Abstract
Nitrogen is quantitatively the most important nutrient that plants acquire from the soil. It is well established that plant roots take up nitrogen compounds of low molecular mass, including ammonium, nitrate, and amino acids. However, in the soil of natural ecosystems, nitrogen occurs predominantly as proteins. This complex organic form of nitrogen is considered to be not directly available to plants. We examined the long-held view that plants depend on specialized symbioses with fungi (mycorrhizas) to access soil protein and studied the woody heathland plant Hakea actites and the herbaceous model plant Arabidopsis thaliana, which do not form mycorrhizas. We show that both species can use protein as a nitrogen source for growth without assistance from other organisms. We identified two mechanisms by which roots access protein. Roots exude proteolytic enzymes that digest protein at the root surface and possibly in the apoplast of the root cortex. Intact protein also was taken up into root cells most likely via endocytosis. These findings change our view of the spectrum of nitrogen sources that plants can access and challenge the current paradigm that plants rely on microbes and soil fauna for the breakdown of organic matter.
Keywords: nitrogen uptake, organic nitrogen, plant nutrition, plant roots, soil protein
Soil organic matter contains nitrogen predominantly as protein, which is considered a nitrogen source exclusively for microbes and animals (1, 2). Despite the importance of protein in soils, little research has been carried out to elucidate the role of complex organic nitrogen as a nitrogen source for plants. The current view is that in ecosystems where the rate of microbial mineralization is slow, such as in boreal forests and heathlands, woody plants rely on ecto- or ericoid mycorrhizal fungal symbioses to break down soil protein, whereas in ecosystems with high microbial activity and mineralization rates, such as grasslands and tropical forests, herbaceous and woody plants use mostly inorganic nitrogen (2–4). However, there is conflicting evidence about the roles and interactions between microbes and plants for converting and accessing nitrogen in soils (1).
Plants take up organic nitrogen compounds of low molecular mass, including amino acids and possibly di- and tripeptides, via membrane transporters into root cells (5). Amino acids are a nitrogen source for plants in natural ecosystems and agricultural systems (6–10), but peptides and proteins have received less attention as potential nitrogen sources for plants. There is limited knowledge about peptides in soils, and protein is considered to be not directly accessible to plants. Previously, it was shown that heathland and forest plants can use protein as a nitrogen source when grown in axenic culture with fungal symbionts, but not when grown without fungi (2–6, 11, 12).
To investigate the possibility of protein utilization by nonmycorrhizal plant species, we studied a woody and a herbaceous plant species from contrasting ecosystems. We hypothesized that Hakea actites is able to access soil protein because (i) soil in its heathland habitat is protein-rich and inorganic nitrogen-poor (13), (ii) most other heathland plants have mycorrhizal symbioses and/or form symbioses with N2-fixing microbes (13), and (iii) Hakea forms cluster roots that have a role in the acquisition of organic nitrogen (14). We further hypothesized that Arabidopsis thaliana is unable to use protein as a nitrogen source because it does not form cluster roots and grows in ruderal habitats that typically contain inorganic nitrogen. With the aid of fluorescent proteins, we show that both plant species, in the absence of microbes, can use protein as a nitrogen source.
Results
Axenically Cultivated H. actites and A. thaliana Use Externally Supplied Protein as a Nitrogen Source for Growth.
