Abstract
Epigenetic regulation through chromatin is thought to play a critical role in the establishment and maintenance of pluripotency. Traditionally, antibody-based technologies were used to probe for specific posttranslational modifications (PTMs) present on histone tails, but these methods do not generally reveal the presence of multiple modifications on a single-histone tail (combinatorial codes). Here, we describe technology for the discovery and quantification of histone combinatorial codes that is based on chromatography and mass spectrometry. We applied this methodology to decipher 74 discrete combinatorial codes on the tail of histone H4 from human embryonic stem (ES) cells. Finally, we quantified the abundances of these codes as human ES cells undergo differentiation to reveal striking changes in methylation and acetylation patterns. For example, H4R3 methylation was observed only in the presence of H4K20 dimethylation; such context-specific patterning exemplifies the power of this technique.
Keywords: electron transfer dissociation, epigenetics, posttranslational modification, histone code, pluripotency
Pluripotency—the ability to differentiate into any specialized lineage—is the hallmark of embryonic stem (ES) cells and the basis for their experimental and therapeutic potential. The precise molecular mechanisms that define pluripotency remain elusive; however, a number of recent works suggest a central role for epigenetic regulation through chromatin (1–6). Either by recruiting or shielding certain factors, modifications on histone proteins modulate a gene's local environment and thereby regulate expression (7–12). Concerted changes in histone modification states occur during differentiation (13). For example, high levels of histone H3 and H4 acetylation are characteristic of pluripotent cells in mice and the abundance of these marks decreases during differentiation (14, 15). Methylation of R2, R17, and R26 of histone H3 by CARM1 also correlates with cell fate and potency (6). Cells with higher levels of methylation, at these residues, were enriched in the embryonic part of the blastocyst. Next, specific patterns of histone H3K4me3 and H3K27me3 are observed at promoter regions of genes that are regulated during differentiation (1, 2). Finally, demethylation of H3K27me3 is required for activation of certain HOX genes essential for proper development (16, 17). The demethylase responsible interacts directly with MLL 2/3 complexes, which methylate histone H3K4 (18). Taken together these experiments have shed new light on the power of epigenetic regulation within ES cells; however, the precise details and role(s) of such combinatorial PTM patterns remain largely unknown.
Technological limitations in our ability to discover and quantify combinatorial histone PTMs has, and continues to, present a major obstacle. Most of our knowledge of epigenetics has been derived by antibody-based approaches. Antibodies require a priori knowledge of individual modifications, are subject to epitope occlusion, and have difficulty distinguishing PTM patterns. Imagine the code as a complete sentence, antibodies can identify letters or sometimes words, but these words and letters lack context. Mass spectrometry-based (MS) sequencing methods, however, are rapidly evolving, have high sensitivity, and can identify and quantitate PTM patterns without a priori knowledge (19–29). Here, we have developed and applied a method for the discovery and quantification of histone H4 combinatorial codes that is based on chromatography and recently developed MS technology. First, intact histone H4 tails are chromatographically separated by using nanoflow high-performance liquid chromatography (nHPLC) wherein the eluate is sampled directly by either a hybrid linear quadrupole ion trap-orbitrap MS (orbitrap) or a linear quadrupole ion trap MS (QLT). The orbitrap records the mass of each eluting histone tail with extraordinary mass accuracy for assignment of overall PTM state, whereas the QLT employs electron transfer dissociation (ETD) to pinpoint the exact residue(s) carrying the individual PTMs. We and others have used a similar approach to study the modifications present on the N-terminal tail of histone H3 (20, 22, 30). Here, we apply this methodology to decipher 74 discrete combinatorial codes occurring on the intact tail of histone H4 from human ES cells. Finally, we quantified the abundances of these codes as human ES cells undergo differentiation, recorded striking changes in global methylation and acetylation patterns, and observed that methylation of H4R3 is only observed in the presence of H4K20 dimethylation.
Results
Cell Lines and Treatments.
