Abstract
The full-genome sequencing of the filamentous fungi Aspergillus nidulans, Aspergillus niger, and Aspergillus oryzae has opened possibilities for studying the cellular physiology of these fungi on a systemic level. As a tool to explore this, we are making available an Affymetrix GeneChip developed for transcriptome analysis of any of the three above-mentioned aspergilli. Transcriptome analysis of triplicate batch cultivations of all three aspergilli on glucose and xylose media was used to validate the performance of the microarray. Gene comparisons of all three species and cross-analysis with the expression data identified 23 genes to be a conserved response across Aspergillus sp., including the xylose transcriptional activator XlnR. A promoter analysis of the up-regulated genes in all three species indicates the conserved XlnR-binding site to be 5′-GGNTAAA-3′. The composition of the conserved gene-set suggests that xylose acts as a molecule, indicating the presence of complex carbohydrates such as hemicellulose, and triggers an array of degrading enzymes. With this case example, we present a validated tool for transcriptome analysis of three Aspergillus species and a methodology for conducting cross-species evolutionary studies within a genus using comparative transcriptomics.
Keywords: Aspergillus nidulans, Aspergillus niger, Aspergillus oryzae, XlnR
The Aspergillus genus of filamentous fungi has a long history as a work horse in the service of humankind. Aspergillus oryzae (koji mold) was first used for the preparation of food stuffs in China almost 2,000 years ago and was used for one of the first commercial preparations of enzymes in the late 19th century (1, 2). Since then, Aspergillus niger has also proven to be a high-yield producer of organic acids and enzymes, and today, both of these fungi are used as hosts for production of heterologous proteins (3). Since the 1950s, Aspergillus nidulans has been used as a model fungus (4) and has advanced the understanding of eukaryotic cellular physiology and genetics. Advancing our knowledge of these fungi as individual species and as a group holds interest for both fundamental biological sciences and applied biotechnology.
With the publication of the genome sequences of these three aspergilli (5–8), genome-wide systems biology studies in the aspergilli have been made a possibility. As a parallel to the Yeast 2.0 GeneChip that allows for transcriptome analysis of both Saccharomyces cerevisiae and Schizosaccharomyces pombe, we have designed a chip to facilitate system-wide studies of A. nidulans, A. niger, and A. oryzae.
Results
Array Design.
An Affymetrix GeneChip was designed for genes from A. nidulans, A. niger, and A. oryzae. The chip has probes for 99.5% of the genes from the three aspergilli. Because of a selection of unique nonoverlapping probes of similar melting point, not all genes have the maximum of 11 probes. Details on the probe distribution can be found in supporting information (SI) Table 2.
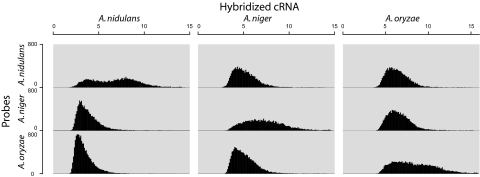
To evaluate the effect of having probes for three species of the same genus on the same chip, we randomly picked one set of transcriptome data from each of the three species (see below for details on the experiments generating these data). For each experiment, the distribution of gene expression values for all three species was examined (Fig. 1). Expression values for the genes specific for the species from which the hybridized cRNA was isolated are higher than expression values for genes from the other two aspergilli. This is the case for samples from all three species. Even though A. niger and A. oryzae are more closely related to each other than A. nidulans, this lineage is not reflected in the shape and levels of the distributions. This reflects that probes not targeting the genes from the species of the hybridized sample are acting in an unspecific manner, much in the same way as probes for unexpressed genes from the same species. Exceptions to this pattern are genes with high expression values in the nonhybridized species. These are composed of constitutively highly expressed and conserved genes, specifically ribosome and histone components. Although this issue influences the expression levels measured, we do not believe this affects evaluation of differential expression between two sets of experiments, because the effect of the probes on the conserved genes will be the same between experiments.
Fig. 1.
Histograms of gene expression values. The distribution of log2-transformed gene expression values of the genes of the three Aspergillus sp. in three experiments. Each column shows an experiment where cRNA from the noted organism was hybridized to the chip. Each row shows the distribution for genes from a specific organism. The analysis was done with the statistical software R (31).
Protein Comparison.
To examine systems regulating transcription conserved in all three aspergilli, genes having homologues in all three species were identified by using a blastp-based comparison (see Materials and Methods). Using this approach, based on bidirectional best hits with an e-value cutoff of 1e-30, 5,561 ORFs were found to be conserved in all three species (tridirectional best hits, SI Table 3). The number of genes with bidirectional, unidirectional, and no hits are shown in SI Fig. 4. The three sets of 5,561 conserved genes (1:1:1 homologues) were used for the further analysis of the transcript data.
Fermentation Results.