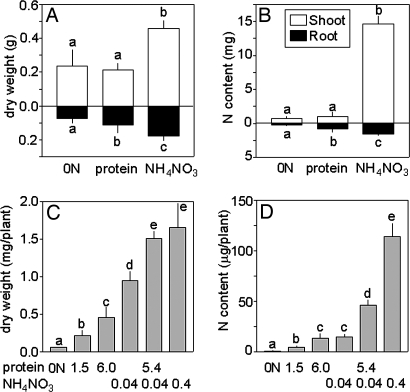
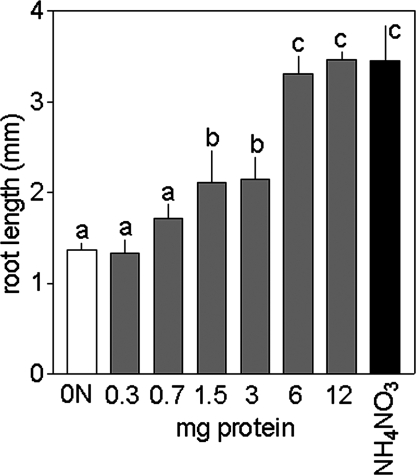
Grown with protein as the sole nitrogen source, Hakea seedlings produced significantly more root biomass and had a greater nitrogen content in roots than plants grown without nitrogen (Fig. 1 A and B). Shoot biomass and shoot nitrogen content, as well as total plant biomass and nitrogen content, were similar in Hakea grown without nitrogen or with protein, and best shoot and root growth was observed with inorganic nitrogen (Fig. 1 A and B). Arabidopsis grown with protein (1.5 or 6 mg BSA per ml of growth medium) as the sole nitrogen source had significantly greater dry weight and nitrogen content than plants grown without nitrogen (Fig. 1 C and D). Arabidopsis grown with a low amount of inorganic nitrogen (0.04 mg NH4NO3 per ml of growth medium) produced more dry weight but had similar nitrogen content as plants grown with 6 mg BSA per ml growth medium. Arabidopsis supplied with a mixture of protein and a low amount of inorganic nitrogen (5.4 mg BSA per ml and 0.04 mg NH4NO3 per ml growth medium) grew significantly better than plants grown with either nitrogen source individually and produced the same dry weight as plants grown with a high amount of inorganic nitrogen (0.4 mg nitrogen per ml growth medium) (Figs. 1 C and D and 2). Greater concentrations of protein in the growth medium led to concentration-dependent increases in root length in Arabidopsis (Fig. 3). Thus, protein as the sole source of nitrogen stimulated root growth in both Hakea and Arabidopsis.
Fig. 1.
Axenic H. actites and A. thaliana use external protein for growth. (A and B) Root and shoot dry weight and nitrogen content of Hakea seedlings grown without nitrogen, with protein, or with inorganic nitrogen; 30 mg of nitrogen was supplied to each plant as protein or inorganic nitrogen (17 mM protein nitrogen as BSA or 17 mM inorganic nitrogen as NH4NO3). (C and D) Arabidopsis dry weight and nitrogen content grown without nitrogen or with nitrogen supplied as protein (1.5 or 6 mg BSA per ml), inorganic nitrogen (0.04 or 0.4 mg NH4NO3 per ml), or protein and inorganic nitrogen combined (5.4 mg BSA per ml and 0.04 mg NH4NO3 per ml). Bars represent averages and SD of five to eight Hakea plants and 7–10 plates with ≈80 Arabidopsis plants per plate (see also Fig. 2). Different letters indicate significant differences at P < 0.05 (Hakea) and P < 0.01 or P < 0.001 (Arabidopsis) (ANOVA, Neuman–Keuls post hoc test; data were log-transformed before analysis to account for different variances between groups).
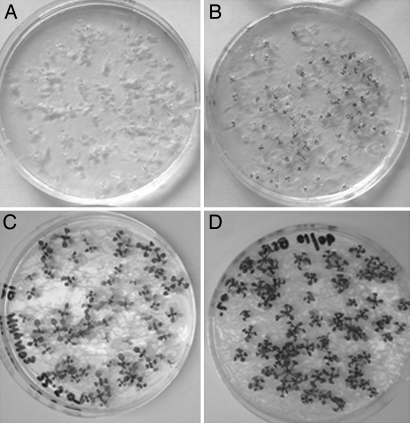
Fig. 2.
Protein and low inorganic nitrogen in combination supported better growth of Arabidopsis than protein or low inorganic nitrogen alone. (A) No nitrogen added. (B) Protein only (6 mg BSA per ml). (C) Inorganic nitrogen only (0.04 mg NH4NO3 per ml). (D) Protein plus inorganic nitrogen (5.4 mg BSA per ml and 0.04 mg NH4NO3 per ml).
Fig. 3.
Root length of Arabidopsis increased in response to increasing protein levels in growth medium. Bars represent averages and SD of two to four plates with ≈20 Arabidopsis plants per plate. Treatments included no nitrogen added to growth medium, protein added (0.3–12 mg BSA per ml), or inorganic nitrogen (0.4 mg NH4NO3 per ml). Different letters indicate significant differences at P < 0.001 (ANOVA, Neuman–Keuls post hoc test).
Roots Have Proteolytic Activity.