Federally registered human ES cell lines H1, H7, and H9 and human fibroblasts IMR90 (a model for fully differentiated, noncancerous cells) were used in this study. Treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA), a potent differentiation agent, induced a rapid (within 24 h) epithelial–mesenchymal transition and a drastic change in ES cell morphology [supporting information (SI) Fig. 4]. In contrast to the control human ES cell sample, single cells are readily distinguished with nuclei that are smaller and darker; further, cells are spread throughout the growth surface rather than remaining in tight colonies. Fibroblast morphology was unchanged (data not shown). To determine on a molecular level whether cells had retained pluripotency, treated and control samples were immunostained by using an anti-Oct4 antibody and analyzed by flow cytometry (Oct4 is an established marker for pluripotency; SI Fig. 4). An anti-IgG antibody was included as a control for nonspecific binding. Untreated human ES cells stained positive for Oct4, whereas TPA-treated cells (72 h) showed no appreciable signal above the anti-IgG control.
Isoform Identification.
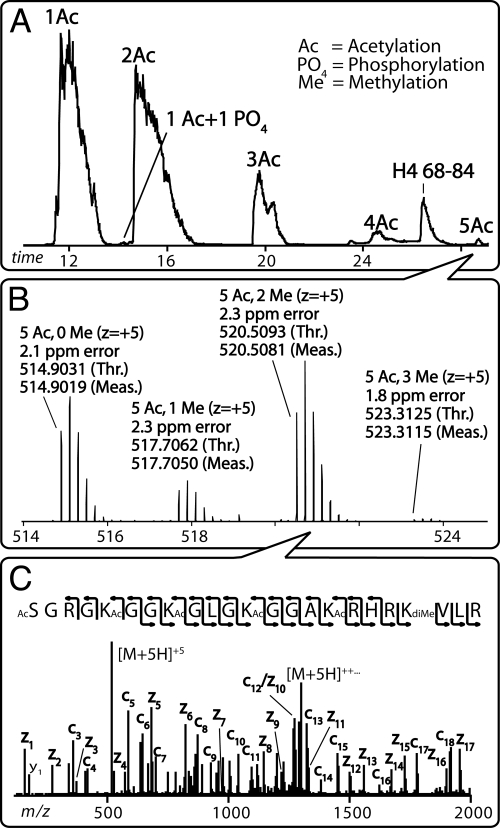
After isolation from cells and HPLC purification, harvested histone H4 was digested by using endoproteinase AspN and loaded onto a nanoflow reversed-phase capillary column (nHPLC) and gradient-eluted into either a linear ion trap-orbitrap hybrid MS (for accurate mass measurement) or an ETD-enabled linear ion trap MS (for PTM site localization). The orbitrap MS achieved high resolution and recorded the mass of the peptide/protein ion to within ≈3 ppm error. Such mass accuracy allowed for both the assignment of the various modified isoforms of H4 and for isoform quantification. Fig. 1A displays the chromatographic separation of the intact N-terminal tail of H4. Several distinctly modified forms of this 23-residue peptide were observed. The major species are mono-, di-, and triacetylated; however, tetra-, penta-, and nonacetylated versions were also detected. Note these chromatographic peaks are broad and seemingly unresolved. The peak broadening is not, however, a result of poor chromatography, instead it results from a number of different positional isoforms eluting within each major group. For example, the diacetylated peak contains peptides with varying location of acetylation (i.e., N terminus plus either K5, K8, K12, or K16).
Fig. 1.
Method for analysis of histone H4 N-terminal tail (residues 1–23) combinational codes. (A) Display of the nHPLC chromatogram and the associated N-terminal peptides. (B) Demonstration of the high mass accuracy mass spectra resulting from the pentaacetylated peak. This low-level peak displays four different isoforms: non-, mono-, di-, and trimethylated versions. (C) Depiction of ETD-MS/MS analysis of the dimethylated, pentaacetylated species. This spectrum reveals that K5, K8, K12, K16, and the N terminus are acetylated but K20 is dimethylated in this peptide.
As these peptides elute, their mass is recorded with high accuracy by using the orbitrap. Fig. 1B displays the peptide ion m/z values that comprise the low-level penta-acetylated peak. Even for this low-abundance isoform, four unique methylation states are observed: non-, mono-, di-, and trimethylated. The mass accuracy afforded by the orbitrap allowed us to distinguish precisely which modifications are present; for example, we could discriminate trimethylation from acetylation, a difference of 0.03638 Da. With this information we determined the overall number of histone H4 isomers, but had no knowledge of where each of these identified modifications reside or how many permutations within each isoform exist. To obtain this information we performed ETD MS/MS. Fig. 1C displays ETD MS/MS single-scan analysis of the penta-acetyl, dimethyl-containing H4 tail (≈500 ms per scan). From the near-complete fragmentation we localized acetylation on K5, K8, K12, and K16 combined with dimethylation of K20. This process was repeated for each isoform until the locations of all PTMs were assigned.
