Abstract
We have characterized the adaptations of Helicobacter pylori to a rarely captured event in the evolution of its impact on host biology—the transition from chronic atrophic gastritis (ChAG) to gastric adenocarcinoma—and defined the impact of these adaptations on an intriguing but poorly characterized interaction between this bacterium and gastric epithelial stem cells. Bacterial isolates were obtained from a single human host colonized with a single dominant strain before and after his progression from ChAG to gastric adenocarcinoma during a 4-year interval. Draft genome assemblies were generated from two isolates, one ChAG-associated, the other cancer-associated. The cancer-associated strain was less fit in a gnotobiotic transgenic mouse model of human ChAG and better able to establish itself within a mouse gastric epithelial progenitor-derived cell line (mGEP) that supports bacterial attachment. GeneChip-based comparisons of the transcriptomes of mGEPs and a control mouse gastric epithelial cell line revealed that, upon infection, the cancer-associated strain regulates expression of GEP-associated signaling and metabolic pathways, and tumor suppressor genes associated with development of gastric cancer in humans, in a manner distinct from the ChAG-associated isolate. The effects on GEP metabolic pathways, some of which were confirmed in gnotobiotic mice, together with observed changes in the bacterial transcriptome are predicted to support aspects of an endosymbiosis between this microbe and gastric stem cells. These results provide insights about how H. pylori may adapt to and influence stem cell biology and how its intracellular residency could contribute to gastric tumorigenesis.
Keywords: microbial pathogenesis, intracellular bacteria, genome sequencing, functional genomics, gnotobiotic mice
Helicobacter pylori is a Gram-negative microaerophilic bacterium that typically establishes a lifelong infection in humans after acquisition during childhood (1). Virtually all hosts develop gastritis but only a small subset progress to chronic atrophic gastritis (ChAG), a condition characterized by loss of acid-producing parietal cells. ChAG is, in certain instances, an antecedent to gastric adenocarcinoma (2).
Using a germ-free transgenic mouse model of ChAG (Atp4b-tox176), we have found that clinical isolates of H. pylori can establish themselves in dividing and nondividing gastric stem cells (3). tox176 mice have a genetically engineered ablation of their parietal cells: ablation leads to an expanded population of gastric epithelial progenitors (GEPs) that express NeuAcα2,3Galβ1,4-containing glycan receptors for a subset of H. pylori adhesins (4). In this mouse model, H. pylori invasion appears to be GEP-specific. For example, internalization is not seen in NeuAcα2,3Galβ1,4-expressing differentiated mucus-producing gastric pit cells descended from GEPs.
Intracellular H. pylori have also been detected by transmission electron microscopy in preneoplastic and neoplastic gastric epithelium recovered by endoscopic biopsy of the stomachs of infected humans (5). Although typically viewed as an extracellular pathogen, a “liaison” between H. pylori and GEPs may have implications for tumorigenesis. Gastric stem cells are long-lived. The cancer–stem cell hypothesis argues for a stem-cell origin of many tumor types (6). The concept that a bacterium, classified as a class I carcinogen (7), can adapt to an intracellular stem cell habitat suggests a potentially novel form of initiation of tumorigenesis and begs the question of how bacterial and host cells establish and coevolve their relationship.
To explore this issue, we have sequenced two H. pylori isolates recovered from the same patient as he progressed from ChAG to gastric adenocarcinoma. These ChAG- and cancer-associated isolates exhibited markedly different fitness in the gnotobiotic transgenic mouse model of ChAG. The interactions of the two isolates with a newly developed cell line derived from mouse gastric stem cells were subsequently characterized by whole-genome transcriptional profiling of host and bacterium, and the results were used to guide analysis of interactions with GEPs in vivo. Isolate-specific molecular correlates of the liaison between GEPs and H. pylori are described and discussed.
Results
A Population-Based Endoscopy Study Yields H. pylori Isolates from a Single Patient Who Progressed from ChAG to Adenocarcinoma.
The original purpose of the now completed “Kalixanda” study, conducted in two northern Swedish cities, was to design an endoscopic survey of the upper gastrointestinal tract for a general adult population and explore whether it biases symptom reporting (8). Biopsies for histology and H. pylori culture were obtained from defined regions of the stomach, including its middle and distal portions (corpus and antrum, respectively). A subset of subjects (n = 289), randomly selected from those who underwent endoscopy (n = 1001) and were not treated for H. pylori infection, were reendoscoped four years later. According to then-prevailing Swedish medical practices and the human studies committee-approved study protocol, ChAG diagnosed at the initial endoscopy was not considered to be an indication for H. pylori eradication.
A single male patient progressed from ChAG (moderate atrophy grade 2) to non-cardia adenocarcinoma (intestinal type) during the four-year interval. Eighteen H. pylori isolates were recovered from the first endoscopy, and 50 isolates were recovered four years later. A random amplified polymorphic DNA (RAPD) screen did not detect any chromosomal differences between the isolates (data not shown). Because this result suggests initial infection with a single strain, we randomly picked one of the 18 ChAG-associated isolates from the first endoscopy (designated Kx1) and one of the adenocarcinoma-associated isolates from the second endoscopy (Kx2) for further characterization. Both isolates were from the corpus region of the patient's stomach.
