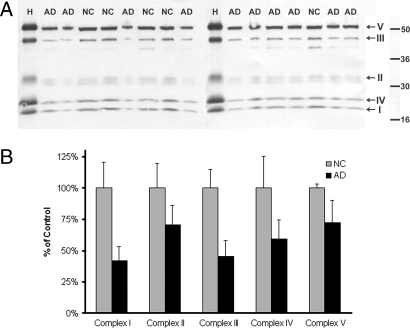
To validate our findings of underexpression of energy metabolic factors, we performed Western blots on the posterior cingulate in controls and AD cases. Concurrent with expression findings, statistically significant decreases were found in the protein levels of each enzyme complex subunit assessed using two-tailed t tests, P < 0.01): complex I (mitochondrially encoded subunit ND6), 41.9 ± 7.0% (mean ± 95% CI) of control, P < 0.006; complex II, 70.6 ± 9.5% of control, P < 0.000005; complex III, 45.3 ± 7.8% of control, P < 0.008; complex IV (mitochondrially encoded subunit COXII), 59.1 ± 9.3% of control, P < 0.002; and complex V, 72.4 ± 11.1% of controls, P < 0.000008 (Fig. 2; human heart was used as positive control)
Fig. 2.
Western blot validation. (A) Western blots using a five-antibody mixture labeling a subunit of each mitochondrial enzyme complex from PCC whole-brain extract. H, human heart for positive control; AD, Alzheimer's patient; NC, normal control. Apparent molecular mass scale is indicated on the right. From highest to lowest are complex V alpha subunit, complex III core 2 subunit, complex II 30-kDa subunit, complex IV subunit II, and complex I 20-kDA subunit. Results are representative of duplicate blots. (B) Western blot-band optical density, expressed as percentage of normal-aged control. Subunit protein expression was significantly lower (two-tailed t test, P < 0.01) for each subunit tested. n = 10 AD; n = 5 NC.
Discussion
This study provides transcriptomic and protein evidence of neuronal metabolic impairments in AD brains, complementing previously established PET evidence of metabolic impairments in persons afflicted by or at risk for this disorder. In our analysis of the nuclear genes influencing mitochondrial energy metabolism (i.e., 80 nuclear genes encoding ETC subunits and 17 nuclear genes encoding mitochondrial translocases responsible for the entry of ETC subunits into the mitochondria), the largest proportion of underexpressed genes was in the PCC, a region which PET studies find to be metabolically affected in the earliest stages of AD. The proportion of underexpressed genes was significantly greater than those in the VC, which is relatively spared in PET studies of AD. The MTG, EC, and HIP, which are also affected in PET studies of AD, had proportions of underexpressed genes in between those in the PCC and VC.
Because our analysis was confined to laser-capture microdissected neurons, our findings suggest that the CMRgI reductions observed in PET studies of AD are at least partly related to molecular processes in neurons themselves, and are thus not solely attributable to a reduction in the activity of perisynaptic glial cells known to influence CMRgI (23). They also suggest that the CMRgI reductions could be at least partly related to molecular processes in neuronal cell bodies (i.e., changes in nuclear gene expression), even though alterations in PET CMRgI measurements have been suggested to be more strongly influenced by the activity of terminal neuronal fields (22). Furthermore, this differential pattern of ETC and metabolic involvement across the cortex highlights the differential vulnerability of various regions to the pathophysiology of AD. Our current transcriptomic findings and previous PET studies raise the possibility that the molecular processes involved in neuronal energy metabolism may be involved in the earliest pathogenesis of AD, possibly preceding the onset of neuritic plaques.
Earlier gene expression and functional activity studies provide confirmatory evidence of this regional pattern of ETC changes. Underexpression was found for mitochondrially-encoded and nuclear-encoded ETC subunits in the middle temporal cortex, but not in motor cortex, of AD patients (38, 39). In a laminar analysis, the PCC also showed the most prominent and significant functional declines across each cortical layer in complex IV activity, whereas the motor cortex was relatively spared (27). Middle temporal, superior temporal, and inferior parietal cortices showed significant layer III activity declines, in agreement with our gene expression data, with the midfrontal cortex showing the smallest decline (40). Further, as the layer III cell bodies from which the expression data were taken project apical dendrites (containing the highest metabolic demand) into the superficial layers of the cortex, these layers (I and II) showed globally, albeit not always significantly, reduced ETC activity that could result from decreased subunit expression in the layer III soma (40).