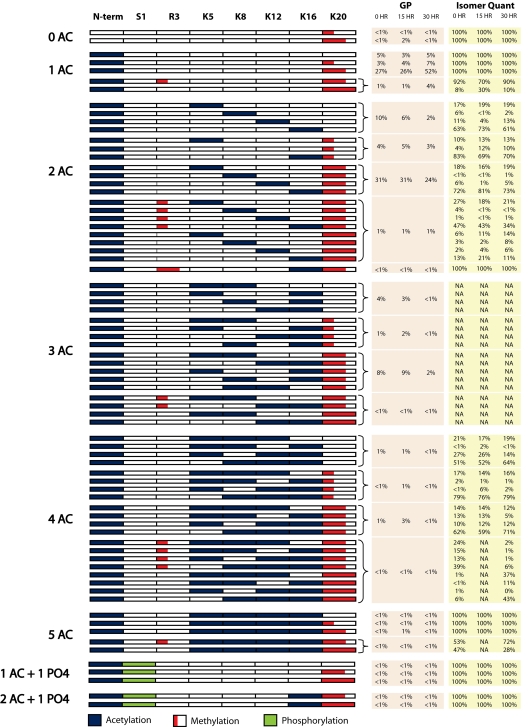
Using this approach, we characterized the histone H4 tail isoforms found in three human ES cell lines (H1, H7, and H9) and one somatic cell line (fibroblast, IMR90). From these data we have identified 74 unique combinatorial PTM codes occurring on histone H4. Fig. 2 presents a graphical display of these codes—to our knowledge it is the most comprehensive list of histone H4 isoforms compiled to date by nearly an order of magnitude (21). The modifications we identified include N-terminal acetylation, phosphorylation of S1, mono- or dimethylation of R3, acetylation of K5, K8, K12, and K16, and mono-, di-, or trimethylation of K20. All of these modifications were previously known to exist; the combinatorial PTM patterns they exhibited were not.
Fig. 2.
Map of the 74 histone H4 combinational codes detected in human ES cells (cell line H1). Bracketed isoforms are positional isomers that coeluted and the corresponding percentages indicate the amount of this whole set (i.e., the global isoform percentage, GP). Shown in the right-hand column (Isomer Quant) is the amount of each positional isomer for each of these subsets. For instance, four diacetylated tails were detected and these four forms constituted 10% of the H4 population. By use of ETD-MS/MS we assigned the percentages of the four forms as 17, 6, 11, and 63. This means that the H4 tail having both the N terminus and K16 acetylated constitutes ≈6.3% of the entire histone H4 population in control human ES cells (0 h). Also shown are the percentages of each form at the 15 and 30 h TPA treatment time points. NA indicates either that (i) that form was present in that particular sample at levels that were too low to acquire reliable ETD data or (ii), in the case of triacetylated forms, that the combinations of modifications make it mathematically impossible to quantify coeluting isomers. All triacetylated isoforms were sequenced manually.
Global Isoform Quantification.
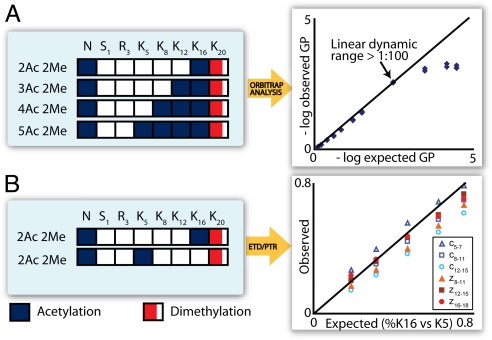
To compare histone H4 isoform populations between many different samples and cell lines we developed a label-free quantification method. Here, peak areas corresponding to individual histone H4 isoforms were determined and divided by the sum of the peak areas corresponding to all H4 isoforms, a methodology pioneered by Kelleher et al. (21). We refer to the resulting percentage as the global isoform percentage (GP). By using GPs it was possible to compare relative amounts of histone H4 isoforms in any number of different samples. Sometimes isomers having the same nominal mass, and thus m/z values, coeluted (e.g., diacetylated tails that vary only in the location of the acetyl groups) so that a single GP represents this entire population. In such cases, we used our ETD experiment to determine what percentage of each coeluting isomer was present (details below). The GP approach was validated by synthesizing histone H4 tails corresponding to the first 23 amino acids of histone H4, the same peptide observed after endoproteinase AspN digestion, that differed only in number of acetylated lysine residues (Fig. 3A). The peptides were then mixed in known ratios and analyzed as described earlier. From Fig. 3A we observe a linear response that correlates well (R2 = 0.9958 for ratios up to and including 1:100) to the expected with a linear dynamic range in excess of 1:100 and a dynamic range of ≈1:10,000 (e.g., one fmol of one form detected in the presence of 10 pmol, each, of three others). When mixed in ratios of 1:1 to 1:100 the average difference between observed and expected values was 3% (standard deviation = 2%). This window of error compares favorably with stable isotope-based quantification methods. For example, isobaric tagging reagents, iTRAQ, are reported to have an error <6%, with standard deviations <23% (31).
