Abstract
Retroposons, such as short interspersed elements (SINEs) and long interspersed elements (LINEs), are the major constituents of higher vertebrate genomes. Although there are many examples of retroposons' acquiring function, none has been implicated in the morphological innovations specific to a certain taxonomic group. We previously characterized a SINE family, AmnSINE1, members of which constitute a part of conserved noncoding elements (CNEs) in mammalian genomes. We proposed that this family acquired genomic functionality or was exapted after retropositioning in a mammalian ancestor. Here we identified 53 new AmnSINE1 loci and refined 124 total loci, two of which were further analyzed. Using a mouse enhancer assay, we demonstrate that one SINE locus, AS071, 178 kbp from the gene FGF8 (fibroblast growth factor 8), is an enhancer that recapitulates FGF8 expression in two regions of the developing forebrain, namely the diencephalon and the hypothalamus. Our gain-of-function analysis revealed that FGF8 expression in the diencephalon controls patterning of thalamic nuclei, which act as a relay center of the neocortex, suggesting a role for FGF8 in mammalian-specific forebrain patterning. Furthermore, we demonstrated that the locus, AS021, 392 kbp from the gene SATB2, controls gene expression in the lateral telencephalon, which is thought to be a signaling center during development. These results suggest important roles for SINEs in the development of the mammalian neuronal network, a part of which was initiated with the exaptation of AmnSINE1 in a common mammalian ancestor.
Keywords: conserved noncoding element, enhancer, evolution, mouse
Retroposons, including SINEs, LINEs, and long-terminal repeat or LTR retrotransposons, propagate within the host genome via RNA intermediates (1–3). Despite the abundance of retroposons in mammalian genomes (e.g., 42% and 37% of human and mouse genomes, respectively) (4, 5), most are nonfunctional and are regarded as genomic parasites or “junk DNA.” However, in several cases, copies of retroposons have been involved in generating new cis-regulatory elements for processes such as alternative splicing, mRNA polyadenylation, and promoter activity (6), all of which are examples of exaptation (7, 8). The idea that retroposons may impact genome evolution and functionality (8–10) has been gradually accepted. However, most examples seem to have only local effects, and no example has been reported where an inserted retroposon plays a pivotal role in higher-order morphological or biological innovations.
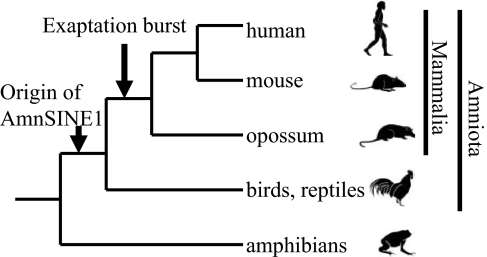
On the other hand, recent genome-scale comparative analyses have identified conserved noncoding elements (CNEs) that are suggested to be essential for host survival (11). CNEs constitute 3–4% of the human genome, whereas protein-coding exons constitute only ≈1.5% (4). Recently, it was reported that 16% of eutherian-specific CNEs are derived from transposable elements (retroposons) (12, 13), which supports the importance of retroposons in evolution (14–18). This observation on CNEs is important because retroposons arose only recently in evolution and therefore may have the potential to acquire new functions through exaptation. We recently identified a new SINE family, AmnSINE1, in the genomes of Amniota (mammals, birds, and reptiles). The ≈100 copies of AmnSINE1s are highly conserved as mammalian-specific CNEs (16), suggesting that these SINEs acquired a function (exapted) during the emergence of mammals (or therians) (Fig. 1). Therefore, these AmnSINE1s are the best available mammalian-specific CNEs to study the evolutionary impact of retroposons on morphological changes among mammals (16).
Fig. 1.
Exaptation of AmnSINE1s in a common ancestor of mammals. The known phylogeny of tetrapods is shown with the time of exaptation of AmnSINE1 sequences. AmnSINE1 originated in a common ancestor of amniotes, and multiple copies acquired function in a common ancestor of mammals.