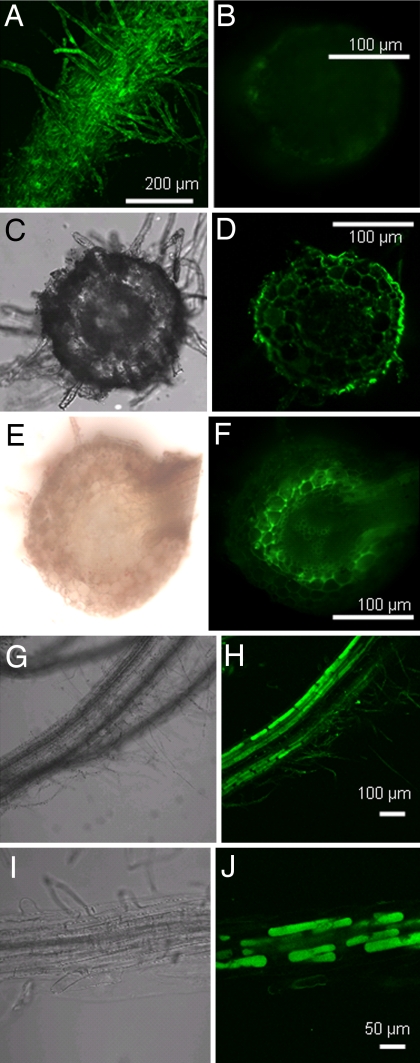
We used a fluorescent protein substrate to examine whether axenic Hakea and Arabidopsis roots exhibit proteolytic activity. The protein–chromophore complex (DQ green BSA) fluoresces upon proteolysis. The diameter of BSA (without the chromophore) is ≈6 nm, and intact BSA does not pass through cell wall pores that have a diameter of ≈4 nm (15). Fluorescence was observed at the root surface of Hakea, indicating that proteases at the root surface cleaved the protein (Fig. 4A). Fluorescence also was observed in the outer and inner root cortex of Hakea roots after incubating roots for 1.5 and 24 h, respectively (Fig. 4 D and F). No fluorescence was observed in the outer cortex of Hakea after 24 h of incubation most likely because substrate depletion prevented further movement of fluorescent peptides into the apoplast because only small amounts of protein were detected in the incubation solution 3 h after commencing the experiment. The results show that proteolysis occurred at the root surface of Hakea, but do not rule out the possibility that it continues throughout the cortical apoplast. Distinct fluorescence was associated with root cortex cells of Arabidopsis (Fig. 4 H and J).
Fig. 4.
Protease activity and protein fragments in roots of axenic Hakea and Arabidopsis after incubation with protein–chromophore complex (DQ green BSA) that fluoresces upon proteolytic degradation. (A) Protease activity resulted in fluorescence at the surface of Hakea cluster roots after 1.5 h of incubation. (B) Root cross-section of negative control with no protein–chromophore complex added. (D and F) Hakea roots incubated with protein–chromophore complex for 1.5 h (D) and 24 h (F) and cross-sectioned. (H and J) Arabidopsis roots incubated for 6 h. (C, E, G, and I) Bright-field images of D, F, H, and J, respectively. Images in B, E, and F were taken with a fluorescence microscope; all others were taken with a confocal microscope.
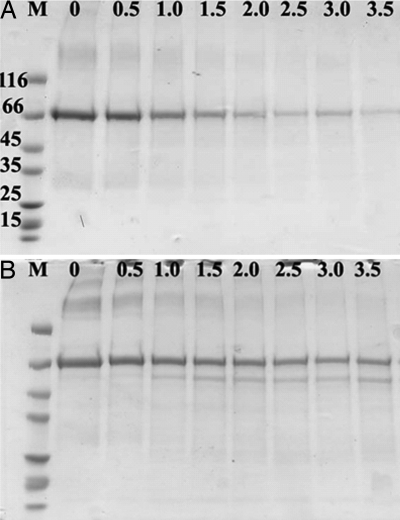
The fate of protein in the liquid medium also was monitored and visualized with gel electrophoresis. Protein content of the incubation solution was strongly reduced over a 3.5-h period in the presence of Hakea roots (Fig. 5A). No protein-degradation products were detected in the incubation solution, indicating that protein was removed as intact protein from the solution, that protein breakdown occurred but that smaller peptides were not visible, or that smaller peptides were taken up or otherwise associated with the roots. In Arabidopsis, the concentration of initial protein in the incubation solution of roots was reduced after 3.5 h incubation, and a peptide of ≈50 kDa increased in abundance over time, indicative of protein degradation (Fig. 5B).
Fig. 5.
Proteolysis and/or depletion of BSA as observed by analyzing protein solution incubated with H. actites (A) and A. thaliana (B) roots. The presence of protein in the incubation solution of intact Hakea roots is strongly reduced over the course of 3.5 h. No peptides of lower molecular mass were observed in the incubation solution of Hakea. The incubation solution of Arabidopsis roots contained decreasing amounts of BSA but also a smaller protein fragment that increased over time. Protein solution was supplied to the root as 50 μg BSA per ml. Numbers on the left side identify peptide standards of 15–116 kDa (BSA has a molecular mass of 66 kDa); numbers across columns indicate hours of incubation.