Fig. 3.
Quantification of histone H4 isoforms. (A) Four synthetic H4 tails varying in the number of acetylation sites were prepared, mixed in known ratios, and analyzed by MS. These data show that a linear response that matches closely with the theoretical (solid line). Three technical replicates of each mixture were performed resulting in three observed GPs for each expected GP. (B) Positional isomer quantification was validated by use of two synthetic tails having the same number of modifications, but only varying in placement. Here, the c- and z-type ion fragments generated by ETD are used to localize and quantify the relative amounts of each. Each data point represents the average of three or four consecutive ion ratios. Although there is a certain amount of systematic error as the ratio of K16Ac to K5Ac is increased, a good linearity is observed as the ratios are varied.
Analysis of GPs revealed that human ES cells from three different cell lines (H1, H7, and H9) were enriched in isoforms bearing activating marks (e.g., acetylation) when compared with somatic control cells (fibroblast, IMR90). Two of these human ES cell lines (H1 and H9) were treated with TPA to induce differentiation and the histone H4 isoforms were examined at variable TPA-treatment time points. As a control we repeated the same TPA treatment time course with fibroblast cells. Across all three human ES cell lines 20.0% (standard error = 1.0%) of the histone H4 population was hyperacetylated (3, 4, or 5 acetylations), whereas only 6.0% (standard error = 0.5%) of the H4 tails exhibited such marks in fibroblasts. Moreover, after 30 h of TPA treatment, the percentage of hyperacetylated histone H4 decreased to levels similar to those seen in the fibroblast samples—8.0% (standard error = 1.7%). Consistent with the acetylation results, all three human ES cell lines exhibited high levels of isoforms that lacked silencing marks (e.g., K20 dimethylation, SI Fig. 5B). Unmethylated isoforms constituted 19.5% (standard error = 0.5%) of the histone H4 population among human ES cells, compared with only 2.09% (standard error = 0.05%) in the fibroblast samples. After TPA treatment the amount of these forms was considerably reduced and closely matched that found in fibroblasts. As the human ES cells progressed through differentiation the abundance of the unmethylated forms continuously decreased and by 75 h of TPA treatment only 0.40% (standard error = 0.01%) of the histone H4 population remained unmethylated. A concomitant increase in the di- and trimethylated isoform of histone H4 was observed (SI Fig. 5 D and E). SI Fig. 5A displays the expression of Oct4, a pluripotency marker, as measured by quantitative PCR during TPA treatment. It is intriguing that the methylation of H4K20 occurs on a time scale that correlates precisely with the decision to exit the pluripotent state.
Quantification of Coeluting, Positional Isomers.
For coeluting isoforms who share the same nominal mass, but only differ in the placement of modifications (i.e., positional isomers), we adapted an MS/MS-based quantification strategy (SI Fig. 6) to accommodate ETD-MS/MS spectra of chromatographed peptides (21). The relative ratios of the unmodified-to-modified peak areas for each fragment ion were used to construct a series of linear equations that were solved simultaneously by least squares regression analysis. We validated this method by using synthetic peptides as depicted in Fig. 3B. Here, two positional isomers of histone H4 tail (H41–23) were mixed in known ratios and fragmented simultaneously by using ETD-MS/MS. Ratios of fragment ions distinguishing the two species were then compared with expected ratios (Fig. 3B); observed ratios matched closely with expected values. The average difference between observed and expected values was 7% (standard deviation = 5%). We used this approach to calculate the abundance of each of the 74 histone H4 isoforms presented in Fig. 2 (Isomer Quant column).