Studying brain development raises two fundamental questions (19). What characteristics of the brain make mammals distinct from other vertebrates? How have these characteristics evolved? In vertebrates, the anterior neural epithelium undergoes morphological subdivisions during development to generate vesicle-like structures known as the forebrain, midbrain, and hindbrain. The forebrain further divides into the telencephalon and the diencephalon. The telencephalon gives rise to the neocortex, basal ganglia, and hippocampus, whereas the diencephalon develops into the thalamus, prethalamus, and pretectum. Several patterning centers used during development have been well characterized and are generated on boundaries or ridges of each region, such as the anterior neural ridge, the zona limitans intrathalamica, and the midbrain–hindbrain boundary. The patterning centers are thought to establish the regional identities by secreting signaling molecules such as Shh, Wnts, and Fgfs (20, 21). The mammalian-specific brain is identified by three characteristics (19, 22): (i) the six-layered neocortex in the dorsal pallium (the pallial area in birds and reptiles); (ii) generation of the Cajal–Retzius cells, one of whose sources is generated in the pallial–subpallial boundary, and migrate tangentially toward the dorsal telencephalon; and (iii) the absence of the dorsal ventricular ridge, which has been commonly observed in the brains of birds and reptiles. The molecular mechanisms that generate these characteristics are largely unknown. In this study we demonstrate a potential role for SINEs in controlling gene expression and activity during forebrain development. Because the mammalian forebrain is evolutionarily unique, SINEs may have played a role in mammalian-specific evolution.
Results and Discussion
Enhancer Activity of the AS071 Locus for FGF8 Expression.
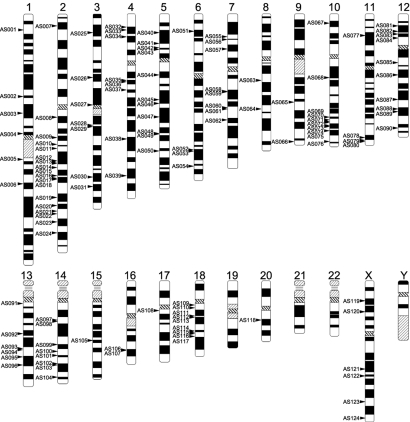
To elucidate the function of the conserved AmnSINE1 loci, we recharacterized conserved AmnSINE1 loci by updating the homology search as described in Methods. We refined 124 AmnSINE1 loci and 974 neighboring genes. The 124 loci are scattered among the human chromosomes (except chromosomes 19, 21, 22, and Y) but are not evenly distributed among the chromosomes (Fig. 2).
Fig. 2.
Positions of the 124 conserved AmnSINE1 loci on the human chromosomes. Each locus is indicated by an arrow.
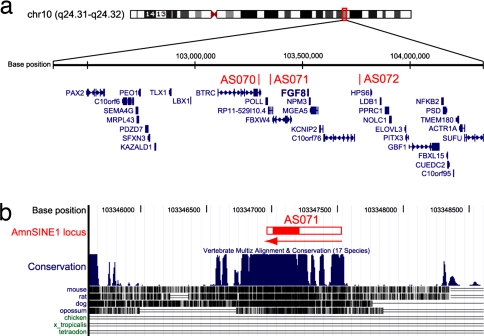
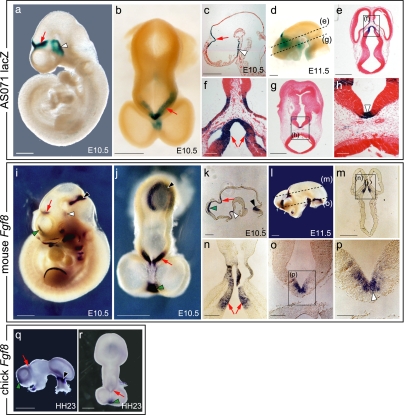
To test whether the conserved AmnSINE1 loci regulate the expression of adjacent genes, we used a β-galactosidase (lacZ) transgenic mouse system. We first focused on locus AS071, located 178 kbp downstream from FGF8 (Fig. 3a). The 570 bp of the putative full-length AmnSINE1 element (red horizontal bar) appears to be conserved among mammals (Fig. 3b). Synteny is conserved between human and mouse in this region (data not shown). The mouse sequence of the AS071 conserved region (600 bp) was cloned upstream of a minimal heat-shock protein 68 (Hsp68) promoter coupled to the lacZ reporter gene [SI Fig. 7 in supporting information (SI) Appendix], and this was injected into a fertilized mouse oocyte. To understand spatial and temporal lacZ expression in the mouse embryo, whole-embryo staining for lacZ activity was performed at embryonic day 10.5 (E10.5), E11.5, and E13.5 (Fig. 4 a–h and SI Table 1 in SI Appendix). Consistent expression of LacZ is observed in independent transgenic embryos in the lateral wall of the diencephalon, dorsal midline of the caudal telencephalon, and the hypothalamus (Fig. 4 a–c and SI Fig. 8 in SI Appendix) at E10.5 (30 embryos of 50), E11.5 (eight of 18), and E13.5 (eight of 17). FGF8 is well established as a signaling molecule, and its expression pattern and function during CNS development in mice and chicks have been reported (23–28). Because the expression of AS071 is restricted to the lateral diencephalon and hypothalamus, we compared the expression pattern of AS071 lacZ with that of Fgf8 by in situ hybridization at E10.5 and E11.5 (Fig. 4 i, j, and l). Interestingly, lacZ expression and Fgf8 expression in the lateral diencephalon (red arrows) as well as in the hypothalamus (white arrowheads) coincided exactly (Fig. 4 a, b, d, i, j, and l). Sagittal (Fig. 4 c and k) and transverse (Fig. 4 e–h and m–p) sections of brain showed detailed expression of lacZ and Fgf8, revealing identical expression in the diencephalon (red arrows) and hypothalamus (white arrowheads). These data suggest the possible role of AS071 as a specific enhancer of Fgf8 in the diencephalon and hypothalamus of the developing forebrain.