The regional pattern of underexpression in the genes encoding ETC subunits, which have been validated here with Western blots, corresponds to the regional pattern of CMRgI reductions observed in PET studies of patients with AD (28, 41–45) such that the PCC and precuneus are preferentially affected in AD (11, 16, 17, 19). Because PET studies have also identified the PCC as showing the earliest metabolic changes in cognitively normal carriers of the APOE ε4 allele (19), these changes may precede downstream development of AD pathologies including amyloid plaque and neurofibrillary tangle formation. Metabolic alterations in the PCC have also been implicated in maintaining the brain's “default” state when it is not engaged in the performance of specific tasks (46) or episodic memory tasks (20), as well as in the predisposition to initial hypometabolic (11–13) and subsequent fibrillar amyloid changes (20, 47, 48) associated with AD. The current study provides new information about the neuronal processes involved in these normal and pathological human behaviors.
This study capitalized on the genome-wide evaluation of gene transcripts from laser-capture microdissected neurons from extremely high-quality brain tissue [mean postmortem interval (PMI) = 2.5 h] in clinically and neuropathologically well-characterized AD cases and age-matched controls, and on the analysis of neurons from brain regions preferentially affected or spared in PET studies of AD. Although this study provides clues about the molecular processes related to the CMRgI reductions observed in persons afflicted by and at genetic risk for AD, it has several limitations. First, given the descriptive nature of our findings, it remains to be clarified whether the changes in the nuclear expression of mitochondrial metabolism genes cause reductions in the density or activity of terminal neuronal fields or instead are a consequence of the reduced metabolic demands associated with terminal neuronal changes in AD. Second, it also remains to be clarified whether or not AD is also associated with underexpression of mitochondrially encoded genes from the same neurons (a technically challenging question to address in laser-capture microdissected cells) or in the nuclear or mitochondrial genes of perisynaptic glial cells. Third, although we controlled for mean age at death and gender, there is a small possibility that other group differences in the agonal state preceding death contributed to some of the AD-related reductions in neuronal gene expression observed in this study. Finally, findings from our study suggest that a high proportion of genes are underexpressed in AD, whether or not they are known to directly influence neuronal metabolism. However, the regional pattern of underexpressed genes regulating mitochondrial metabolism appear to correspond to the pattern of CMRgI changes in AD.
Noting the pattern of metabolic changes in our PET studies of AD, Roses et al. have suggested that insulin sensitizers may be helpful in the treatment and prevention of AD (49, 50). Findings from our neurotranscriptomic study could provide clues about the molecular mechanisms that may be involved in the earliest pathogenesis of this disorder and potential targets at which to aim new disease-slowing and prevention therapies.
Conclusion
This study provides transcriptomic and protein evidence that the neuronal nuclear genes influencing mitochondrial energy metabolism are underexpressed in AD, particularly in brain regions like the PCC, which are found to be preferentially affected in PET studies of AD patients and cognitively normal persons at genetic risk for this disorder. In doing so, this study provides information about the molecular processes involved in the pathogenesis of AD and potential therapeutic targets at which to aim disease-slowing and prevention therapies.
Materials and Methods
Tissue Collection.