Analysis of the ChAG-Associated Kx1 and Cancer-Associated Kx2 Genomes.
To examine how H. pylori adapts to changes in the gastric ecosystem associated with the transition from ChAG to adenocarcinoma, we sequenced the Kx1 and Kx2 genomes and compared them to the three reported, finished H. pylori genomes: strain HPAG1 from a patient with ChAG (9), strain 26695 from a patient with gastritis (10), and strain J99 from a patient with duodenal ulcer disease (11).
Kx1 and Kx2 lacked 31, 64, and 24 genes that were present in HPAG1, 26695, and J99, respectively; 91% of these genes are hypothetical or encode components of restriction–modification systems [supporting information (SI) Table 1]. Three genes were present in at least one of the HPAG1, 26695, and J99 strains and in the Kx1 isolate, but not in the cancer-associated Kx2 strain; one of these (JHP1117) has high homology (90% sequence identity) to a gene encoding an exported protein that, when mutated, results in reduced H. pylori motility and the absence of flagella (12); the other two are hypothetical proteins (SI Table 2). No genes were identified as being present in the Kx2, HPAG1, 26695, and J99 strains and absent in Kx1. No genes were identified as being under positive selection between Kx1 and Kx2. (See SI Table 3 and SI Text for details of five genes found to be under positive selection in the Kx1 and Kx2 versus the other three strains.)
Phenotypic Differences Between Kx1 and Kx2 in Vivo.
To test whether Kx1 and Kx2 exhibit differences in their fitness, we inoculated one or the other strain into germ-free tox176 mice (single gavage of 2 × 108 CFU per strain per mouse) and killed the animals 4 weeks and 6 months later (n = 4–6 mice per bacterial strain per time point sampled). tox176 transgenic mice express an attenuated diphtheria toxin A fragment (tox176) under the control of a parietal cell-specific promoter (nucleotides −1,035 to +24 of mouse Atp4b), resulting in loss of differentiated members of the two epithelial lineages that are also lost in humans with ChAG (parietal cells and digestive enzyme-secreting zymogenic cells). As noted in the introduction, ablation of these lineages is associated with a concomitant amplification of GEPs (3, 13, 14). Germ-free rather than conventionally raised animals were used because the latter have a complex gastric microbiota, reflecting their coprophagy, that could confound interpretation of the phenotypes produced by infection with different H. pylori isolates.
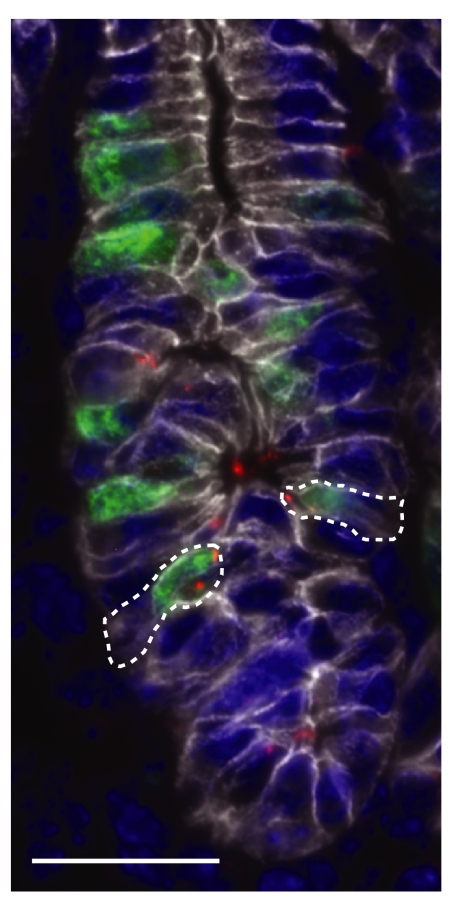
The ChAG-associated Kx1 isolate colonized the stomachs of all germ-free mice at both time points (10 of 10 animals; range, 3 × 102 to 2 × 105 CFUs per stomach). In contrast, only 2 of 10 animals contained viable Kx2 cells, one killed after 4 weeks and one killed after 6 months (SI Fig. 4A). Immunohistochemistry revealed foci of Kx1 and Kx2 associated with dividing GEPs (Fig. 1). This statistically significant difference in colonization efficiency (Fisher's exact test, P < 0.001) indicated that the Kx2 cancer strain has a reduced fitness in a ChAG-like gastric environment.
Fig. 1.
Association of H. pylori with gastric epithelial progenitors. In this multilabel immunohistochemical analysis, bacteria (red) are seen in association with BrdU-labeled GEPs (green) in the epithelium of a gnotobiotic transgenic tox176 mouse colonized with the cancer-associated Kx2 isolate. The epithelial marker, E-cadherin, appears white, and nuclei are stained blue with DAPI. Dashed lines indicate Kx2-associated progenitor cells that were targeted by n-LCM for transcriptional assays (see SI Text for details). (Scale bar: 25 μm.)