Evaluation of coeluting positional isomers from human ES cells (H1) over the TPA treatment time course revealed that the ratio of trimethylated K20 (K20me3) to monomethylated R3 (R3me1) increased with TPA treatment—that is, the elevation in trimethylated histone H4 (SI Fig. 5E) is primarily due to the deposition of an additional methyl group at K20me2 rather than at R3. Further, R3 methylation was never observed in the absence of K20 dimethylation (SI Fig. 7)—a strong indication that a functional relationship between the two modification sites exists. These data suggest that K20me2 is a prerequisite for methylation of R3.
Discussion
Human ES Cell Epigenetics.
Untreated human ES cells exhibited high levels of activating marks (acetylation of residues K5, K8, K12, and K16) and low levels of repressive marks (K20 dimethylation). On differentiation, the histone methylation and acetylation patterns closely match those seen in fibroblasts. These findings are consistent with previous studies and indicate that ES cells exhibit more permissive chromatin overall and that differentiation is associated with a move toward more highly silenced chromatin (4, 15, 32–35). Both the presence of highly permissive chromatin and the changes observed during differentiation support the hypothesis that these modification states are required for the maintenance of pluripotency. The striking correlation between K20 methylation and the loss of pluripotency (SI Fig. 5) is intriguing. We reasoned that these changes were likely the result of differentiation, but previous work has suggested that methylation of H4K20 may be cell cycle-dependent (36–38). Because stem cells divide more rapidly than other cell lines (with ≈50% of the population in S phase at any given time), it is possible that cell cycle dependencies for histone methylation may play a role in the observed changes. By using flow cytometry analysis we determined that TPA treatment does induce changes in cell cycle state. After TPA treatment, the population of human ES cells in S phase (≈50%) was substantially reduced within 30 h of TPA treatment (≈10%). The number of cells in G2 increased initially, but declined after 30 h of TPA treatment, whereas the population of cells in G1 continued to climb throughout the 75-h treatment window (from ≈30% to ≈80%, see SI Fig. 8). Rice et al. (36) have shown that H4K20 methylation is most abundant during the G1 phase of the cycle. Because synchronization of human ES cells is not possible at present, it is difficult to determine whether the methylation changes we observe relate to differentiation, cell cycle changes, or both.
Histone H4 Combinatorial Codes.
Among the ≈3 million possible combinatorial codes present on the histone H4 N terminus (based on the known modification sites) we have identified only 74. Beyond that, we have discovered intriguing nuances that reveal sequential mechanisms by which these modifications are encoded. For example, R3 methylation (mono- or di-) was only observed in the presence of K20 dimethylation, suggesting that K20Me2 is requisite for methylation of R3. Interestingly, R3Me1 is associated with transcriptional activation, whereas K20 methylation has been linked to transcriptionally inactive regions of heterochromatin (39–41). It has not escaped our notice that this combination of modifications closely resembles the bivalent signature exhibited by methylation of residues K4 and K27 of histone H3. In both cases the “active” mark is located near the N termini and the “repressive” mark ≈20 residues downstream. Here, we provide direct evidence that (i) both R3 methylation and K20 dimethylation are present on the same histone H4 tail, (ii) R3 methylation exists at detectable levels only in the presence of K20 dimethylation, and (iii) significant changes arise in the abundance of K20 methylation during human ES cell differentiation. We also discovered that a small percentage (<1%) of the histone H4 tails exhibited nonacetylated N termini, and furthermore, acetylation at other residues was never observed in the absence of N-terminal acetylation. Thereafter, K16 was the most commonly acetylated residue followed by K5. However, these two modifications rarely occur on the same histone H4 molecule, indicating that acetylation at these sites is, to some extent, mutually exclusive (Fig. 2). In a very real sense, these combinatorial PTM maps will serve as a foundation for numerous follow-up experiments and provide a roadmap for targeted ChIP-chip experiments.