As a model example of experiments that can be conducted by using the presented microarray, cultures of A. nidulans, A. niger, and A. oryzae were prepared in well controlled bioreactors. All cultivations were batch cultures grown on defined salt medium with glucose or xylose as carbon sources. Each species had its own specific cultivation medium. For each of the three species, triplicate cultivations were performed on each carbon source. Fig. 2 presents a summary of the six sets of triplicates. Dispersed filamentous growth was observed throughout the fermentation for all cultivations.
Fig. 2.
Summary of fermentation parameters. For each of the Aspergillus sp., a profile of a representative replicate is shown. (□): Sugar concentration (g/liter). (▵): Biomass concentration (g dry weight/liter). The vertical line shows the average time of sampling for transcriptome analysis. Time, sampling time for transcriptome analysis; Biomass, biomass concentration at the sampling time; μmax, maximum specific growth rate. For all three values are shown average and standard deviation for the three replicates.
Transcriptome Analysis.
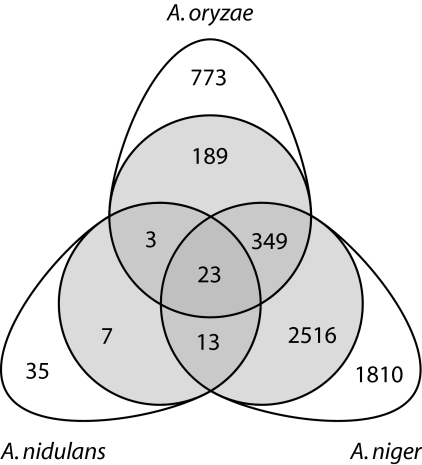
For all three sets of glucose/xylose fermentations, statistical transcriptome analysis was performed. The significantly regulated genes in all three species were compared with the list of the 5,561 conserved genes and with each other (Fig. 3). This resulted in the identification of 23 conserved genes (Table 1) that are differentially regulated in all three species and 365 genes that are differentially expressed in only two of the aspergilli (Fig. 1). The 23 genes that are significant in all three can be seen as a conserved response across the Aspergillus genus.
Fig. 3.
Venn diagram of differentially expressed genes. The gray circles contain the genes that are significantly differentially expressed and conserved in all three Aspergillus species. The numbers on a white background are not conserved in all three species, but still differentially expressed in a single species.
Table 1.
Twenty-three differentially expressed genes conserved in A. nidulans, A. niger, and A. oryzae
| A. nidulans | A. oryzae | A. niger | A. niger annotation | Regulation | Motif A | Ng | O |
|---|---|---|---|---|---|---|---|
| AN0250 | AO090001000069 | JGI55668 | Sugar transporter | Up | NdNgO | ||
| AN0280 | AO090005000767 | JGI55419 | Glucosyl hydrolase | Up | |||
| AN0423 | AO090003000859 | JGI51997 | d-xylose reductase (xyrA) | Up | NdNgO | 12 | |
| AN0942 | AO090005001078 | JGI46405 | l-arabitol dehydrogenase | Up | NdNgO | ||
| AN10124 | AO090003000497 | JGI213437 | β-glycosidase | Up | Ng | ||
| AN10169 | AO090038000426 | JGI177736 | Short-chain dehydrogenase | Up | NdNgO | ||
| AN1677 | AO090023000688 | JGI54541 | Short-chain dehydrogenase | Up | |||
| AN2359 | AO090005000986 | JGI205670 | β-xylosidase (xlnD/xylA) | Up | NdNgO | 10, 13 | 11, 14 |
| AN3184 | AO090012000809 | JGI55604 | Aldose 1-epimerase | Up | Ng | ||
| AN3368 | AO090010000208 | JGI212893 | Glycoside hydrolase | Up | NdNg | ||
| AN3432 | AO090020000042 | JGI56084 | Aldose 1-epimerase | Up | NdNgO | ||
| AN4148 | AO090009000275 | JGI205766 | Sugar transporter | Up | NgO | ||
| AN4590 | AO090011000483 | JGI180923 | Sugar transporter | Up | NgO | ||
| AN5860 | AO090026000494 | JGI197162 | Monosugar-transporter (mstC) | Down | |||
| AN7193 | AO090023000264 | JGI55928 | Aldo/keto reductase | Up | NdNgO | ||
| AN7610 | AO090012000267 | JGI48811 | XlnR | Up | NgO | ||
| AN8138 | AO090010000684 | JGI212736 | α-galactosidase | Up | Ng | ||
| AN8400 | AO090020000324 | JGI199510 | Sugar transporter | Up | NgO | ||
| AN8790 | AO090020000603 | JGI209771 | d-xylulokinase (xkiA) | Up | NdNgO | 15 | |
| AN9064 | AO090038000631 | JGI203198 | Xylitol dehydrogenase (xdhA) | Up | NdNgO | ||
| AN9173 | AO090010000063 | JGI194438 | Sugar transporter | Up | NdNgO | ||
| AN9286 | AO090026000127 | JGI56619 | α-glucuronidase (aguA) | Up | NdNgO | 13, 16 | |
| AN9287 | AO090701000345 | JGI54859 | Lipolytic enzyme | Up | NdNgO |
Genes marked with ″Up″ are up-regulated on xylose medium. The presence of motif A (5′-GGNTAAA-3′) in the promoter region of the A. nidulans, A. niger, and A. oryzae genes is marked with Nd, Ng, and O, respectively. Columns ″Ng″ and ″O″ give references to studies of XlnR induction in A. niger and A. oryzae. No references were found for A. nidulans. VanKuyk et al. (15) show that xkiA is induced on d-xylose in A. niger but does not depend on XlnR. The remaining references describe XlnR induction of the genes on d-xylose.