Brain samples were collected at three Alzheimer's Disease Centers (Washington University, Duke University, and Sun Health Research Institute) from clinically and neuropathologically classified late-onset AD-afflicted individuals (15 males and 18 females) with a mean age of 79.9 ± 6.9 years. Tissue collection of healthy elderly controls was published in a previous report (51); Consortium to Establish a Registry for Alzheimer's Disease (CERAD) neuritic plaque density was infrequent and Braak stages ranged from I to II. All individuals were Caucasian and were matched as closely as possible for gender and mean age at death. Subjects in the AD group had a Braak stage of V or VI (47) with a CERAD score of moderate or frequent (52). Samples were collected (mean PMI of 2.5 h) from six brain regions that are either histopathologically or metabolically relevant to AD. These include the EC (BA 28 and 34), SFG (BA 10 and 11), HIP, VC (BA 17), MTG (BA 21 and 37), and PCC (BA 23 and 31). The EC and HIP are preferentially affected by intracellular neurofibrillary tangles, the MTG and PCC are preferentially affected by metabolism and extracellular β-amyloid plaques, the SFG is preferentially affected by aging, and the VC is relatively spared from both aging and AD pathologies. After dissection, samples were frozen, sectioned (10 μm), and mounted on glass slides.
As previously described (53), brain sections were stained with a combination of thioflavin-S (Sigma) and 1% neutral red (Fisher Scientific), pyramidal neurons were identified by their characteristic size, shape, and location within the region of interest, and tangles were identified by the bright green fluorescence of thioflavin-S staining. In the EC, the large stellate neurons lacking thioflavin-S staining were collected from layer II, and pyramidal cells lacking thioflavin-S staining were collected from CA1 of the HIP. The CA1 region was selected for study because this area is the most, and earliest, affected region in terms of tangle formation, and it has already been expression profiled in neurologically healthy elderly individuals. In all other regions, cortical layer III pyramidal neurons lacking thioflavin-S staining were collected (for all collected neurons, cell bodies were extracted). For each individual, ≈500 histopathologically normal pyramidal neurons were collected from the six brain regions by using laser-capture microdissection (Veritas automated laser-capture microdissection system; Arcturus). Cells were collected onto Arcturus CapSure Macro LCM caps and extracted according to the manufacturer's instructions. Total RNA was isolated from the cell lysate by using the Arcturus PicoPure RNA isolation kit with DNase I treatment using the Qiagen RNase-free DNase set.
Expression Profiling.
Expression profiling was performed as previously described (51). Isolated total RNA was double-round amplified, cleaned, and biotin labeled with the Affymetrix GeneChip two-cycle target labeling kit with a T7 promoter and the Ambion MEGAscript T7 high-yield transcription kit according to the manufacturer's instructions. Amplified and labeled cRNA was quantitated on a spectrophotometer and run on a 1% TAE gel to check for an evenly distributed range of transcript sizes. Twenty micrograms of cRNA was fragmented to ≈35–200 bp by alkaline treatment (200 mM Tris-acetate, pH 8.2; 500 mM KOAc; 150 mM MgOAc) and run on a 1% TAE gel to verify fragmentation. Separate hybridization cocktails were made by using 15 μg of fragmented cRNA from each sample according to Affymetrix's instructions.
Microarray Analysis.
Two hundred microliters of each mixture was separately hybridized to an Affymetrix Human Genome U133 Plus 2.0 array for 16 h at 45°C in the Hybridization Oven 640. The Affymetrix human genome arrays measure the expression of over 47,000 transcripts and variants, including 38,500 characterized human genes. Hybridization cocktails for nine EC samples (normal neurons from AD brains) previously collected following the same profiling methodology used in this project (53) were reanalyzed on the U133 Plus 2.0 array to be evaluated in this study; a 10th EC sample was also separately processed for this sample group. Arrays are washed on the Affymetrix upgraded GeneChip Fluidics Station 450 by using a primary streptavidin phycoerythrin (SAPE) stain, subsequent biotinylated antibody stain, and secondary SAPE stain. Arrays are scanned on the Affymetrix GeneChip Scanner 3000 7G with AutoLoader. Scanned images obtained by the Affymetrix GeneChip Operating Software (GCOS) v1.2 were used to extract raw signal intensity values per probe set on the array and calculate detection calls (absent, marginal, or present). Assignment of detection calls is based on probe-pair intensities for which one probe is a perfect match of the reference sequence and the other is a mismatch probe for which the 13th base (of the 25-oligonucleotide reference sequence) is changed. All raw chip data were scaled to 150 in GCOS to normalize signal intensities for interarray comparisons. Reports generated by GCOS were reviewed for quality control; we looked for at least 20% present calls, a maximum 3′/5′ GAPDH ratio of 30, and a scaling factor <10. Twenty arrays that failed to pass these standards were not included in further analyses.