Fig. 3.
Characterization of the AS071 locus. (a) The 2-Mb region containing three AmnSINE1 elements in human chromosome 10. Coordinate (Top), AmnSINE1-inserted loci (Middle), and gene annotations (Bottom) are based on the hg18 assembly in the UCSC genome browser (http://genome.ucsc.edu). (b) The 3.0-kb region around the AS071 locus and conservation among vertebrate species. The 570-bp region of the putative full-length AmnSINE1 element (red horizontal bar) appears to be conserved among mammals, including opossum. Within the bar, only 200 bp (red fill) can be aligned with the AmnSINE1 consensus sequence. The sequences indicated by the open bar regions of the SINE have decayed and thus cannot be precisely aligned. The arrow indicates the direction of the element (5′ to 3′).
Fig. 4.
Comparison of the lacZ expression pattern in an AS071 transgenic mouse (a–h) with the FGF8 mRNA expression pattern in mouse (i–p) or chick (q and r) embryos. Lateral (a, i, and q) and dorsal (b, j, and r) views of embryos are shown. For hybridization and staining of the head, surface ectoderm and mesenchyme were removed. (c and k) Sagittal section of the head region. (d and l) Lateral view of the head. Transverse sections (e, g, m, and o) and corresponding close-up views (f, h, n, and p) are shown. lacZ and FGF8 mRNA expression in the lateral diencephalon and hypothalamus are indicated by red arrows, and those in the ventral diencephalon are indicated by a white arrowhead. FGF8 mRNA expression at the midbrain–hindbrain boundary and the commissural plate are indicated by black and green arrowheads, respectively. The dashed lines indicates where the embryos were sectioned for the staining shown in e, g, m, and o. [Scale bars: 500 μm (a–e, g, i–m, q, and r), 100 μm (f, h, n, and o), and 50 μm (p).]
Function of FGF8 Expression in Diencephalon.
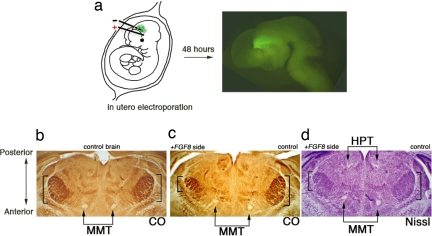
To elucidate the function of FGF8 in diencephalon, we used in utero electroporation (24) to overexpress FGF8 in the developing diencephalon at E10.5 (Fig. 5a). With the possibility that FGF8 could be involved in area patterning of the diencephalon through the anterior/posterior axis, we tested the pattern of thalamic nuclei at postnatal day 6 (P6) by cytochrome oxidase staining and Nissl staining (Fig. 5 b–d). The somatosensory nucleus in the thalamus, which represents the pattern of whiskers on the animal's snout (barreloid), was easily distinguished in both sides of the thalamus in the control animal (Fig. 5b). Unilateral FGF8 electroporation of the lateral diencephalon caused one side of the barreloid to shift and shrink as compared with the control (Fig. 5c, bracket). Nissl staining revealed a shifted mammillothalamic tract, which runs through the boundary of the thalamus and prethalamus. Conversely, the habenulopeduncular tract, which runs through the pretectum and thalamus, was not altered, suggesting that repatterning upon FGF8 overexpression occurred specific to the thalamus but not to the pretectum (Fig. 5d). During CNS development, the diencephalon dives down in between the telencephalon, and its bent longitudinal axis causes its true anterior/posterior axis to rotate 90° (T. Shimogori, unpublished data). Accordingly, the shift of the barreloid upon FGF8 expression is toward the posterior, which is linking to the role of this protein in other parts of the CNS (24, 27). These data demonstrate that the expression of FGF8 in the diencephalon controls patterning of thalamic nuclei.