A further inspection of the expression values of the 23 common genes revealed that the homologues are regulated in the same direction, with 22 of the genes being up-regulated on the xylose medium and only one gene being down-regulated.
The function of the genes in the less-annotated A. nidulans and A. oryzae was inferred from the well annotated A. niger genome sequence, based on the conserved sequences and responses (Table 1). The majority of the 23 common genes are enzymes and sugar transporters. Specifically, the entire d-xylose degradatory pathway (see SI Fig. 5 for an overview) was induced in all three species. A low-affinity glucose transporter (mstC) (9) was down-regulated, implying that this transporter has a higher affinity for glucose than xylose in all three species.
Interestingly, the xylanolytic transcriptional activator XlnR, previously described only in A. niger (10) and in A. oryzae as AoXlnR (11), has a homologue in A. nidulans (AN7610) that is significantly induced on xylose as well. This suggests that XlnR regulation is present in A. nidulans and functions in a manner similar to that reported for A. niger and A. oryzae.
cis-Acting Elements.
In recognition that one or more conserved transcriptional regulators might be active in all three species to produce the conserved response of the 23 genes, statistical promoter analysis was performed for all three species sets of 22 genes up-regulated on xylose. A 5′-GGNTAAA-3′ motif (motif A) was found to be significant (P = 3.6e-28) and present 102 times in the promoters of 46 of the 3 × 22 genes (Table 1). In 12 of the 22 conserved genes, the motif was present in the promoter region of all three species. Included in these 12 sets of genes is the xylose catabolic pathway. For some of the genes (l-arabitol dehydrogenase and d-xylose reductase), the motif is found at the same distance from the start codon for all three homologues (within 5–20 bp) and with a different third base in each of the species. A preference for a specific third base in any of the species across promoters could not be observed. This indicates evolutionary pressure for maintaining the motif but not the third base. Details on the location and sequence of the motifs are in SI Table 4.
The 5′-GGCTAAA-3′ motif has been reported to be the binding site for XlnR from A. niger and A. oryzae (10, 11). However, based on the statistical analysis, we are proposing that the 5′-GGNTAAA-3′ motif is indeed the XlnR motif that is conserved in A. nidulans, A. niger, and A. oryzae.
A separate analysis of the promoters that did not contain the XlnR-motif was carried out, but neither statistical analysis nor manual inspection for syntenic regions revealed any conserved domains. A similar analysis was performed for the down-regulated gene, but no significant motif was found.
Known XlnR-Induced Genes.
As a further validation of the method and the quality of the array, we examined the expression of genes known to be induced by XlnR/AoXlnR on d-xylose. Genes found among the 22 sets of homologues have been marked with a reference in Table 1. Details for all genes are in SI Table 5. A multitude of genes are known from A. niger: xylanase B (xlnB/xynB) (10, 13), arabinoxylan arabinofuranohydrolase (axhA) (13), acetyl xylan esterase (axeA) (13), ferulic acid esterase A (faeA) (13, 17), endoglucanase A (eglA) (13), α- and β-galactosidase [lacA (18), and (aglB) (18)], cellobiohydrolase A (cbhA) (19), d-xylose reductase (xyrA) (12), β-xylosidase (xlnD) (10, 13), and α-glucuronidase (aguA) (13, 16). Additionally, d-xylulokinase (xkiA) (15) has been reported to be induced on xylose but not regulated by XlnR. All were found to be significantly induced in this study. Endoglucanase B and C (eglB/C) and cellobiohydrolase B (cbhB) are known to be induced by XlnR in A. niger but not when grown on d-xylose (13, 19, 20). These were not found to be significantly induced.
Fewer genes have been reported to be induced by XlnR on d-xylose in A. oryzae: β-xylosidase (xylA) (11, 14), endoxylanases F1 (xynF1) (14, 11, 21), G1 (xynG1) (21), and G2 (xynG2) (21). All were significantly induced in A. oryzae. Endoglucanases A and B (celA/B) and cellobiohydrolases C and D (celC/D) are known to be induced by AoXlnR but not when A. oryzae is grown on d-xylose (21). These genes were not significantly induced.