To verify these results and determine whether there was a differential adaptive immune response to the ChAG and cancer isolates, we collected sera from all mice at their time of killing. ELISA was used to measure levels of IgM, IgG1, and IgG2b isotypes that react with Kx1 and Kx2. Age-matched germ-free tox176 mice served as negative controls. Both Kx1 and Kx2 elicited a comparable IgM response (SI Fig. 4B), indicating that both had been able to initially colonize the stomachs of tox176 hosts. However, a significantly greater induction of IgG1 and IgG2b occurred in Kx1- compared with Kx2-exposed mice at the 6-month time point (P < 0.05, Student's t test; SI Fig. 4 C and D). The reduced IgG1 and IgG2b levels are consistent with the reduced ability of Kx2 to persist in the ChAG-like gastric ecosystem of tox176 mice.
Establishing a Mouse Gastric Epithelial Progenitor (mGEP) Cell Line.
We subsequently turned to the question of whether the clinical isolates evoke different transcriptional responses in GEPs. To do so, we generated a GEP-like cell line that could be cultured and infected under defined (controlled) conditions. GEPs were amplified in FVB/N transgenic mice by expressing SV40 TAg under the control of the same Atbp4 regulatory elements used to direct expression of tox176. This resulted in progenitor entrapment/amplification because of a block in the differentiation of oligo-potential preparietal progenitors to mature parietal cells. A cloned cell line was then established from the stomachs of these mice (see refs. 14 and 15 and Materials and Methods for details).
Several lines of evidence indicated that this mGEP cell line resembled GEPs present in the intact mouse stomach. First, transmission EM studies established that mGEPs had morphologic features of preparietal cell progenitors present in the isthmal stem cell niche of gastric units that line the stomachs of normal (nontransgenic) conventionally raised and germ-free mice (SI Fig. 5A). Second, immunohistochemical studies established that mGEPs express readily detectable levels of a number of biomarkers that are enriched in gastric stem cells in vivo (16) (e.g., SI Fig. 5 B and C). Third, an Ingenuity Pathways Analysis (IPA) (www.ingenuity.com)-based comparison of the transcriptomes of mGEPs (“present” calls by Affymetrix GeneChip software) and a control nonprogenitor Gastric Epithelial Cell (npGEC) line that expresses a temperature-sensitive SV40 TAg under the control of transcriptional regulatory elements from the IFN-gamma (Inf-γ) gene [shifting these cells to 37°C in the absence of Inf-γ, inactivates SV40 TAg (17)] revealed that the 978 transcripts uniquely represented in mGEPs are significantly enriched for a number of canonical signaling pathways (Wnt/β-catenin, G protein-coupled receptor, PI3K/Akt, Pdgf, Igf-1, Sapk/Jnk and integrin); these pathways are also enriched in laser capture microdissected (LCM) GEPs harvested from germ-free tox176 mice compared with their differentiated lineage descendants (16) (SI Tables 4 and 5).
Assessing the Impact of Kx1 and Kx2 on the Transcriptional Profile of mGEPs.
Low-passage-number mGEP cells were grown to 70% confluency and subsequently infected with the ChAG-associated Kx1 strain, or the cancer-associated Kx2 isolate for 24 h (the time point was selected based on a pilot time course study described in SI Fig. 6 and SI Text). In additional studies, we determined that there was no statistically significant difference in the number of Kx1 versus Kx2 cells that attached to mGEP cells (SI Fig. 7A). Both strains elicited similar levels of interleukin-6, a marker of NFkB activation by H. pylori (SI Fig. 7B, P = 0.22 in Student's t test).
We were unable to use gentamycin-protection assays to directly compare the relative efficiency of invasion of mGEPs by the Kx1 and Kx2 isolates because the latter isolate was a 1,000-fold more resistant to killing by this antibiotic (data not shown). Therefore, invasion was scored by using a multilabel immunohistochemical assay in which H. pylori antibodies labeled with one fluorescent tag were used to stain infected mGEPs before treatment with saponin, followed by addition of this cell-permeabilizing agent plus the same H. pylori antibody but labeled with a second tag. The results revealed no detectable intracellular bacteria after Kx1 infection, whereas the cancer-associated Kx2 strain was invasive (two independent experiments; SI Fig. 8).
We also infected the npGEC line with the ChAG-associated Kx1 and cancer-associated Kx2 isolates, and subsequently compared the responses of mGEPs and npGECs to Kx1 and Kx2 to identify progenitor-specific transcriptional responses differentially elicited upon infection with one or the other isolate (one GeneChip per infected cell line per bacterial strain per experiment; n = 3 replicate infections per experiment plus uninfected controls; total of 18 GeneChips).
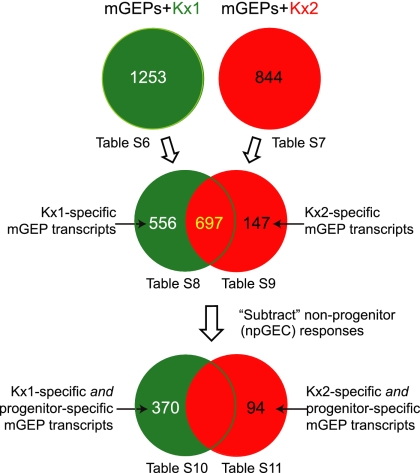
Twenty-four hours after exposure to Kx1, 1,253 mGEP transcripts were defined as being differentially up- or down-regulated compared with uninfected mGEP controls. [See Fig. 2 for experimental design and SI Table 6 for a list of genes that satisfied our selection criteria (defined in Materials and Methods), plus their fold-difference in expression.] Using the same selection criteria, a total of 844 transcripts were defined as significantly changed after infection with Kx2 (Fig. 2 and SI Table 7).