Root Cells Take Up Protein.
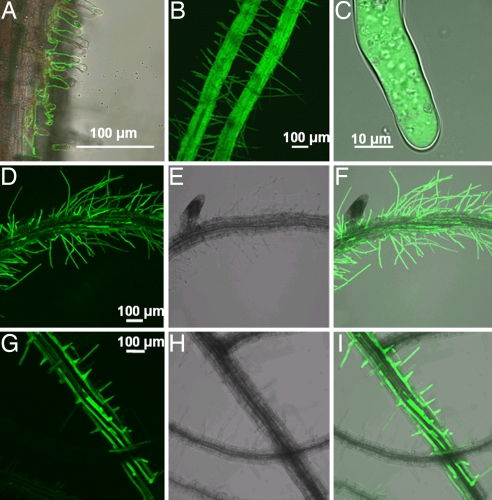
The absorption of intact protein by roots was investigated by incubating intact roots of axenic Hakea and Arabidopsis in liquid medium with GFP. GFP was observed on the surface of Hakea and Arabidopsis roots (Fig. 6A, B, F, and I) and inside intact root hairs (Fig. 6C), where it was associated with cytoplasmic streaming [supporting information (SI) Movie 1]. GFP was observed in root cortex cells (Fig. 6 F and I), providing further evidence that intact GFP entered root cells. Acquisition of intact protein as shown by GFP fluorescence was not observed in newly formed lateral roots that lacked root hairs (data not shown), suggesting that protein uptake depends largely on the presence of root hairs in Hakea and Arabidopsis.
Fig. 6.
Roots of axenic Hakea and Arabidopsis seedlings incubated with GFP show intact protein associated with root surfaces, root hairs, and root cortex cells. (A) GFP was associated with Hakea root hairs. (B) The whole root surface of Arabidopsis. GFP also was observed inside Arabidopsis root hairs and cortex cells. (C, F, and I) SI Movie 1 shows cytoplasmic streaming in C. (E and H) Bright-field images of D and G, respectively, and combined in F and I. Fluorescent images were taken with a confocal microscope.
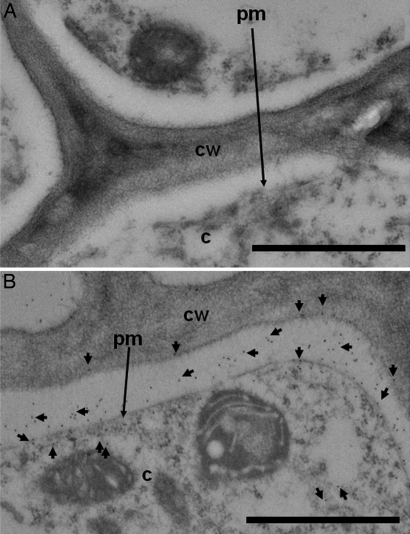
These findings were further supported by the localization of GPF in Hakea roots by using immunogold labeling for EM. Gold particles indicative of intact GFP and/or its cleaved fragments were found in apoplast and cytoplasm of root cortex cells (Fig. 7B).
Fig. 7.
EM of Hakea root-transverse sections show GFP in apoplast and root cortex cells. (A and B) Roots were incubated without GFP (A, negative control) and with GFP (B) and probed with immunogold labeled anti-GFP. GFP was detected in the apoplast, cell wall (cw), and cytoplasm (c) of root cortex cells. Gold labeling is marked with short arrows. The plasma membrane (pm) also is indicated. (Scale bars: 1 μm.)
Discussion
This article provides evidence that plants can assimilate protein without assistance from soil organisms. Whether the ability to use protein as a nitrogen source is limited to nonmycorrhizal plant species or is more widespread in the plant kingdom has to be established. In the experimental systems used here, protein did not support plant growth to the same extent as inorganic nitrogen sources. The slower growth of plants supplied with protein alone suggests that there may be metabolic bottlenecks associated with protein catabolism in the absence of inorganic nitrogen sources. In natural conditions, soil contains a combination of organic and inorganic nitrogen forms (16), and the addition of inorganic nitrogen significantly increased the ability of Arabidopsis to use protein as a nitrogen source. Our study shows that protein as a sole nitrogen source does not support plant growth to the same extent as inorganic nitrogen, but that protein can supplement plant nitrogen demand. The interaction between protein and inorganic nitrogen use by plants should be a focus of future investigations. Contrary to our initial hypothesis, Hakea seedlings did not have a greater capacity than Arabidopsis to use protein as a sole nitrogen source. However, the early growth of Hakea seedlings occurs after fire when inorganic nitrogen is more abundant (13), and the ability to use protein could improve in older plants. We excluded microbes from our study, and future research should address the capacity of plants to directly use protein in the presence of microbes. The techniques presented here may be useful in this respect.