The MS-based technology described here provides a sensitive and rapid methodology to detect and quantify the combinatorial PTM codes present on the intact termini of histone H4. We validated this method by using synthetic peptides and calculated an overall estimate of experimental error. We have applied the technology to identify 74 distinct combinatorial PTM patterns occurring on histone H4 and tracked their abundances in human ES cell lines progressing through TPA-induced differentiation. These data provide evidence that ES cells have unique epigenetic signatures and that these codes are imparted in a sequential fashion. Finally, we note this procedure should be applicable to other core histones and any other protein of interest containing a high degree of variable modifications.
Methods
Cell Culture and TPA Treatment.
Human embryonic stem cells (lines H1, H7, and H9) were maintained in feeder-independent media (TeSR) as described in ref. 42. Cells were treated for 0, 3, 15, 30, 60, and 75 h with 12-O-tetradecanoylphorbol-13-acetate (TPA) (Sigma–Aldrich) at a final concentration of 50 ng/ml. Human fibroblasts (line IMR90) were cultured according to American Type Culture Collection recommendations and were likewise treated with 50 ng/ml TPA for 0, 3, 15, 30, 60, and 75 h. Cells were individualized for 10 min with an adequate volume of prewarmed (37°C), 0.05% Trypsin-EDTA to cover the culture surface. After cell detachment, an equivalent volume of ice-cold growth medium (10% FBS in DMEM) was used to neutralize the trypsin before pelleting. Cell pellets were subsequently washed twice in ice-cold PBS and stored at −80°C.
Histone Purification and Digestion.
Starting with ≈2 × 107 cells, nuclei were collected as previously described, except hypotonic lysis buffer contained 5 mM Tris·HCl (pH 8.0), 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose, 2 mM sodium vanadate, 1 mM DTT, 0.6% Nonidet P-40, and 2.5 μg/ml each of the protease inhibitors, leupeptin, pepstatin, antipain, chymostatin, and aprotinin (Sigma–Aldrich) (43). After 5 min on ice, nuclei were pelleted at 960 × g for 5 min. The resulting pellets were washed twice in the hypotonic lysis buffer, resuspended in 1.5 ml of 0.4 N H2SO4, and incubated for 2 h at 4°C with agitation. The sample was then centrifuged at 4°C for 15 min at 20,800 × g. The supernatant was collected and histones were precipitated overnight at 4°C with 20% trichloroacetic acid. The next day, samples were centrifuged at 20,800 × g, washed once in 1.5 ml of acetone, and resuspended in 0.1% trifluoroacetic acid. Histone proteins were then separated as previously described, dried to remove organic solvent, and resuspended in 0.1% TFA acid (43). A small sample was removed from each fraction for quantification by BCA assay (Pierce) and histone identities were confirmed by gel electrophoresis and Coomassie staining. Purified histone H4 samples were digested overnight with Asp-N (Roche) (1:20) at a pH 8 before MS analysis. Synthetic peptides were generated at the UW Biotechnology Center (Madison, WI) and resuspended in either 0.1% acetic acid or 30% acetonitrile with 0.1% acetic acid before analysis.
Instrumentation.
Chromatographic peptide separations were performed on a reversed-phase self-prepared capillary column as described in ref. 44. Online peptide separations were performed by using an Agilent 1100 Series binary HPLC system that was coupled to either an ETD-enabled Finnigan LTQ mass spectrometer or a Finnigan LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). The LTQ mass spectrometer was operated in a targeted fashion by using a parent mass list. First, a full-scan (300–2,000 m/z) mass spectrum was acquired and then five ETD/PTR scans were acquired on the most intense peak from the parent mass list. An isolation window of 2 m/z and a precursor target value of 80,000 were used. ETD reactions were carried out by mutual storage of isolated cations and fluoranthene radical anions for 30 ms in the LTQ. This was followed by 150 ms reaction with benzoic acid anions (PTR) before mass analysis. The LTQ Orbitrap was operated only in MS1 mode with a target value of 500,000 and a resolving power of 60,000. LTQ Orbitrap data were deconvoluted by using Xtract software (Thermo Fisher Scientific) with a signal-to-noise (S/N) threshold of 2 and a fit factor of 0. A selected ion chromatogram was constructed for the most abundant isotopic peak of each isoform (±0.02 m/z) and areas were selected manually. Tandem mass spectra were exported to.csv file and processed by software written in-house. First, the percentage of K20 methylation was determined by peak areas corresponding to modified and unmodified z4 fragment ions. Next, the percentage of N-terminal acetylation and percentage of R3 methylation was determined by peak areas corresponding to c3 fragment ions. Then all peaks were use to determine acetylation composition and location. The software extracted peak areas (±2 m/z) for each expected c- and z-type ion. Percentages of nonacetylated to acetylated values were used to construct equations summing to 100%. Matlab (The Mathworks) was used to solve the system of linear equations by using a least squares regression analysis.