Fig. 5.
Gain-of-function analysis of FGF8 in developing diencephalons. (a) Scheme for in utero electroporation. Plasmid solution was injected in the third ventricle of E10.5 embryos in utero, followed by insertion of needle-type electrodes. A series of three square-wave current pulses (7 V, 100 ms, three times) was delivered, resulting in gene transfection into a restricted region in the unilateral diencephalons (the region is indicated by +, and − indicates the region where gene transfection was not performed). Gene transfer was visualized by GFP after 48 h of electroporation, demonstrating localized expression of the transgene. (b–d) Coronal section of P6 brains processed for cytochrome oxidase (CO) histochemistry (b and c) and Nissl staining (d). The control brain shows the symmetric position of the barreloid (b), whereas the FGF8-electroporated brain shows a posteriorly shifted and shrunken barreloid on the electroporated side (c) as well as mammillothalamic tract (MMT). Nissl staining reveals the habenulopeduncular tract (HPT), which is not altered by FGF8 electroporation (d).
FGF8 Expression in Diencephalon Is Mammalian-Specific.
Because conservation of the AS071 locus is observed only in mammals, the enhancer function of this locus is expected to be mammalian-specific. Based on our results that AS071 is likely to enhance FGF8 expression, we investigated FGF8 expression in chick and performed in situ hybridization for Fgf8 in Hamburger–Hamilton (HH) stage-23 chick brain to compare with the mouse results (Fig. 4 q and r). Although the chick diencephalon showed expression of Fgf8, it was weaker than that of the mouse (compare Fig. 4 i and j with q and r, red arrows), whereas strong expression in the anterior telencephalon (green arrowheads) was observed in both mouse and chick embryos (Fig. 4 i, j, q, and r). This result suggests the existence of mammal-specific enhanced expression of FGF8 in diencephalon, which organizes mammalian thalamic patterning differently from other vertebrates. Although overexpression of FGF8 in chick embryos changes regional identity (29), the different organization of thalamic nuclei between chicks and mice makes direct comparison difficult. However, our findings suggest local gain of function of specific protein by SINE insertion, possibly changing the functional area identity in specific region of the brain. This patterning change may also have the chance to alter the pattern of its wired region of the brain.
The functional significance of the dorsal forebrain, where FGF8 is expressed, has not been well examined. Based on our results, we propose a role for FGF8 in thalamic pattern formation, and such function may have been introduced by enhancer activity of the AS071 locus. In addition, we confirmed the expression of signaling molecules such as Bmp4, noggin, and Wnt in the dorsal forebrain and observed that Otx2 (30) expression coincided with that of Fgf8 (SI Fig. 9 in SI Appendix), supporting the assertion that this region constitutes a patterning center for the diencephalon. How the regional identity of the thalamus and thalamic patterning are independently achieved in the diencephalon using signals from this center remains unclear.
Enhancer Activity of the AS021 Locus.