We identified two mechanisms by which Hakea and Arabidopsis access protein. First, root-derived proteases break down protein. A smaller protein (≈50 kDa) was generated in the incubation solution of Arabidopsis roots when roots were supplied with a larger protein (66 kDa), and a protein–chromophore complex was cleaved by root-derived proteases of Hakea and Arabidopsis.
The Arabidopsis genome encodes 828 proteases (http://merops.sanger.ac.uk) (17), and secreted proteases have been detected in the apoplast of several plant species including Arabidopsis (18, 19). Cluster roots of H. undulata exude acid phosphatases (20), and evidence for root-secreted proteases has been recently reported in several crop and wild species (21). Digestive glands of carnivorous plant Nepenthes alata also are known to secrete aspartic proteinases into the liquid of the pitcher (22). Our results confirm the notion that cluster roots of Hakea are active in enzyme exudation (20, 23) and show that exudation of enzymes is not restricted to cluster roots. Root-derived proteases should now be isolated and characterized to determine the contribution of root proteases to protein breakdown in the rhizosphere. In an evolutionary context, such a strategy would only be advantageous if nitrogen gain from accessing protein outweighed nitrogen loss in the form of proteases exuded from roots or, alternatively, if protein breakdown products played important signaling roles in the rhizosphere.
The second mechanism of protein acquisition observed was the uptake of intact protein. Although the uptake of protein into roots has not been considered previously, integrated endocytotic and secretory networks have been described in tip-growing root hairs (24). It is likely that protein enters root hairs via endocytosis and that root cells subsequently catabolize the acquired protein, but other possibilities, including membrane transport, cannot be ruled out. Protein uptake was visualized with GFP that entered into root hair cells and root cortex cells. In Hakea, the presence of GFP was analyzed with immunogold labeling that confirmed the presence of GFP in apoplast and cells of the root cortex. Immunogold labeling does not allow distinguishing between intact or degraded protein because only small peptides (more than five amino acids) are required to selectively bind the GFP antibody. We did not study whether in planta proteins or large peptides are transported through roots via the symplastic pathway, but it is well documented that proteins can move cell to cell through plasmodesmata (25). In animals, so-called TAP transporters import degraded protein, 6–59 amino acids in length, into the lumen of the endoplasmic reticulum (26), but GFP is considerably larger (238 amino acids) (27). The Arabidopsis genome encodes three TAP-like members of this transporter subfamily, but their function has not been established (5). It appears likely that, apart from the suggested uptake of protein via endocytosis, the protein of a sufficiently small size to move through the root apoplast could be degraded by apoplastic proteases. Evidence for this suggestion exists because in Arabidopsis a serine-protease was detected that is exclusively localized in intercellular space (19).
The ability of Hakea and Arabidopsis to digest and take up intact protein without assistance from microbes has far-reaching implications. Microbes and soil fauna have been considered crucial for converting soil protein into nitrogen compounds of low molecular mass, and microbes compete with plants for soil nitrogen (1). It is not well understood how nonmycorrhizal plant species like Hakea and Arabidopsis compete for nitrogen with mycorrhizal plants, but root-derived proteases may function similarly to proteases of ecto- and ericoid mycorrhizal fungi (2, 12). Our initial hypothesis was that Hakea uses specialized cluster roots for acquisition of protein similar to what has been described for other soil nutrients (23). However, Arabidopsis lacks cluster roots, but also is able to degrade, take up, and assimilate protein. Digestion and uptake of protein may be widespread in the plant kingdom and may be crucially important for the 10% of plant species that do not form mycorrhizal symbioses.
Taken together, the findings change our view of nitrogen cycling in soils and question the concept of the soil microbial loop, the cycling of nitrogen and carbon between soil and microbial pools (28, 29). Previously observed effects of plants on nitrogen cycling in soils (28) may be due to the ability of plants to be actively involved in the turnover of soil organic matter. Our study is a step toward a broader view of plant nitrogen relations that also may offer opportunities for developing sustainable agriculture based on organic nitrogen sources (30).
Materials and Methods
Axenic Plant Culture.