Fig. 2.
Flow-chart showing the approach used to define isolate-specific and progenitor-specific mGEP transcriptional responses to infection with the Kx1 and Kx2 isolates.
Components of the elicited mGEP transcriptional responses were then identified as being unique to infection with the Kx1 ChAG isolate (556 transcripts; Fig. 2 and SI Table 8) versus infection with the cancer-associated Kx2 isolate (147 transcripts; Fig. 2 and SI Table 9). By eliminating transcripts that were also differentially expressed in npGECs upon infection with Kx1 or Kx2, we reduced the list to 370 mGEP-specific transcripts that were significantly changed only upon infection with the Kx1 strain (Fig. 2; SI Table 10) and 94 mGEP-specific transcripts that were significantly changed only upon Kx2 infection (Fig. 2 and SI Table 11).
Signaling and metabolic pathways significantly over-represented in the mGEP- and Kx1- or mGEP- and Kx2-specific transcriptional response datasets were identified with IPA software. The results disclosed that the mGEP- and Kx1-specific response was significantly enriched in components of Ppar-, Tgf-β-, p38 Mapk-, and ephrin receptor-signaling pathways (SI Table 12). A number of the genes that were uniquely and differentially expressed upon infection with the Kx1 isolate (SI Table 10) are known to regulate the proliferative activity of stem cells and/or to be associated with gastrointestinal tumorigenesis. They include (i) ephrin receptor signaling pathway components [ephrin-B3 (induced 3.1-fold compared with uninfected mGEPs; regulates epithelial progenitor cell proliferation and migration in the intestine; ref. 18); ephrin receptor A2 (+2.4-fold) and the pathway's downstream Mapkkkk-4; (+3.5-fold)]; (ii) enzymes involved in extracellular matrix remodeling, such as matrix metalloproteinase 10 [+7.2-fold; up-regulated in intestinal adenomas (19)]; (iii) the Gata binding protein 4 transcription factor [+4.0-fold; expression levels and copy number reduced in gastric cancer (20)]; (iv) tumor suppressors Cdkn1a (+3.4-fold) and Kangai1 [+5.0-fold; Fig. 3; higher expression of Kangai1 correlates with a better prognosis in gastric cancers (21)]; and (v) ApoE [+14.1-fold; increased in gastric cancers compared with normal tissue (22)].
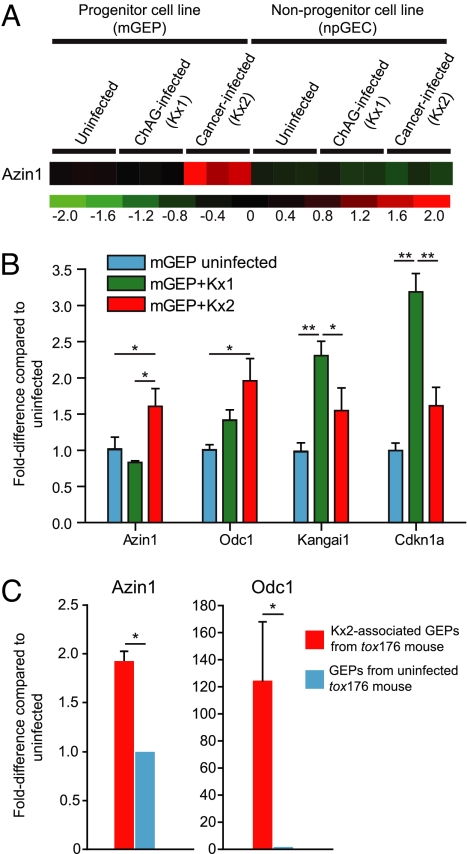
Fig. 3.
Kx2-associated up-regulation of antizyme inhibitor 1 expression in gastric epithelial progenitors in vitro and in vivo. (A) Mouse GeneChip analysis showing mGEP-specific up-regulation of antizyme inhibitor 1 (Azin1) after infection with the cancer-associated Kx2 strain. Three independent experiments for each condition were performed. Numbers at the bottom indicate standard deviations above (red) or below (green) the mean level of expression (black) of the Azin1 gene. (B) qRT-PCR of mGEP responses to Kx1 and Kx2 infection. *, P < 0.05; **, P < 0.01 in a two-sample two-tailed Student's t test with three biological replicates per condition for each mGEP transcript. (C) Up-regulation of Azin1 and Odc1 in gastric epithelial progenitor cells recovered by n-LCM from a tox176 mouse that had been colonized for 6 months with the Kx2 strain (mean values ± 1 SD for two to three technical replicates are plotted). *, P < 0.05.