Pyramidal Cell Quality Control.
To ensure neuronal cell purity in the samples, we evaluated expression of GFAP, an astrocyte cell marker. Fourteen samples that had GFAP expression greater than one standard deviation from the mean were removed from statistical analyses.
Statistical Analysis.
Data for samples from neurologically healthy elderly controls were generated in a previous study (51). Microarray data files of the normal samples are available on the Gene Expression Omnibus site at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE5281 (project accession no. GSE5281), and regional analyses are posted at www.tgen.org/neurogenomics/data/private3.
We compared AD cases and controls in the neuronal expression of 80 nuclear genes encoding subunits of the mitochondrial ETC in each of our sampled brain regions. The nuclear genes included 39 complex I genes coding for NADH dehydrogenase (NDUFA1–A13, NDUFAF1, NDUFAB1, NDUFB1–B11, NDUFC1 and -C2, NDUFS1–S8, and NDUFV1–V3), all 4 complex II genes coding for succinate dehydrogenase (SDHA, SDHB, SDHC, and SDHD), 9 complex III genes coding for ubiquinol-cytochrome c reductase (UCRC, UQCR, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, CYC1, and CYCS), 13 complex IV genes coding for cytochrome c oxidase (COX4I1, COX5A, COX5B, COX6A1, COX6A2, COX6B1, COX6C, COX7A2L, COX7B, COX7B2, COX7C, COX8A, and COX8C), and 15 complex V genes coding for ATP synthase (ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5G1–G3, ATP5H, ATP5I, ATP5J, ATP5J2, ATP5L, and ATP5O). TIMM and TOMM expression was also evaluated (TIMM8A, TIMM8B, TIMM9, TIMM10, TIMM13, TIMM17A, TIMM17B, TIMM22, TIMM23, TIMM44, TIMM50, TOMM7, TOMM20, TOMM22, TOMM34, TOMM40, and TOMM70A). Mitochondrial DNA-encoded subunits were not assessed in this study.
Direct comparisons between neurologically healthy and AD-afflicted brains were performed in all six regions to analyze expression differences in the above-listed ETC subunits. For each analysis, genes that did not demonstrate at least ≈10% present calls across all transcripts profiled for each region-specific comparison were removed by using Genespring GX 7.3 Expression Analysis software (Agilent Technologies). A two-tailed unpaired t test, assuming unequal variances (with a multiple testing correction using the Benjamini and Hochberg false discovery rate), was applied to each comparison for all genes that passed the 10% present-call criterion to locate genes that were statistically significant in differentiating expression between healthy brains and AD brains. (After the present-call filter, 32,153 genes in the PCC were used for P value correction, 30,897 genes in the EC, 32,265 genes in the MTG, 31,496 genes in the HIP, 32,482 genes in the VC, and 32,118 genes in the SFG.) For each analysis comparing AD expression levels with control levels, genes that had a corrected P value ≤ 0.01 were collected, and those genes whose average AD signal and average control signal were both below a threshold of 150 were removed. Fold-change values were determined by calculating the ratio between the average scaled expression signal (for all affected samples) for a gene from the AD sample region and the average scaled expression signal for the same gene from the normal samples across all regional comparisons.
Using this approach, we evaluated ETC and translocase genes that were differentially expressed in the AD cases versus controls for each of the six brain regions of study. Proportions of ETC and translocase genes demonstrating under- and overexpression were determined by calculating the number of subunits (at least ≈10% present calls with corrected P < 0.01) showing such changes over the total number of nuclear-encoded subunits (at least ≈10% present calls with corrected P < 0.01) on the Affymetrix human genome array. Evaluation of all human genes demonstrating underexpression in each region focused on only those genes with statistical significance (P < 0.01 with multiple testing corrections, after present-call filters) for each regional AD versus controls comparison; underexpression percentages were calculated based on a ratio of underexpressed statistically significant genes over all genes.
Data Posting.