Surface-sterilized H. actites seeds were grown individually in sterile polycarbonate containers with 20 g of vermiculite and 125 ml of liquid growth medium (14) because H. actites does not grow well in agar culture and does not produce cluster roots. Seedlings were grown without added nitrogen, with protein, or with inorganic nitrogen. Both nitrogen treatments received 30 mg of nitrogen (17 mM nitrogen in nutrient solution) per container. The protein treatment received 0.24 mg high-purity protein nitrogen per ml liquid growth medium (≈99% purity BSA, 193.2 mg of BSA per container; Sigma–Aldrich). To eliminate any minor contamination, BSA was solubilized in sterile distilled water, sterile filtered (0.22 μm; Millipore), and twice subjected to dialysis at 4°C against distilled water (1:100 vol/vol) for 12 h each time. The dialysis tubing (Spectra/Por; Spectrum Laboratories) has a nominal molecular weight cutoff of 25 kDa to remove traces of nitrogen ions, amino acids, or small peptides. The resulting protein solution was analyzed for ammonium and amino acid by liquid chromatography (Acquity, UPLC; Waters) equipped with a BEH C18 1.7 μm 2.1 × 50 mm column and tunable UV detector at 254 nm. Protein solution (40 μl) was mixed with 120 μl of borate buffer (Waters) and 40 μl of Acqutag-derivatizing reagent (Waters). No ammonium or amino acids were detected (detection limit 100 pmoles/ml). After dialysis, the solution was sterile filtered (0.22-μm filter) and added to the growth medium. The inorganic nitrogen treatment received 0.24 mg of inorganic nitrogen per ml (85.7 mg NH4NO3 per container). Plants were cultivated in a growth cabinet (30°C, 60% humidity, 1,000 μmol per m2/s light intensity, 18-h/6-h day/night). At 12 weeks, verified axenic plants were analyzed for dry weight and nitrogen content. Visibly contaminated containers were discarded throughout the experiment. To verify sterility, samples of vermiculite and growth solution were taken from each container at 6 and 12 weeks in a laminar air flow and plated on LB nutrient agar. If no microbial growth occurred after 7 days of incubation at 30°C, containers were considered axenic. Phosphorus was supplied at 5 μM because this concentration does not induce cluster root formation in H. actites with adequate nitrogen supply (14). Here Hakea produced cluster roots in all treatments. Additional axenic plants grown without nitrogen addition to the nutrient solution were incubated with fluorescing proteins GFP or DQ green BSA (Molecular Probes).
Surface-sterilized seeds of A. thaliana Columbia were sown on Petri dishes (80 seeds per dish) with 25 ml of nitrogen-free Murashige and Skoog (MS) (31) basal salt solution (M0529; Sigma–Aldrich) supplemented with 10 g of sucrose, 3 mM CaCl2, 1.5 mM MgSO4·7H2O, 1.25 mM KH2PO4, and 0.3% phytagel (pH 5.3) (Phytotechnologies). Nitrogen was added as protein (1.5 or 6 mg of BSA per ml growth medium, 16.1 or 64.4 mM protein nitrogen), inorganic nitrogen (0.04 or 0.4 mg of NH4NO3 per ml, 1 mM or 10 mM inorganic nitrogen), or a combination of protein and inorganic nitrogen (5.4 mg of BSA per ml and 0.04 mg NH4NO3 per ml). Plants were incubated in a cold room for 3 days and then transferred to a growth room (21°C, 16-h/8-h day/night, 150 μmol per m2/s). GFP or DQ green BSA was applied to additional plates of axenic plants grown with inorganic nitrogen (0.4 mg of NH4NO3 per ml).
Axenic Hakea seedlings were separated into roots and shoots while entire Arabidopsis plants were rinsed and cleaned three times in 0.5 mM CaCl2 to remove nitrogen from plant surfaces. Plants were dried at 60°C for 2 days, weighed, homogenized, and analyzed for nitrogen content with a flash combustion elemental analyzer (Thermo Finnigan EA 1112 series; CE Instruments).
Root Length Measurement.
Twenty sterile A. thaliana seeds were sown per plate on nitrogen-free MS medium amended with 0, 0.75, 3, and 12 mg/ml BSA or inorganic nitrogen (0.4 mg of NH4NO3/ml). Plates were kept in a cold room for 3 days and then transferred and placed vertically in a growth cabinet with 21°C, 16-h/8-h day/night, 150 μmol per m2/s. Root length was measured 11 days after sowing.
Proteolysis of BSA and Analysis on SDS/PAGE.