The smaller set of genes whose expression was significantly changed in a mGEP-specific fashion upon infection with the cancer-associated Kx2 isolate (SI Table 11) included components of IPA pathways (SI Table 12) involved in amino acid metabolism (tryptophan, arginine, proline, and ornithine). Ornithine decarboxylase 1 (Odc1; EC 4.1.1.17), the rate limiting enzyme in polyamine biosynthesis (decarboxylates l-ornithine to putrescine), was up-regulated in mGEPs infected with Kx2 compared with uninfected cells. Interestingly, Odc is a known biomarker of gastric intestinal metaplasia (a precancerous condition), and increases in expression during the transition from ChAG to gastric adenocarcinoma (23). In addition, expression of antizyme inhibitor 1 (Azin1) was significantly increased in Kx2 versus Kx1-infected mGEPs (2.8-fold-difference in a direct comparison of these samples), whereas there was no significant difference in expression between Kx1 versus Kx2 infected npGECs (Fig. 3A). Antizymes are enzyme inhibitors that function at the posttranscriptional level (24). The antizyme inhibitor Azin1 increases the level and activity of Odc (25). Azin1 is highly up-regulated in human gastric cancer compared with adjacent normal gastric tissue (26).
The higher levels of Odc and Azin1 expression, documented by GeneChip studies of mGEPs after infection with the cancer-associated Kx2 versus ChAG-associated Kx1 strains, were validated by qRT-PCR (Fig. 3B). We also confirmed that Odc1 and Azin1 were up-regulated in GEPs in vivo after infection with the Kx2 isolate. To do so, we prepared serial gastric cryosections from the one tox176 mouse that had viable Kx2 after 6 months of infection (judged by CFUs of stomach homogenates). Navigated laser capture microdissection (n-LCM) was used to capture H. pylori-associated cycling progenitor cells marked by BrdU administered 90 min before killing (see SI Text). RNA was isolated from ≈30 n-LCM H. pylori- and BrdU-positive progenitor cells (Fig. 1). Using qRT-PCR, Odc1 and Azin1 mRNA levels in these cells were compared with levels in n-LCM BrdU-positive progenitors harvested from an age-matched germ-free tox176 control. The results revealed statistically significant elevations of both transcripts in Kx2-associated GEPs (Fig. 3C).
Genome-Wide Transcriptional Profiling of the ChAG- and Cancer-Associated Strains upon Infection of mGEPs.
To address the question of whether Kx1 and Kx2 genes were differentially expressed after a 24-h infection of the mGEP cell line, RNA was prepared from bacteria harvested from the same mGEP cultures that were used to define the host-cell transcriptome, and from Kx1 and Kx2 harvested after a 24-h incubation in cell culture medium alone (n = 2–3 parallel incubations with and without mGEP cells). cRNA targets were generated from each RNA preparation and hybridized to separate custom-designed Affymetrix GeneChips that contained probesets to 1,530 of the 1,536 chromosomal protein-coding genes present in the finished genome of the ChAG-associated HPAG1 strain (9). Control hybridizations of labeled Kx1 and Kx2 genomic DNA to these HPAG1 GeneChips (9) identified 218 probesets that were associated with “absent” calls in either of these two isolates; these probesets were eliminated from our subsequent analysis.
We focused on transcripts that were uniquely regulated by Kx1 or Kx2 upon infection of the mGEP cell line. Sixty-one bacterial genes (SI Table 13) were identified as being significantly up- or down-regulated in mGEP-infecting Kx1 bacteria compared with Kx1 cultured in medium alone [see Materials and Methods for selection criteria]. Seventeen of these genes (SI Table 14) were defined as uniquely changed in the ChAG-associated Kx1 isolate after infection of mGEPs (i.e., expression of these genes changed <1.2-fold in the comparison of Kx2 infection of mGEPs versus Kx2 cultured in medium alone; note that this approach circumvents potential strain-specific hybridization biases to probesets present in our HPAG1-based GeneChip).
The uniquely up-regulated transcripts in Kx1 included a gene encoding cysteine-rich protein A (hcpA), which induces release of Th1-biasing inflammatory cytokines (Inf-γ, interleukin-12, ref. 27) associated with evasion of the host immune response (28). This bacterial response is consistent with our observation that the Kx1 strain is able to better colonize germ-free tox176 mice than the Kx2 strain. Three other genes uniquely up-regulated in Kx1 are involved in cell wall and envelope biosynthesis: (i) HPAG1_0209 is a predicted lipopolysaccharide 1,2-glycosyltransferase [glycosyltransferase family 8 in the carbohydrate-active enzymes (CAZy) database (www.cazy.org)]; (ii) HPAG1_1094 (UDP-N-acetylglucosamine lipid transferase) also has predicted glycosyltransferase activity (CAZy family 28; EC 2.4.1.227); and (iii) HPAG1_1028 (spore coat polysaccharide biosynthesis protein C; EC 2.6.1.50) is a predicted aminotransferase. The differential regulation of these genes in the Kx1 versus Kx2 isolates could produce differences in their surface structures that may, in turn, affect the isolates' capacity to persist in the gastric environment of ChAG mice. Another Kx1-specific up-regulated gene, aliphatic amidase E (amiE), is involved, together with urease and arginase, in the production of ammonia and thus should help promote adaptation to a ChAG gastric environment that contains scattered foci of residual acid-producing parietal cells.