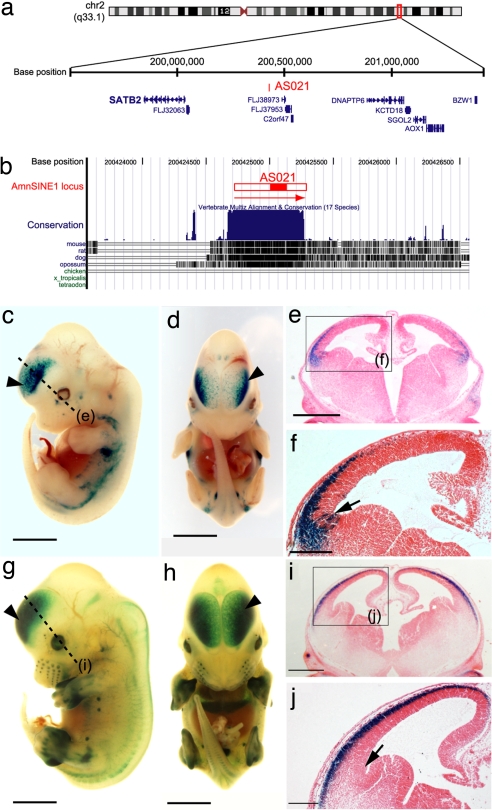
Given the above results, we examined other loci, looking for enhancer activities among the 124 conserved AmnSINE1 loci. We found that locus AS021 in human chromosome 2 (Fig. 6a) contains a 600-bp region of AmnSINE1 highly conserved among mammals (Fig. 6b). To test its function as an enhancer of gene expression, we used the same method as above. We assayed the enhancer activity in AS021 locus at E10.5, E11.5, E12.5, and E13.5 (SI Table 1 in SI Appendix). At E10.5 we could not observe any consistent lacZ expression patterns. The first consistent lacZ expressions were observed in two of 10 embryos in the dorsolateral side of telencephalon at E11.5 (SI Fig. 10a in SI Appendix, arrowhead). At E12.5 the expression patterns in the dorsolateral side of telencephalon became more noticeable (SI Fig. 10 b and c in SI Appendix). In the later stage, at E13.5, 13 of 23 embryos consistently showed lacZ expression in the neocortex of telencephalon (Fig. 6 c–j and SI Fig. 10 d–h in SI Appendix). Judging from the differences of limb development of embryos, we can discriminate embryos at relatively early phase (Fig. 6 c–f) from those at late phase (Fig. 6 g–j) even at E13.5. It appears that the lacZ expression patterns in the telencephalon expanded from the restricted area of telencephalon (dorsolateral side, Fig. 6 c–f) to the entire of neocortex (Fig. 6 g–j) as the developmental stage progressed. We found that locus AS021 is located 390 kbp upstream from the gene SATB2 (Fig. 6a), which was recently shown to be a multifunctional determinant of craniofacial patterning and osteoblast differentiation (31). SATB2 is also expressed in the telencephalon at E13.5 (32), suggesting a role in the developing telencephalon. Because SATB2 is multifunctional and is expressed ubiquitously, including in the brain and skeleton, it is difficult to prove that locus AS021 enhances neocortical subexpression of SATB2 based on a lacZ expression pattern. Circumstantial evidence, however, suggests that it does. First, SATB2 is the only gene involved in development in the AS021-neighboring region we searched (2 Mb in total), which includes 10 mouse genes (SI Table 2 in SI Appendix). Second, LacZ staining in the neocortex was first observed at E11.5–E12.5. (SI Table 1 in SI Appendix). This developmental expression pattern is consistent with that reported for SATB2 (31) (SI Fig. 10 in SI Appendix). Third, a brain section study showed that lacZ staining was first observed in the cortical plate at E13.5 (Fig. 6 c–f) and then expanded into other neocortex areas (the intermediate zone; Fig. 6 g–j), a developmental expression pattern consistent with that of SATB2 (32) (SI Fig. 10 in SI Appendix). Furthermore, detailed expression of lacZ was evident in sectioned brain tissue, which revealed restricted expression in the lateral telencephalon (Fig. 6 e and f).
Fig. 6.
Characterization of the AS021 locus. (a) The 2-Mb region containing an AmnSINE1 element in the AS021 locus in human chromosome 2. Coordinate (Top), AS021 locus (Middle), and gene annotations (Bottom) are based on the hg18 assembly in the UCSC genome browser (http://genome.ucsc.edu). (b) The 3.0-kb region around the AS021 locus and conservation among vertebrate species. The red horizontal bar indicates the putative full-length AmnSINE1 element. Within the bar, the red fill indicates an element that can be aligned with the AmnSINE1 consensus sequence. The display form is in accord with Fig. 3b. (c–j) lacZ expression patterns in an AS021 transgenic mouse embryos (E13.5). Lateral (c and g) and frontal (d and h) views of lacZ-stained whole-mount embryo are shown. The dashed line indicates where the embryo was sectioned for the staining shown in the coronal section (e and i). Arrowheads indicate lacZ expression in the neocortex. Arrows indicate the antihem (the border of the pallium and subpallium). [Scale bars: 2.0 mm (c, d, g, and h), 0.5 mm (e and i), and 0.2 mm (f and j).]