Axenic H. actites seedling were grown in sterile polycarbonate containers without nitrogen (see Axenic Plant Culture above). A. thaliana plants were grown axenically on N-free MS media amended with 5 mM NH4NO3 (see Axenic Plant Culture). After 8 (Hakea) and 5 (Arabidopsis) weeks, plants were carefully removed from containers and incubated in sterile inorganic N-free nutrient solution with 50 mg BSA/ml for 3.5 h in a laminar flow. Samples of the incubation solution were taken every 30 min. Protein was concentrated with TCA, resuspended, and loaded on SDS/10% PAGE. Gels were stained with Coomassie brilliant blue R-250.
Fluorescence Imaging.
Proteolysis releases protein fragments of DQ green BSA that contain dequenched fluorophores to emit green fluorescence. Hakea roots were incubated for 1.5 or 24 h in 50 μg/ml DQ green BSA. Root cross-sections were washed with growth medium, and images were taken with a fluorescence microscope (Eclipse E600; Nikon) at excitation and emission wave lengths of ≈505 nm and ≈515 nm, respectively, or a confocal microscope (see Confocal Microscopy). Axenic Arabidopsis roots were incubated in 50 μg/ml DQ green BSA for 6 h and imaged with a confocal microscope.
GFP was expressed from proviral vectors in tobacco leaves (32) and purified by using anion exchange chromatography. Protease inhibitors (5 μg/ml leupeptine, 5 μg/ml aprotinin, pepstatin 5 μg/ml, and 5 mM PMSF) were added to purified GFP solution (50 μg of GFP per ml) to limit degradation of GFP by root-derived proteases. Axenic roots of Hakea and Arabidopsis were incubated for 2 h in GFP solution and washed with PBS buffer before confocal microscopy.
Confocal Microscopy.
A Zeiss LSM501 Meta (Carl Zeiss) confocal laser scanning microscope was used with 10× dry and 20× water-immersion objectives, as well as ×40 and ×60 oil-immersion objectives. GFP and DQ green BSA were visualized by excitation with an argon laser at 488 nm and detection with a 505–530 nm band-path filter.
Immunocytochemistry.
Hakea root tissues were incubated in the presence or absence (negative control) of GFP solution for 4 h. Freshly excised tissues were fixed in 8% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer (pH 6.8) and stored at 4°C until further processing. Roots were washed three times in 0.1 M phosphate buffer. Root samples of ≈200-μm diameter were cut behind root caps. Tissues were dehydrated for 1 h in a graded series of ethanol solution (30%, 50%, 70%, and 100%) at progressively lowered temperatures. Tissues were then embedded in Lowicryl HM20 resin. The UV-polymerization step was performed at −50°C and 20°C for 48 h each. Tissue samples were sectioned with a diamond knife to 60-nm thickness and incubated with anti-GFP (Invitrogen), diluted in PBS at 1:100 for 30 min. PBS contains as a blocking agent 0.2% fish skin gelatin, 0.2% BSA, and 20 mM glycine. Tissue for negative controls was not incubated in GFP solution. Tissue was incubated in protein A/gold diluted in PBS/FBG (diluted 1:60) for 30 min. Sections were examined on a transmission electron microscope (JEOL 1010; JEOL Limited) at 80 kV, and images were recorded on a Megaview III digital camera (Soft Imaging Systems).
Supplementary Material
Acknowledgments.
We thank S. R. Mudge, A. L. Rae, and M. Jackson for suggestions and Icon Genetics for proviral vectors. This work was supported by Australian Research Council (ARC) Discovery Grant DP0557010 (to D.R. and S.S.) and the ARC Centre of Excellence for Integrative Legume Research (B.J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712078105/DC1.