Minimum information about a microarray experiment (MIAME)-compliant microarray data files for control samples are located on the Gene Expression Omnibus (GEO) site at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5281 (project accession no. GSE5281). Fold changes and corrected and uncorrected P values for all ETC and translocase genes on the Affymetrix Human Genome Array for each of the six regional comparisons are available online at www.tgen.org/neurogenomics/pcc. Posted lists show respective genes that have at least ≈10% present calls across regional samples (no fold change thresholds have been applied on these lists). Additional expression results will be published in a separate report.
Western Blotting Validation.
Based on availability, 10 profiled AD PCC cases and 5 profiled healthy control cases were collected for Western blot validation. Blots were run by researchers blind to subject condition. Sections were washed from slides by using PBS, and 35 μg of protein per lane was loaded into 10–20% 1.5 mm Tris-glycine 15-well minigels (Invitrogen) with Novex Tris-glycine SDS 2× sample buffer. Gels were run in Novex minicell (Invitrogen) for 90 min at constant 150 V with Novex Tris-glycine SDS running buffer. Proteins were transferred to Immobilon P (Millipore) PVDF membrane, fully submerged in CAPS transfer buffer (10 mM 3-[cyclohexylamino]-1-propane sulfonic acid, pH 11 with NaOH, 10% methanol) by using constant 150 mA for 2 h. Membranes were blocked overnight in 5% Carnation dry milk in PBS at 4°C and then probed with the total OXPHOS detection kit (MS601; 1:1,000 in 1% dry milk; Mitosciences) for 2 h at room temperature with gentle rocking. This five-antibody mixture marks the 20-kDa subunit of complex I (ND6; mitochondrially encoded), the 30-kDa iron–sulfur subunit of complex II, the core 2 subunit of complex III, COX II of complex IV (mitochondrially encoded), and the F1α subunit of ATP synthase. Goat anti-mouse alkaline phosphatase (AP)-conjugated secondary antibody (1:10,000 in 1% dry milk; Santa Cruz Biotechnology) was used (2 h at room temperature with rocking), followed by application of an AP substrate detection kit (Bio-Rad), according to the manufacturer's instructions. Membranes were dried overnight, then simultaneously scanned on a Mustek A3 EP flatbed scanner. All blots were performed in duplicate. The image was imported into Optimas image analysis software (Media Cybernetics), and average OD measures of each band were taken by using a sampling window of constant size. Groups were compared by using Student's two-tailed t tests, uncorrected for multiple comparisons, and 95% confidence intervals for each complex were calculated in Excel.
Acknowledgments.
We thank Roger Higdon and Daniel McKeel for their support through Alzheimer's Disease Research Centers, Lucia Sue (Sun Health Research Institute) for assistance with clinical data, Nick Lehmans (Translational Genomics Research Institute) for setting up the supplementary data site, the National Institute on Aging's Alzheimer's Disease Centers (ADC) program, and the National Alzheimer's Coordinating Center for help in obtaining samples for analysis. This project was funded by the National Institute on Aging Grants K01AG024079 (to T.D.), 1-RO1-AG023193 (to D.A.S.), P30 AG19610 (to E.M.R. for Arizona ADC), P50 AG05681 (to J.C.M.), P01 AG03991 (to J.C.M.), AG05128 (for the Duke University Alzheimer's Disease Research Center), National Alzheimer's Coordinating Center Grant U01AG016976, the state of Arizona (E.M.R.), and the Barrow Neurological Foundation Women's Board (J.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5281).