Using the same statistical criteria, we identified 101 Kx2 genes that were either up- or down-regulated with mGEP infection (SI Table 15); 44 of these changed <1.2-fold in the same direction upon infection with the Kx1 isolate and thus were classified as genes uniquely changed in the Kx2 cancer isolate after infection of the mGEP cell line (SI Table 16). Interestingly, this group included several genes encoding outer membrane proteins: hopG (3.2-fold up-regulated compared with Kx2 incubated without mGEPs), hofA (3.0-fold up-regulated), and hopZ [a known adhesin (ref. 29; 4.2-fold down-regulated)]. Two genes induced uniquely in Kx2 are involved in amino acid biosynthesis: homoserine kinase (HPAG1_0397; EC 2.7.1.39) and a predicted ketol-acid reductoisomerase (HPAG1_0334; EC 1.1.1.86). The latter enzyme is thought to be involved in valine and isoleucine biosynthesis. However, the pathways for valine and isoleucine production are incomplete in Helicobacter, producing auxotrophy for these amino acids (30). Thus, Kx2 may be better able to use host cell-derived substrates to fulfill this auxotrophic requirement than the Kx1 strain and as a result may be better equipped to function inside a GEP.
The tetA(P) multidrug efflux transporter (HPAG1_1104) is also uniquely up-regulated (3.4-fold) in the cancer-associated Kx2 isolate. Yeast two-hybrid screens have shown that the product of tetA(P) interacts with a urea transporter (UreI) (31). Interestingly, tetA(P) is under positive selection in both of the Kx isolates (SI Table 3). This observation and the finding that its expression in the cancer-associated strain is greater during infection of mGEPs compared with the Kx1 strain makes this gene a potential biomarker of bacterial adaptation to progression from ChAG to cancer.
Discussion
We have been able to characterize adaptations of H. pylori to a rarely captured event in the evolution of its effect on host biology: the transition from chronic atrophic gastritis (ChAG) to gastric adenocarcinoma. Our analysis focused on the interaction between this bacterium and gastric epithelial stem cells, based on the belief that such an interaction is worthy of investigation both from the standpoint of how bacteria are able to adapt to and influence gastric epithelial stem cell biology and how intracellular residency may contribute to initiation of gastric tumorigenesis.
The experimental design attempted to control several variables that could affect our ability to gain insights into the effects of the ChAG-to-cancer transition on bacterial genome microevolution, and the functional effects that observed genomic changes may have on the progenitor cell-bacterial interaction. First, RAPD analysis of 68 H. pylori isolates obtained from a single host before and after his progression from ChAG to gastric cancer suggested that by the time he developed ChAG, a single H. pylori strain dominated his gastric microbiota. Two isolates were then randomly selected from the panel of ChAG-associated isolates and cancer-associated isolates for deep draft genome sequencing. This approach was designed to control for the large interstrain variations that have been noted among H. pylori isolates obtained from different individuals (32) and to allow the characterization of genomic changes associated with disease progression. Second, phenotypic differences between the sequenced ChAG and cancer isolates were assayed in a gnotobiotic transgenic mouse model of ChAG, where the gastric microbiota and host genotype could be held constant, and in a mouse gastric epithelial progenitor cell line (mGEP) grown under well defined conditions that support attachment, internalization, and intracellular survival of H. pylori. Isolate-specific and progenitor-specific molecular phenotypes were identified from comparisons of infection-related changes in bacterial transcriptomes and the transcriptomes of the clonally derived mGEP cell line and a control mouse gastric epithelial cell line.
Several observations emerge from this study. The two isolates exhibit marked differences in fitness: The ChAG-associated Kx1 strain is a significantly better colonizer compared with the cancer-associated Kx2 strain. The IgG1 and IgG2b host response is reduced in Kx2-colonized mice, whereas the IgM response to both isolates is similar, indicating that, although both strains are able to initially colonize the stomach, Kx1 is more able to sustain a persistent population. The observed difference in fitness, documented at 4 weeks and 6 months after gavage of germ-free mice with the Kx1 and Kx2 strains, could reflect the adaptation of Kx1 to the patient's ChAG environment and changes in the selective pressures placed on Kx2 as cancer evolves.
Our studies also support the notion that the cancer-associated Kx2 strain is more adapted to an intracellular habitat than the ChAG-associated strain. Kx2 has a far greater capacity to invade mGEPs. The increased expression of bacterial ketol-acid reductoisomerase observed upon mGEP infection by the Kx2 compared with Kx1 strain suggests that Kx2 may be more capable of overcoming its auxotrophic requirements for valine and isoleucine by establishing residency within stem cells.
The cancer stem cell hypothesis argues for an adult stem cell origin of several types of tumors (6). Given the strong association between H. pylori infection and gastric cancer (7), it is intriguing to conceptualize the observed bacterial-progenitor cell interaction as a dynamic coevolving relationship that provides an intracellular microhabitat for H. pylori but also affects the risk for malignant transformation. Our comparative studies of the Kx1 and Kx2 strains support this concept. The cancer-strain induces higher levels of expression of ornithine decarboxylase (Odc1) and antizyme inhibitor (Azin1) (an inhibitor of ornithine decarboxylase's inhibitor) in cultured mGEPs and up-regulates these transcripts in GEPs recovered by navigated laser capture microdissection from the stomachs of colonized gnotobiotic transgenic tox176 mice. There are no proteins encoded in the five sequenced H. pylori genomes (Kx1, Kx2, HPAG1, J99, and 26695) that have homologies to Odc, agmatinase or agmatine deiminase, which produce putrescine. However, all have a spermidine synthase (EC 2.5.1.16), which uses putrescine and S-adenosylmethioninamine as substrates to produce spermidine. Polyamines such as spermidine stimulate growth of bacteria (33) and a variety of cultured mammalian cell types and human tumors (34). Thus, regulation of polyamine availability by intracellular H. pylori could affect the proliferative status of GEPs. Intriguingly, Odc exhibits increased expression in gastric adenocarcinoma compared with tissue without metaplasia (23).