Flow Cytometry.
ES cells were labeled with bromodeoxyuridine (BrdU) by adding BrdU to a concentration of 10 μM 30 min before harvest. Cells were individualized and collected as described earlier (≈5 × 106 cells per experiment). After the wash steps, the cell pellet was resuspended in 100 μl of PBS. To fix the cells, ice-cold 70% ethanol was added dropwise while vortexing. Cells were pelleted and resuspended in 1 ml of cold 0.1 M HCl/50% Triton X-100. After a 10-min incubation on ice, the cells were centrifuged, resuspended in 2 ml of water, boiled for 10 min, and placed on ice for 5 min. Then 5 ml of PBS/0.5% Triton was added, followed by centrifugation and resuspension in 100 μl of PBS containing 5 μg/ml of anti-BrdU-FITC antibody and 0.1% BSA. The cells were incubated in the dark for 30 min at room temperature. Next, 5 ml of PBS was added, cells were pelleted and resuspended in 500 μl of PBS containing 5 μg/ml propidium iodide and 200 μg/ml RNase. After incubation for 30 min at 37°C, cells were analyzed by using an Aria flow cytometer (BD Biosciences).
Supplementary Material
Acknowledgments.
We thank April Jue, Guokai Chen, Ron Stewart, Victor Ruotti, Chuhu Yang, Gheorghe Craciun, and Beatrix Ueberheide for helpful discussions. This work was supported by the University of Wisconsin-Madison, Thermo Fisher, the Beckman Foundation, Eli Lilly, National Institutes of Health (NIH) Grant 1R01GM080148 (to J.J.C.), Genomic Sciences Training Program Predoctoral Fellowship NIH 5T32HG002706 (to D.H.P.), Biotechnology Training Program Predoctoral Fellowship NIH 5T32GM08349 (to J.B.), and a National Science Foundation graduate research fellowship (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710515105/DC1.
References
- 1.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Guenther MG, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meshorer E, Misteli T. Opinion—Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 5.Passeri D, et al. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26:5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pray-Grant MG, et al. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 8.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 9.Margueron R, Trojer P, Reinberg D. The key to development: Interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 11.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 12.Pokholok DK, et al. Genome-wide map of nucleosorne acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Lin WC, Dent SYR. Functions of histone-modifying enzymes in development. Curr Opin Genet Dev. 2006;16:137–142. doi: 10.1016/j.gde.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Hart SRL, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 16.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 17.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 18.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 19.Garcia BA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 20.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 21.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: Human histone H4. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 22.Taverna SD, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3N termini. Proc Natl Acad Sci USA. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: A new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyne MT, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histories in the H2A gene family by top down mass spectrometry. J Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 25.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: A bird's eye view. J Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 26.Siuti N, et al. Gene-specific characterization of human histone H2B by electron capture dissociation. J Proteome Res. 2006;5:233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 27.Medzihradszky KF, et al. Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS). Mol Cell Proteomics. 2004;3:872–886. doi: 10.1074/mcp.M400041-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang LW, Freitas MA. Comparison of peptide mass mapping and electron capture dissociation as assays for histone posttranslational modifications. Int J Mass Spectrom. 2004;234:213–225. [Google Scholar]
- 29.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coon JJ, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Martens JHA, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keohane AM, et al. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 34.Francastel C, Schubeler D, Martin DIK, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 35.Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- 36.Rice JC, et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepherd GR, Hardin JM, Noland BJ. Methylation of lysine residues of histone fractions in synchronized mammalian cells. Arch Biochem Biophys. 1971;143:1–5. doi: 10.1016/0003-9861(71)90180-9. [DOI] [PubMed] [Google Scholar]
- 38.Thomas G, Lange HW, Hempel K. Kinetics of histone methylation in vivo and its relation to cell-cycle in Ehrlich ascites tumor-cells. Eur J Biochem. 1975;51:609–615. doi: 10.1111/j.1432-1033.1975.tb03963.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang HB, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 40.Huang SM, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishioka K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 43.Hake SB, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McAlister GC, et al. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.