No genes have been reported to be induced by XlnR in A. nidulans, but the xylanases X22 (xlnA) (22, 23), X24 (xlnB) (22, 23), and xylanase X34 (xlnC) (24) are known. Xylanases X22 and X24 are specific for acidic and alkaline medium, respectively, and only xylanase X24 was found to be induced. It had a P value of 0.0527 and was thus not included among the significant 81 genes. Xylanase X34 has been tested only for induction on xylan (24) but was found to be significantly induced on d-xylose.
Interestingly, xylanases are found to be significantly induced in all three species, but none of them are tridirectional best hits and are therefore not found in the core response of 22 genes. It thus seems that each species has a unique set of xylanases.
Discussion
In an application of the presented high-density microarray, we identify a carbon source-based response conserved in three aspergilli. The design of the study involving three different species, grown on three different defined minimal media, at three different values of pH, increases the likelihood of the found genes to be the true conserved “core” response to growth on xylose and not responses relying on an extra factor in addition to xylose (with the possible exception of abundant oxygen). We also believe this approach validates our argument that the xylanolytic transcriptional activator XlnR is a conserved system, even though it has not previously been studied in A. nidulans. Backed by the finding that the 5′-GGNTAAA-3′ motif is present and in some cases conserved as syntenic regions in all three species, we propose that the motif is indeed a XlnR motif and conserved in A. nidulans, A. niger, and A. oryzae. As a point of interest, a study of the homologous genes and their promoter regions in A. fumigatus, a known degrader of dead organic matter, showed a XlnR homologue to be present (Afu2g15620) and the presence of the 5′-GGNTAAA-3′ motif in several of the other homologues, including the entire xylose catabolic pathway (SI Table 6).
Upon further examination of the function of the up-regulated genes in A. nidulans, A. niger, and A. oryzae, it is interesting that the induction of l-arabitol dehydrogenase is found as a part of the core response. Because both the laboratory of Ronald de Vries (personal communication) and our laboratory have found minuscule amounts of arabinose in the commercial preparations of xylose, it might be an artifact. However, a further examination of the data shows that l-arabinose reductase (ORFs AN1679, JGI46249, and AO090009000031), the first step in l-arabinose degradation (see SI Fig. 5) is not significantly induced in any of the three species. Additionally, the XlnR motif is found in the promoter of l-arabitol dehydrogenase in all three homologues of the gene. This implies that this induction is not an artifact and is indeed triggered by xylose. One hypothesis might be that l-arabitol dehydrogenase has an affinity for xylitol as well. Another hypothesis is based on the observation that in nature, xylose is seldom encountered alone, but is a constituent of hemicellulose, along with arabinose, galactose, glucuronic acid, mannose, and other sugars (25, 26). It is thus likely that fungi have evolved coupled responses. This also poses an explanation for the conserved induction of the multitude of sugar transporters, glucuronidase, epimerases, α-galactosidase, and an array of glucoside hydrolases. It thus seems that the conserved xylose response is tailored to degrade complex carbohydrates such as hemicellulose, and xylose triggers this.
Another point of interest is that the promoter analysis suggests xlnR is not autoinducible in A. nidulans. Additionally, promoter analysis shows three sugar transporters in the core response to have only the XlnR motif in A. niger and A. oryzae. This might imply that another less- or non-conserved system is coregulating the xylose/xylanolytic response in A. nidulans along with XlnR. Another possibility is that XlnR can bind to variations of the motif and induces more of the genes than the statistical analysis indicates. van Peij et al. (13) demonstrate that for A. niger, the last base of the motif can vary. de Vries et al. (16) describe induction via a 5′-GGCTAR-3′ motif in A. niger. Marui et al. (11) describe the binding site to be 5′-GGCTA/GA-3′ for A. oryzae. It is thus likely that each species has versions of the motif that are not statistically significant because of the increased variation, and these facilitate induction by XlnR.
The low number of differentially expressed genes in A. nidulans might indicate a lower sensitivity. However, manual inspection of the expression values shows this is because of a higher level of variation between the replicates of the A. nidulans cultivations. Other in-house experiments with A. nidulans and the presented arrays have identified >2,500 significantly regulated genes and >1,000 in a single comparison (data not shown). Additionally, the validation of the array using statistical analysis is confirmed by the expression patterns of hemicellulytic genes known to be induced on d-xylose and/or by XlnR. These are in perfect accordance with the transcriptional analysis of this study.
With this study, we give access to a validated platform for analyzing transcription response in three different Aspergillus sp. This array allows transcriptome analysis of A. oryzae, which was previously unavailable, and is a publicly available Affymetrix-based platform for transcriptome studies in any of the three species. We hope this publication will spur an increase of transcriptome analysis in the individual fungi, thus adding to our knowledge base of this interesting genus of fungi. However, although we acknowledge the multitude of aspects that can be elucidated by traditional single-organism transcriptome analysis, we believe the biggest potential of the herein-presented microarray lies in studies of the multispecies type described in this study. We have demonstrated that data-analysis strategies, such as the blast-based strategy presented here, can add strength to conclusions and help identify systems and responses that are conserved across a genus. This possibility of studying the evolutionary depth of transcriptional regulation adds a new dimension to comparative transcriptomics.