Additional factors likely affect the outcome of this intimate association between H. pylori and gastric epithelial stem cells. Compared with the ChAG-associated Kx1 strain, Kx2 infection of mGEPs results in lower levels of expression of several tumor suppressors, including Kangai1: Its lower expression correlates with poorer prognosis in human gastric cancers (21). At the same time, Kx2 produces relatively lower levels of expression of components of the ephrin-receptor signaling pathway—a pathway that affects the proliferative status of gut stem cells (18). The distinct regulation of sets of genes involved in mGEP cellular proliferation by the ChAG-associated Kx1 and cancer-associated Kx2 isolate lends credence to the notion that the two strains, isolated 4 years apart as their human host progressed from ChAG to cancer, may make different contributions to initiation versus progression and maintenance of tumorigenesis.
Materials and Methods
mGEPs (passages 4–7) were seeded at 4 × 105 cells per T75 flask (Corning) in RPMI medium 1640 (Sigma) supplemented with 10% FBS (Hyclone) and grown for 3 days at 37°C to 70% confluency. Medium was then removed. H. pylori, which had been grown to log phase in Brucella broth (BB) plus 5% FBS and 1% IsoVitaleX (Becton Dickinson; adjusted to pH 7.0), then spun down, washed in PBS, and resuspended in fresh cell culture medium, was added to the flasks (3–6 × 108 bacteria per flask). After a 24-h incubation at 37°C, supernatants were collected for IL-6 assays (mouse IL-6 Eli-pair kit; Diaclone Research). Residual medium and nonattached bacteria were washed off from mGEPs, using PBS, and the cells were harvested by trypsinization [5 min at 37°C; 0.05% trypsin (Sigma) and 0.02% EDTA]. After neutralization with ice-cold medium and a PBS wash, cells were flash-frozen in liquid nitrogen. The infection protocol used for npGECs was identical, but the cells were shifted from 33°C to 37°C for 3 days before infection to inactivate the ts-SV40 TAg.
Uninfected mGEPs or npGEC controls were incubated with fresh RPMI medium 1640 (supplemented with 10% FBS) for 24 h. Bacteria-alone controls were incubated for 24 h in cell culture medium under the same conditions and underwent identical treatments as the infected samples. Experiments were performed in triplicate (mGEP and npGEC profiling) or in duplicate or triplicate for studies involving transcriptional profiling of H. pylori.
Total cellular RNA was extracted by using the RNeasy miniprep kit (Qiagen). Mammalian cRNA targets were generated as follows: First-strand cDNA synthesis was primed from total cellular RNA, using oligod(T)24 with an attached anti-sense T7 promoter. In vitro transcription was then performed with T7 RNA polymerase and biotinylated NTPs. The biotinylated and fragmented cRNAs were hybridized to Moe430_2 Affymetrix Gene Chips for mouse cell profiling. Raw data were scaled with the Affymetrix Microarray Suite software, Version 5.0, to an intensity of 500 and subsequently analyzed with dChip (35). The selection criteria for differentially expressed genes were: (i) fold-difference in expression (lower bound 90% confidence interval) > 2, (ii) P < 0.05 (t test), (iii) 100% present call in the condition with the higher expression, (iv) minimal absolute intensity difference > 100, and (v) false-discovery rate < 0.5%.
Bacterial biotinylated cDNA targets were generated from RNA samples, using random primers (Invitrogen) and hybridized to custom-designed HPAG1 Affymetrix Gene Chips (9) according to the manufacturer's recommendations. An uninfected mGEP cRNA control was used to correct for signal derived from mammalian cellular RNA. Raw GeneChip data were scaled to 1,500, based on the intensity of spiked-in transcripts (Affymetrix). The scaled signal from the “mGEP-only” GeneChip was subtracted from the infected samples and the resulting intensities (infected and bacteria-only samples) were normalized in dChip.
Additional experimental details can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Maria Karlsson, David O'Donnell, Janaki Guruge, Sabrina Wagoner, Daniel Peterson, Jung Oh, and Helene Kling-Backhed for valuable suggestions during the course of this work; Justin Sonnenburg, Eric Martens, and Andrew Goodman for critically reading the manuscript; Jian Xu, Lucinda Fulton, and our other colleagues at the Washington University Genome Sequencing Center for assistance with genome sequencing; Laura Kyro for graphics help; and Uma Krishna (Vanderbilt University, Nashville, TN) and Richard Peek, Jr. (Vanderbilt University) for generously providing the npGEC cell line. This work was supported by National Institutes of Health Grants DK58529, DK63483, DK52574, AI068362, and DK64540; the Swedish Cancer Foundation; and the Terry Fox Funds for Cancer Research. M.G. is a member of the Washington University Medical Scientist Training Program, which is funded by National Institutes of Health Grant GM07200.
Footnotes
The authors declare no conflict of interest.