References
- 1.Kaye JP, Hart SC. Competition for nitrogen between plants and soil microorganisms. Trends Ecol Evol. 1997;12:139–143. doi: 10.1016/s0169-5347(97)01001-x. [DOI] [PubMed] [Google Scholar]
- 2.Read DJ. Mycorrhizas in ecosystems. Experientia. 1991;47:376–391. [Google Scholar]
- 3.Smith SE, Read DJ. Mycorrhizal Symbiosis. London: Academic; 1997. [Google Scholar]
- 4.Schmidt S, Handley LL, Sangtiean T. Effects of nitrogen source and ectomycorrhizal association on growth and delta δ15N of two subtropical Eucalyptus species from contrasting ecosystems. Funct Plant Biol. 2006;33:367–379. doi: 10.1071/FP05260. [DOI] [PubMed] [Google Scholar]
- 5.Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Chapin FS, Moilanen L, Kielland K. Preferential use of organic nitrogen for growth by a non-mycorrhizal arctic sedge. Nature. 1993;361:150–153. [Google Scholar]
- 7.Kielland K, McFarland J, Olson K. Amino acid uptake in deciduous and coniferous taiga ecosystems. Plant Soil. 2006;288:297–307. [Google Scholar]
- 8.Näsholm T, Huss-Danell K, Högberg P. Uptake of organic nitrogen in the field by four agriculturally important plant species. Ecology. 2000;81:1155–1161. [Google Scholar]
- 9.Schmidt S, Stewart GR. Glycine metabolism by plant roots and its occurrence in Australian plant communities. Aust J Plant Physiol. 1999;26:253–264. [Google Scholar]
- 10.Näsholm T, et al. Boreal forest plants take up organic nitrogen. Nature. 1998;392:914–916. [Google Scholar]
- 11.Abuzinadah RA, Read DJ. The role of proteins in the nitrogen nutrition of Ectomycorrhizal plants. 4. The utilization of peptides by Birch (Betula pendula L) infected with different mycorrhizal fungi. New Phytol. 1989;112:55–60. [Google Scholar]
- 12.Bajwa R, Abuarghub S, Read DJ. The biology of mycorrhiza in the Ericaceae. 10. The utilization of proteins and the production of proteolytic-enzymes by the mycorrhizal endophyte and by mycorrhizal plants. New Phytol. 1985;101:469–486. doi: 10.1111/j.1469-8137.1985.tb02853.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt S, Stewart GR. Waterlogging and fire impacts on nitrogen availability and utilization in a subtropical wet heathland (wallum). Plant Cell Environ. 1997;20:1231–1241. [Google Scholar]
- 14.Schmidt S, Mason M, Sangtiean T, Stewart GR. Do cluster roots of Hakea actites (Proteaceae) acquire complex organic nitrogen? Plant Soil. 2003;248:157–165. [Google Scholar]
- 15.Carpita N, Sabularse D, Montezinos D, Delmer DP. Determination of the pore-size of cell-walls of living plant-cells. Science. 1979;205:1144–1147. doi: 10.1126/science.205.4411.1144. [DOI] [PubMed] [Google Scholar]
- 16.Chapin FS. New cog in the nitrogen-cycle. Nature. 1995;377:199–200. [Google Scholar]
- 17.Rawlings ND, Morton FR, Barrett AJ. MEROPS: The peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tornero P, Conejero V, Vera P. Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: Similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA. 1996;93:6332–6337. doi: 10.1073/pnas.93.13.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton JMU, Simpson DJ, Hyman SC, Ndimba BK, Slabas AR. Ara12 subtilisin-like protease from Arabidopsis thaliana: Purification, substrate specificity and tissue localization. Biochem J. 2003;370:57–67. doi: 10.1042/BJ20021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinkelaker B, Hengeler C, Marschner H. Distribution and function of proteoid roots and other root clusters. Bot Acta. 1995;108:183–200. [Google Scholar]
- 21.Godlewski M, Adamczyk B. The ability of plants to secrete proteases by roots. Plant Physiol Biochem. 2007;45:657–664. doi: 10.1016/j.plaphy.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 22.An CI, Fukusaki E, Kobayashi A. Aspartic proteinases are expressed in pitchers of the carnivorous plant Nepenthes alata Blanco. Planta. 2002;214:661–667. doi: 10.1007/s004250100665. [DOI] [PubMed] [Google Scholar]
- 23.Neumann G, Martinoia E. Cluster roots: An underground adaptation for survival in extreme environments. Trends Plants Sci. 2002;7:162–167. doi: 10.1016/s1360-1385(02)02241-0. [DOI] [PubMed] [Google Scholar]
- 24.Samaj J, Read ND, Volkmann D, Menzel D, Baluska F. The endocytic network in plants. Trends Cell Biol. 2005;15:425–433. doi: 10.1016/j.tcb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Lough TJ, Lucas WJ. Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- 26.Stacey G, Koh S, Granger C, Becker JM. Peptide transport in plants. Trends Plants Sci. 2002;7:257–263. doi: 10.1016/s1360-1385(02)02249-5. [DOI] [PubMed] [Google Scholar]
- 27.Tsien RY. The green florescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 28.Clarholm M. Interactions of bacteria, protozoa and plants leading to mineralization of soil-nitrogen. Soil Biol Biochem. 1985;17:181–187. [Google Scholar]
- 29.Bonkowski M. Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 2004;162:617–631. doi: 10.1111/j.1469-8137.2004.01066.x. [DOI] [PubMed] [Google Scholar]
- 30.Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. doi: 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- 31.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 32.Marillonnet S, et al. In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.