Methods
Strains.
The strains used were A. nidulans A4, A. niger BO-1, and A. oryzae A1560, obtained from Novozymes.
The A. nidulans stock culture was maintained on Sigma potato-dextrose-agar (PDA) at 4°C. A. niger was maintained as frozen spore suspensions at −80°C in 20% glycerol. A. oryzae stock culture was maintained on Cove-N-gly agar at 4°C.
Growth Media.
A. nidulans batch cultivation medium: 15 g/liter (NH4)2SO4, 3 g/liter KH2PO4, 2 g/liter MgSO4·7H2O, 2 g/liter NaCl, 0.2 g/liter CaCl2, and 1 ml/liter trace element solution. Trace element solution: 14.3 g/liter ZnSO4·7H2O, 13.8 g/liter FeSO4·7H2O, and 2.5 g/liter CuSO4·5H2O. Carbon sources used were xylose or glucose monohydrate (20 g/liter). A. niger complex medium: 2 g/liter yeast extract, 3 g/liter tryptone, 10 g/liter glucose monohydrate, 20 g/liter agar, 0.52 g/liter KCl, 0.52 g/liter MgSO4·7H2O, 1.52 g/liter KH2PO4, and 1 ml/liter of trace elements solution. Trace element solution: 0.4 g/liter CuSO4·5H2O, 0.04 g/liter Na2B4O7·10H2O, 0.8 g/liter FeSO4·7H2O, 0.8 g/liter MnSO4·H2O, 0.8 g/liter Na2MoO4·2H2O, 8 g/liter ZnSO4·7H2O. A. niger batch cultivation medium: mineral base, 1.0 g/liter MgSO4·7H2O, 1 g/liter NaCl, 0.1 g/liter CaCl2·2H2O, 0.05 ml/liter antifoam 204 (Sigma), and 1 ml/liter trace element solution. Trace element solution composition: 7.2 g/liter ZnSO4·7H2O, 0.3 g/liter NiCl2·6H2O, 6.9 g/liter FeSO4·7H2O, 3.5 g/liter MnCl2·4H2O, and 1.3 g/liter CuSO4·5H2O. Carbon sources used were xylose or glucose monohydrate (20 g/liter). Nitrogen, sulfur and phosphate sources were 2.5 g/liter (NH4)2SO4, 0.75 g/liter KH2PO4 (glucose medium), or 7.3 g/liter (NH4)2SO4, 1.5 g/liter KH2PO4 (xylose medium). Concentrations were higher on the xylose medium to avoid nitrogen starvation. A. oryzae spore propagation medium (Cove-N-gly): 218 g/liter sorbitol, 10 g/liter glycerol 99.5%, 2.02 g/liter KNO4, 25 g/liter agar and 50 ml/liter salt solution. Cove-N-gly salt solution: 26 g/liter LiCl, 26 g/liter MgSO4·7H2O, 76 g/liter KH2PO4, 50 ml/liter trace element solution. Cove-N-gly trace element solution: 40 mg/liter Na2B4O7·10H2O, 400 mg/liter CuSO4·5H2O, 800 mg/liter FePO4·2H2O, 800 mg/liter MnSO4·H2O, 800 mg/liter NaMoO4·2H2O, 8 g/liter ZnSO4·7H2O. A. oryzae medium for precultures (G2-GLY): 18 g/liter yeast extract, 24 g/liter glycerol 87%, 1 ml/liter pluronic PE-6100. A. oryzae batch cultivation medium: 2.4 g/liter MgSO4·7H2O, 3.6 g/liter K2SO4, 1.2 g/liter citric acid monohydrate, 2.4 g/liter KH2PO4, 3 g/liter (NH4)2HPO4, 1.2 g/liter pluronic acid (PE-6100) and 0.6 ml/liter trace element solution. Trace element solution: 14.3 g/liter ZnSO4·7H2O, 8.5 g/liter MnSO4·H2O, 13.8 g/liter FeSO4·7H2O, 2.5 g/liter CuSO4·5H2O, 3 g/liter citric acid monohydrate (as a chelating agent), and 0.5 g/liter NiCl2·6H2O. Carbon sources used were xylose or glucose monohydrate (15 g/liter).
Preparation of Inoculum.
A. nidulans A4 fermenters were inoculated with spores to a final concentration of 6 × 109 spores per liter. The spores were cultivated on PDA at 37°C for 4–5 days and harvested by adding 20 ml of distilled water.
A. niger BO1 fermentations were initiated by spore inoculation to a final concentration of 2 × 109 spores/liter (glucose cultivations) or 5.7 × 109 spores/liter (xylose cultivations). The spores were propagated on complex media plates and incubated for 7–8 days at 30°C before harvest with 10 ml of Tween 80 0.01%.