References
- 1.Alexander GE, Chen K, Pietrini P, Rapoport SI, Reiman EM. Longitudinal PET evaluation of cerebral metabolic decline in dementia: A potential outcome measure in Alzheimer's disease treatment studies. Am J Psychiatry. 2002;159:738–745. doi: 10.1176/appi.ajp.159.5.738. [DOI] [PubMed] [Google Scholar]
- 2.Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: Power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- 3.Jack CR, Jr, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minoshima S, et al. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 5.Thal LJ, et al. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Leon MJ, et al. Prediction of cognitive decline in normal elderly subjects with 2-[18F]fluoro-2-deoxy-d-glucose/positron-emission tomography (FDG/PET). Proc Natl Acad Sci USA. 2001;98:10966–10971. doi: 10.1073/pnas.191044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosconi L, et al. Hippocampal hypometabolism predicts cognitive decline from normal aging. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiol.aging.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santi S, et al. Hippocampal formation glucose metabolism and volume losses in MCI, AD. Neurobiol Aging. 2001;22:529–539. doi: 10.1016/s0197-4580(01)00230-5. [DOI] [PubMed] [Google Scholar]
- 9.Desgranges B, et al. The neural substrates of memory systems impairment in Alzheimer's disease. A PET study of resting brain glucose utilization. Brain. 1998;121:611–631. doi: 10.1093/brain/121.4.611. [DOI] [PubMed] [Google Scholar]
- 10.Stein DJ, Buchsbaum MS, Hof PR, Siegel BV, Jr, Shihabuddin L. Greater metabolic rate decreases in hippocampal formation and proisocortex than in neocortex in Alzheimer's disease. Neuropsychobiology. 1998;37:10–19. doi: 10.1159/000026471. [DOI] [PubMed] [Google Scholar]
- 11.Reiman EM, et al. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 12.Reiman EM, et al. Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer's disease. Proc Natl Acad Sci USA. 2001;98:3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiman EM, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci USA. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small GW, et al. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer's disease. Proc Natl Acad Sci USA. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small GW, et al. Apolipoprotein E type 4 allele and cerebral glucose metabolism in relatives at risk for familial Alzheimer disease. J Am Med Assoc. 1995;273:942–947. [PubMed] [Google Scholar]
- 16.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 17.Saunders AM, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 18.Tang MX, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. J Am Med Assoc. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 19.Reiman EM, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci USA. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckner RL, et al. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mintun MA, et al. [11C]PIB in a nondemented population: Potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz WJ, et al. Metabolic mapping of functional activity in the hypothalamo-neurohypophysial system of the rat. Science. 1979;205:723–725. doi: 10.1126/science.462184. [DOI] [PubMed] [Google Scholar]
- 23.Magistretti PJ, Pellerin L. Cellular bases of brain energy metabolism and their relevance to functional brain imaging: Evidence for a prominent role of astrocytes. Cereb Cortex. 1996;6:50–61. doi: 10.1093/cercor/6.1.50. [DOI] [PubMed] [Google Scholar]
- 24.Piert M, Koeppe RA, Giordani B, Berent S, Kuhl DE. Diminished glucose transport and phosphorylation in Alzheimer's disease determined by dynamic FDG-PET. J Nucl Med. 1996;37:201–208. [PubMed] [Google Scholar]
- 25.Mark RJ, Pang Z, Geddes JW, Uchida K, Mattson MP. Amyloid beta-peptide impairs glucose transport in hippocampal and cortical neurons: Involvement of membrane lipid peroxidation. J Neurosci. 1997;17:1046–1054. doi: 10.1523/JNEUROSCI.17-03-01046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibanez V, et al. Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer's disease. Neurology. 1998;50:1585–1593. doi: 10.1212/wnl.50.6.1585. [DOI] [PubMed] [Google Scholar]
- 27.Valla J, Berndt JD, Gonzalez-Lima F. Energy hypometabolism in posterior cingulate cortex of Alzheimer's patients: Superficial laminar cytochrome oxidase associated with disease duration. J Neurosci. 2001;21:4923–4930. doi: 10.1523/JNEUROSCI.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minoshima S, Foster NL, Kuhl DE. Posterior cingulate cortex in Alzheimer's disease. Lancet. 1994;344:895. doi: 10.1016/s0140-6736(94)92871-1. [DOI] [PubMed] [Google Scholar]