Data deposition: The GeneChip data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE10261 and GSE10262). Whole Genome Shotgun projects have been deposited in NNA Data Bank of Japan, European Molecular Biology Laboratory Nucleotide Sequence Database, and GenBank [accession nos. ABJO00000000 (H. pylori Kx1) and ABJP00000000 (H. pylori Kx2)]. Genome assemblies used in this paper are the first versions [accession nos. ABJO01000000 (Kx1) and ABJP01000000 (Kx2)]. The raw sequence data used in this paper have been deposited in the GenBank Short Read Archive [accession nos. SRA000264 (Kx1) and SRA000265 (Kx2)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0800668105/DC1.
References
- 1.Mitchell HM, et al. A low rate of re-infection following effective therapy against Helicobacter pylori in a developing nation (China). Gastroenterology. 1998;114:256–261. doi: 10.1016/s0016-5085(98)70475-5. [DOI] [PubMed] [Google Scholar]
- 2.Ohata H, et al. Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. Int J Cancer. 2004;109:138–143. doi: 10.1002/ijc.11680. [DOI] [PubMed] [Google Scholar]
- 3.Oh JD, Karam S, Gordon JI. Intracellular Helicobacter pylori in gastric epithelial progenitors. Proc Natl Acad Sci USA. 2005;102:5186–5191. doi: 10.1073/pnas.0407657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syder AJ, et al. Helicobacter pylori attaches to NeuAc alpha 2,3Gal beta 1,4 glycoconjugates produced in the stomach of transgenic mice lacking parietal cells. Mol Cell. 1999;3:263–274. doi: 10.1016/s1097-2765(00)80454-2. [DOI] [PubMed] [Google Scholar]
- 5.Necchi V, et al. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology. 2007;132:1009–1023. doi: 10.1053/j.gastro.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 8.Aro P, et al. Valid symptom reporting at upper endoscopy in a random sample of the Swedish adult general population: the Kalixanda study. Scand J Gastroenterol. 2004;39:1280–1288. doi: 10.1080/00365520410008141. [DOI] [PubMed] [Google Scholar]
- 9.Oh JD, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: Evolution during disease progression. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 11.Alm RA, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 12.Odenbreit S, Till M, Haas R. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol Microbiol. 1996;20:361–373. doi: 10.1111/j.1365-2958.1996.tb02623.x. [DOI] [PubMed] [Google Scholar]
- 13.Mills JC, Andersson N, Hong CV, Stappenbeck TS, Gordon JI. Molecular characterization of mouse gastric epithelial progenitor cells. Proc Natl Acad Sci USA. 2002;99:14819–14824. doi: 10.1073/pnas.192574799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syder AJ, et al. The impact of parietal cells on Helicobacter pylori tropism and host pathology: an analysis using gnotobiotic normal and transgenic mice. Proc Natl Acad Sci USA. 2003;100:3467–3472. doi: 10.1073/pnas.0230380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farook VS, Alkhalaf M, Karam SM. Establishment of a gastric epithelial progenitor cell line from a transgenic mouse expressing the Simian virus 40 large T antigen gene in the parietal cell lineage. Cell Proliferation. 2008 doi: 10.1111/j.1365-2184.2008.00522.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakis M, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- 17.Franco AT, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmberg J, et al. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 19.Martinez C, Bhattacharya S, Freeman T, Churchman M, Ilyas M. Expression profiling of murine intestinal adenomas reveals early deregulation of multiple matrix metalloproteinase (Mmp) genes. J Pathol. 2005;206:100–110. doi: 10.1002/path.1755. [DOI] [PubMed] [Google Scholar]
- 20.Yang S, et al. Identification of genes with correlated patterns of variations in DNA copy number and gene expression level in gastric cancer. Genomics. 2007;89:451–459. doi: 10.1016/j.ygeno.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Lee HS, Lee HK, Kim HS, Yang HK, Kim WH. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol. 2003;200:39–46. doi: 10.1002/path.1288. [DOI] [PubMed] [Google Scholar]
- 22.Oue N, et al. Gene expression profile of gastric carcinoma: Identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397–2405. doi: 10.1158/0008-5472.can-03-3514. [DOI] [PubMed] [Google Scholar]
- 23.Miao XP, et al. Expression of ornithine decarboxylase in precancerous and cancerous gastric lesions. World J Gastroenterol. 2007;13:2867–2871. doi: 10.3748/wjg.v13.i20.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 25.Mangold U. Antizyme inhibitor: Mysterious modulator of cell proliferation. Cell Mol Life Sci. 2006;63:2095–2101. doi: 10.1007/s00018-005-5583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung MH, et al. Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics. 2000;69:281–286. doi: 10.1006/geno.2000.6338. [DOI] [PubMed] [Google Scholar]
- 27.Deml L, et al. Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent. Infect Immun. 2005;73:4732–4742. doi: 10.1128/IAI.73.8.4732-4742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Elios MM, et al. T helper 1 effector cells specific for Helicobacter pylori in the gastric antrum of patients with peptic ulcer disease. J Immunol. 1997;158:962–967. [PubMed] [Google Scholar]
- 29.Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doig P, et al. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rain JC, et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 32.Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida M, et al. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
- 34.Gerner EW, Meyskens FL. Polyamines and cancer: Old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.