A. oryzae A1560 fermenters were inoculated with ≈60 g of broth of A. oryzae A1560 cultured at 30°C for 24 h on G2-GLY liquid medium in shake flasks at 250 rpm (7 × g). The precultures were inoculated with 5 ml of spore solution harvested from mycelium grown on Cove-N-gly agar at 34°C for 3–4 days. Spores were harvested with Tween 80 0.1%.
Batch Cultivations.
A. nidulans batch cultivations were performed in 1.5-liter bioreactors with a working volume of 1.2 liters. The bioreactors were equipped with two Rushton four-blade disk-turbine impellers rotating at 350 rpm. The pH was kept constant at 5.5 by addition of 2 M NaOH or HCl, and the temperature was maintained at 30°C. Air was used for sparging the bioreactor at a constant flow rate of 1 volume of gas per volume of liquid per minute (vvm).
A. niger batch cultivations on glucose medium were performed in 2-liter Braun fermentors with a working volume of 1.6 liters, equipped with three Rushton four-blade disk turbines. The bioreactor was sparged with air, and the concentrations of oxygen and carbon dioxide in the exhaust gas were measured in a gas analyzer. The temperature was maintained at 30°C. The pH was controlled by automatic addition of 2 M NaOH. Agitation and aeration were controlled throughout the cultivations. For inoculation of the bioreactor, the pH was set to 2.5, stirring rate to 100 rpm, and aeration to 0.1 vvm. After germination, the stirring rate was increased to 300 rpm and the air flow to 0.5 vvm. Eleven to 12 h after inoculation, the stirring rate was increased to 600 rpm and the air flow to 1 vvm. When the CO2 in the exhaust gas reached a value of 0.1%, the stirring rate was set to 1,000 rpm, and the pH was gradually increased to 4.5.
A. niger batch cultivations on xylose medium were carried out in 5-liter reactors with a working volume of 4.5 liters. The bioreactors were equipped with two Rushton four-blade disk turbines, pH and temperature control, and no baffles. Inlet air was controlled with a mass flowmeter. The temperature was maintained at 30°C, and the pH was controlled by automatic addition of 2 M NaOH. The pH was initially set to 3.0 to prevent spore aggregation; only when spores started to germinate was the pH gradually increased to 4.5. Similarly, the stirring speed was initially set to 200 rpm and the aeration rate to 0.05 vvm. After germination, these parameters were progressively increased to 600 rpm and 0.89 vvm and kept steady throughout the rest of the fermentation.
A. oryzae batch cultivations were done in 2-liter fermenters with a working volume of 1.2 liters. The stirrer speed was kept at 800 rpm during the first 4 h and then increased to 1,100 rpm. The pH was controlled at 6 by addition of 4 M NaOH and 4 M HCl, and the temperature was maintained at 34°C. The aeration flow rate was set at 1.2 vvm. Dissolved oxygen tension was initially calibrated at 100%.
The concentrations of oxygen and carbon dioxide in the exhaust gas were monitored with a gas analyzer (1311 Fast Response Triple Gas, Innova combined with multiplexer controller for Gas Analysis MUX100, Braun Biotech).
Sampling.
Cell dry weight was determined by using nitrocellulose filters (pore size 0.45 μm, Gelman Sciences). The filters were predried in a microwave oven at 150 W for 15 min or at 100°C for 24 h, cooled in a desiccator, and subsequently weighed. A known volume of cell culture was filtered, and the residue was washed with distilled water or 0.9% NaCl and dried on the filter for 15 min in a microwave oven at 150 W or at 100°C for 24 h and cooled in a desiccator. The filter was weighed again, and the cell mass concentration was calculated. These values were used to calculate maximum specific growth rates. For gene expression analysis, mycelium was harvested at the mid-late exponential phase by filtration through sterile Mira-Cloth (Calbiochem). At this point, A. niger mycelium was washed with a PBS buffer (8 g/liter NaCl, 0.20 g/liter KCl, 1.44 g/liter Na2HPO4, and 0.24 g/liter KH2PO4 in distilled water). The mycelium was quickly dried by squeezing and subsequently frozen in liquid nitrogen. Samples were stored at −80°C until RNA extraction.
Quantification of Sugars and Extracellular Metabolites.
The concentrations of sugar in the filtrates were determined by using HPLC on an Aminex HPX-87H ion-exclusion column (BioRad). The column was eluted at 60°C with 5 mM H2SO4 at a flow rate of 0.6 ml/min. Metabolites were detected with a refractive index detector and a UV detector.
Extraction of Total RNA.
A. nidulans and A. niger: 40–50 mg of frozen mycelium was placed in a 2 ml of microcentrifuge tube, precooled in liquid nitrogen containing three steel balls (two balls with a diameter of 2 mm and one ball with a diameter of 5 mm). The tubes were then shaken in a Retsch Mixer Mill, at 5°C for 10 min, until the mycelia were ground to powder. Total RNA was isolated from the powder using the Qiagen RNeasy Mini Kit, according to the protocol for isolation of total RNA from plant and fungi.