- 29.Angelie E, et al. Regional differences and metabolic changes in normal aging of the human brain: proton MR spectroscopic imaging study. AJNR Am J Neuroradiol. 2001;22:119–127. [PMC free article] [PubMed] [Google Scholar]
- 30.Convit A, et al. Volumetric analysis of the pre-frontal regions: Findings in aging and schizophrenia. Psychiatry Res. 2001;107:61–73. doi: 10.1016/s0925-4927(01)00097-x. [DOI] [PubMed] [Google Scholar]
- 31.Ivancevic V, et al. Regional cerebral glucose metabolism in healthy volunteers determined by fluordeoxyglucose positron emission tomography: appearance and variance in the transaxial, coronal, and sagittal planes. Clin Nucl Med. 2000;25:596–602. doi: 10.1097/00003072-200008000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Loessner A, et al. Regional cerebral function determined by FDG-PET in healthy volunteers: Normal patterns and changes with age. J Nucl Med. 1995;36:1141–1149. [PubMed] [Google Scholar]
- 33.Moeller JR, et al. The metabolic topography of normal aging. J Cereb Blood Flow Metab. 1996;16:385–398. doi: 10.1097/00004647-199605000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 35.Paschen SA, Neupert W. Protein import into mitochondria. IUBMB Life. 2001;52:101–112. doi: 10.1080/15216540152845894. [DOI] [PubMed] [Google Scholar]
- 36.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 37.Berthold J, et al. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 38.Chandrasekaran K, et al. Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Brain Res Mol Brain Res. 1994;24:336–340. doi: 10.1016/0169-328x(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 39.Chandrasekaran K, Hatanpaa K, Rapoport SI, Brady DR. Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res Mol Brain Res. 1997;44:99–104. doi: 10.1016/s0169-328x(96)00191-x. [DOI] [PubMed] [Google Scholar]
- 40.Valla J, Schneider LE, Small AS, Gonzalez-Lima F. Cytochrome oxidase histochemistry in human Alzheimer's disease and animal models. J Histotechnology. 2007;30:1–30. [Google Scholar]
- 41.McGeer EG, et al. 18Fluorodeoxyglucose positron emission tomography studies in presumed Alzheimer cases, including 13 serial scans. Can J Neurol Sci. 1990;17:1–11. doi: 10.1017/s0317167100029930. [DOI] [PubMed] [Google Scholar]
- 42.Mielke R, Herholz K, Grond M, Kessler J, Heiss WD. Clinical deterioration in probable Alzheimer's disease correlates with progressive metabolic impairment of association areas. Dementia. 1994;5:36–41. doi: 10.1159/000106692. [DOI] [PubMed] [Google Scholar]
- 43.Minoshima S, Frey KA, Koeppe RA, Foster NL, Kuhl DE. A diagnostic approach in Alzheimer's disease using three-dimensional stereotactic surface projections of fluorine-18-FDG PET. J Nucl Med. 1995;36:1238–1248. [PubMed] [Google Scholar]
- 44.Pietrini P, et al. Pattern of cerebral metabolic interactions in a subject with isolated amnesia at risk for Alzheimer's disease: A longitudinal evaluation. Dementia. 1993;4:94–101. doi: 10.1159/000107349. [DOI] [PubMed] [Google Scholar]
- 45.Smith GS, et al. Topography of cross-sectional and longitudinal glucose metabolic deficits in Alzheimer's disease. Pathophysiologic implications. Arch Neurol. 1992;49:1142–1150. doi: 10.1001/archneur.1992.00530350056020. [DOI] [PubMed] [Google Scholar]
- 46.Raichle ME, et al. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 48.Mirra SS, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 49.Risner ME, et al. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer's disease. Pharmacogenomics J. 2006;6:246–254. doi: 10.1038/sj.tpj.6500369. [DOI] [PubMed] [Google Scholar]
- 50.Roses AD, et al. Complex disease-associated pharmacogenetics: Drug efficacy, drug safety, and confirmation of a pathogenetic hypothesis (Alzheimer's disease). Pharmacogenomics J. 2007;7:10–28. doi: 10.1038/sj.tpj.6500397. [DOI] [PubMed] [Google Scholar]
- 51.Liang WS, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKhann G, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 53.Dunckley T, et al. Gene expression correlates of neurofibrillary tangles in Alzheimer's disease. Neurobiol Aging. 2006;27:1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: A reference data set. Physiol Genomics. 2008 doi: 10.1152/physiolgenomics.00242.2007. 10.1152-physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]