A. oryzae.
Total RNA was purified by using the Promega RNAgents Total RNA Isolation system according to the protocol. For purification, ≈1 g of frozen mycelium was ground to a fine powder under liquid nitrogen using a ceramic mortar and pestle.
For all samples, the quality of the total RNA extracted was determined by using a BioAnalyzer 2100 (Agilent Technologies) and the quantity determined by using a spectrophotometer (Amersham Pharmacia Biotech, GE Healthcare Bio-Sciences). Total RNA was stored at −80°C until further processing.
Preparation of Biotin-Labeled cRNA and Microarray Processing.
Fifteen micrograms of fragmented biotin-labeled cRNA was prepared from 5 μg of total RNA and hybridized to the 3AspergDTU GeneChip (available from Affymetrix on request, order no. 520520F) according to the Affymetrix GeneChip Expression Analysis Technical Manual (30).
cRNA was quantified in a spectrophotometer (as above). cRNA quality was assessed by using a BioAnalyzer. A GeneChip Fluidics Station FS-400 (fluidics protocol FS450_001) and a GeneChip Scanner 3000 were used for hybridization and scanning.
The scanned probe array images (.DAT files) were converted into.CEL files by using the GeneChip Operating Software (Affymetrix).
Analysis of Transcriptome Data.
Affymetrix CEL-data files were preprocessed by using the statistical language and environment R (31) version 2.5. The probe intensities were normalized for background by using the robust multiarray average method (32) by using only perfect match (PM) probes. Normalization was performed subsequently by using the quantiles algorithm (33). Gene expression values were calculated from the PM probes with the medianpolish summary method (32). All statistical preprocessing methods were used by invoking them through the affy package (34).
Statistical analysis was applied to determine genes subject to differential transcriptional regulation. The limma package (35) was used to perform moderated Student's t tests between the two carbon sources for each of the three species. Empirical Bayesian statistics were used to moderate the standard errors within each gene and Benjamini–Hochberg's method (36) to adjust for multitesting. A cutoff value of adjusted P < 0.05 was set to assess statistical significance.
Array Design.
Initial probe design was done by using the OligoWiz 2.0 software (27, 28) from the CDS sequences of predicted ORFs from the genome sequences of A. nidulans FGSC A4 (5), A. niger ATCC 1015 (6), and A. oryzae RIB40 (8). For each gene, a maximum of 11 nonoverlapping perfect match probes were calculated by using the OligoWiz standard scoring of cross-hybridization, melting temperature, folding, position preference, and low complexity. A 3′ position preference for the probes was included in the computations. The probes were designed separately for each genome.
Pruning of the probe sequences to comply with Affymetrix recommendations was done by removing duplicate probe sequences and shortening probes that were not possible to synthesize in full length.
Also included on the chip were a number of Affymetrix standard controls, custom controls, an A. oryzae EST collection (courtesy of Novozymes), and probes for ORFs from the Streptomyces. coelicolor A3 (2, 29) genome.
Comparison of Protein Sequences.
The amino acid sequences of the predicted ORFs from each of the three genomes were compared with those of the two others by using blastp (37) with an e-value cutoff of 1e-30. For each protein query sequence that gave one or more positive hits, the best hit was selected based on score (a unidirectional best hit). Bidirectional best hits were found by comparing the lists of best hits for two species against each other and selecting genes where the best hits paired up, thus giving a conservative set of 1:1 homologues for all pairwise comparisons. Tridirectional best hits were found by comparing the lists of bidirectional hits for all comparisons, and selecting the genes that had a 1:1:1 relationship in all comparisons between all three species (see SI Fig. 4).
Detection of Conserved Motifs.
Conserved motifs were identified by using R 2.5 (31) with the cosmo package (38). Default settings were used with the following exceptions: a background Markov model was computed by using the intergenic regions from scaffold 1 of the A. niger ATCC genome sequence. Intergenic regions containing unknown bases (Ns) were pruned from the training set leaving 1.7 Mb in 1,214 sequences. The two-component-mixture model was used to search for conserved motifs. The maximum number of sites were increased to include all 102 sites. For all query sequences, 1,000 bp up-stream of the start codon of the gene was used, or, in the case of some A. niger genes, 1,000 bp upstream of the predicted transcription start. Only 120 bp was available of the AN4590 promoter.
P values were calculated as P(X≤n), with X being a Poisson-distributed stochastic variable with λ = 0.418 and n being the number of motifs found per kilobase. lambda was calculated as the number of the conserved motif found per kilobase of the intergenic training set.
Acknowledgments.
We thank Rasmus Wernersson for help with the addition of the data to the OligoWiz database, Michael L. Nielsen for revision of the manuscript, Scott E. Baker for good scientific discussions, and Lene Christiansen and Tina Johansen for indispensable technical assistance. This work was partially funded by Chr. Hansen, Novozymes, and the Lundbeck Foundation (funding of array design); Danish Research Agency for Technology and Production (M.R.A. and L.L.); Novozymes Bioprocess Academy and the Technical University of Denmark (W.V.); National Council of Research Conacyt-Mexico and the Mexican Ministry of Public Education SEP (M.P.S.); and the Danish Research Council for Technology and Production Sciences (G.P.). We thank the reviewers for their comments, which significantly improved the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The normalized and raw data values reported in this paper have been deposited with Gene Express Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9298).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709964105/DC1.
References
- 1.Baker SE, Bennett JW. The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods. In: Osmani SA, Goldman GH, editors. Boca Raton, FL: CRC Press; 2008. pp. 3–13. [Google Scholar]
- 2.Hjort CM. In: Genetically Engineered Food. Heller KJ, editor. Wennheim, Germany: Wiley, VCH; 2003. pp. 86–99. [Google Scholar]
- 3.Nevalainen KH, Te'o VSJ, Bergquist P. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 2005;23:468–474. doi: 10.1016/j.tibtech.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Pontecorvo G. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 5.Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 6.Baker S. Aspergillus niger genomics: past, present and into the future. Med Mycol. 2006;44:S17–S21. doi: 10.1080/13693780600921037. [DOI] [PubMed] [Google Scholar]
- 7.Pel HJ, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 8.Machida M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 9.Jørgensen TR, et al. Glucose uptake and growth of glucose-limited chemostat cultures of Aspergillus niger and a disruptant lacking MstA, a high-affinity glucose transporter. Microbiology. 2007;153:1963–1973. doi: 10.1099/mic.0.2006/005090-0. [DOI] [PubMed] [Google Scholar]
- 10.van Peij N, Visser J, de Graaff L. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 11.Marui J, et al. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet Biol. 2002;35:157–169. doi: 10.1006/fgbi.2001.1321. [DOI] [PubMed] [Google Scholar]
- 12.Hasper AA, Visser J, de Graaf LH. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol Microbiol. 2000;36:193–200. doi: 10.1046/j.1365-2958.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 13.van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukagoshi N, Kobayashi T, Kato M. Regulation of the amylolytic and (hemi-)cellulolytic genes in aspergilli. J Gen Appl Microbiol. 2001;47:1–19. doi: 10.2323/jgam.47.1. [DOI] [PubMed] [Google Scholar]
- 15.vanKuyk PA, de Groot MJL, Ruijter GJG, de Vries RP, Visser J. The Aspergillus niger d-xululose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on d-xylose and l-arabinose. Eur J Biochem. 2001;268:5414–5423. doi: 10.1046/j.0014-2956.2001.02482.x. [DOI] [PubMed] [Google Scholar]
- 16.de Vries RP, van de Vondervoort P, Hendriks L, van de Belt M, Visser J. Regulation of the α-glucuronidase-encoding gene (aguA) from Aspergillus niger. Mol Genet Genom. 2002;268:96–102. doi: 10.1007/s00438-002-0729-7. [DOI] [PubMed] [Google Scholar]
- 17.de Vries RP, Visser J. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl Environ Microbiol. 1996;65:5500–5503. doi: 10.1128/aem.65.12.5500-5503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries RP, et al. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Aspergillus niger. Appl Environ Microbiol. 1996;65:2453–2460. doi: 10.1128/aem.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gielkens MM, Dekkers E, Visser J, de Graaff LH. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1996;65:4340–4345. doi: 10.1128/aem.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaf LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marui J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N. Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett. 2002;528:279–282. doi: 10.1016/s0014-5793(02)03328-8. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Gonzalez JA, de Graaff LH, Visser J, Ramón D. Molecular cloning and expression in Saccharomyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl Environ Microbiol. 1996;62:2179–2182. doi: 10.1128/aem.62.6.2179-2182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacCabe AP, Orejas M, Pérez-González JA, Ramón D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacCabe AP, Fernández-Espinar MT, de Graaff LH, Visser J, Ramón D. Identification, isolation and sequence of the Aspergillus nidulans xlnC gene encoding the 34-kDa xylanase. Gene. 1996;175:29–33. doi: 10.1016/0378-1119(96)00116-3. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen RC. In: The Chemistry of Solid Wood. Rowell RM, editor. Washington, DC: Am Chem Soc; 1984. pp. 57–126. [Google Scholar]
- 26.Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen H, Wernersson R, Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wernersson R, Nielsen H. OligoWiz 2.0–integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley S, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 30.Affymetrix . Santa Clara, CA: Affymetrix; 2007. GeneChip Expression Analysis Technical Manual, P/N 702232. Rev. 2. [Google Scholar]
- 31.R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 32.Irizarry R, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad B, Irizarry R, Åstrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Gautier L, Cope L, Bolstad B, Irizarry R. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 35.Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 37.McGinnis S, Madden T. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bembom O, Keles S, van der Laan M. Supervised detection of conserved motifs in DNA sequences with cosmo. Stat Appl Genet Mol Biol. 2007;6 doi: 10.2202/1544-6115.1260. Article 8. [DOI] [PubMed] [Google Scholar]