The cerebral cortex has a laminar organization, which shows a birth date-dependent migration manner (i.e., early-born/inside, late-born/outside). Correct ordering of lamina formation requires Reelin expression and several sources of Reelin-expressing Cajal–Retzius cells (33). One of the sources for a subpopulation of Reelin-positive Cajal–Retzius cells is from the border of the pallium and subpallium (33, 34) (antihem), where cells stained by lacZ (arrows in Fig. 6f) appeared to be first generated. Cells generated from the antihem, as labeled by Dbx1-LacZ expression, migrate tangentially toward the dorsal telencephalon, which seems to be lacking in the chick telencephalon (ref. 33 and A. Pierani, personal communication). This result strongly suggests a mammal-specific contribution of locus AS021 activity to the development of a mammal-specific fundamental structure (19). However, further study is required to grasp a total understanding of this phenomenon.
SINEs as Distal Enhancers.
To date, we have examined 10 AmnSINE1 loci and demonstrated that the two loci, AS071 and AS021, function as distal transcriptional enhancers in developing mouse embryos. It should be noted that both enhancers are specific to the developing forebrain and have functions that are mammalian-specific. Although we speculate that these enhancers contribute to mammalian-specific brain formation, it is not certain whether insertions of these SINEs first triggered formation of the unique mammalian brain in a common ancestor of mammals, because the redundancy of enhancers is frequently observed. From the conservation of the enhancer sequence at the AS071 or AS021 locus among mammals, however, we conclude that they contributed to mammalian-specific brain formation through natural selection regardless of the possibility that future knockout experiments may not produce phenotypic alterations (35).
When we assessed the distribution of all 124 AmnSINE1 loci, we discovered that one-fourth (32 of 124 loci) are located near genes involved in brain development. It should also be mentioned that Bejerano's group (36) recently discovered >10,000 transposons conserved among mammals that are located near developmental genes in the human genome. Accordingly, we speculate that the enhancers in loci AS071 and AS021 play important roles in brain development as a part of the neuronal networks generated upon insertion of AmnSINE1s in a common mammalian ancestor, which led to the unique evolution of the mammalian brain.
The fact that many CNEs are derived from transposable elements [e.g., 16% in Eutheria (12)] leads to the speculation that other SINEs in various organisms may also play important biological roles such as enhancers. AmnSINE1 belongs to the Deu-SINE superfamily, having a conserved domain, the Deu domain, in the middle of the SINE. Members of this group include coelacanth LmeSINE1, zebrafish SINE3, and amphioxus BflSINE1 (16). Because they share this conserved domain, we speculate that these SINEs may have important functions. Furthermore, other superfamilies of SINEs, such as V-SINEs (37) and CORE-SINEs (38, 39), have been reported. These SINEs also have a highly conserved domain in the middle of their sequences, even though the overall sequences are unique. We anticipate that functions for these SINEs with the conserved domain will be elucidated in the near future (40).
Methods
Screening the AmnSINE1 Loci.
We searched AmnSINE1 sequences in the human [University of California, Santa Cruz (UCSC) hg18, National Center for Biotechnology Information Build 36.1] and mouse (UCSC mm8, National Center for Biotechnology InformationBuild 36) genomes using the FASTA program. SINE conservation was evaluated based on the phastCons Conserved Elements and the phastCons scores available in the UCSC database (http://genome.ucsc.edu). We searched the refFlat data in the UCSC database for genes having a transcription start site within 1 Mbp (for both strands) from the midpoint of a conserved AmnSINE1 sequence.
Mouse Transgenic Enhancer Assay.
The AS071 and AS021 loci were amplified from MCH mouse DNA by PCR, and each amplified fragment was subcloned into the HSF51 vector containing the mouse hsp68 basic promoter and the bacterial LacZ coding region (41). The subcloned construct (10 ng/μl), digested with ScaI, was purified and injected into pronuclei as described (41). Fertilized eggs were collected from pregnant B6C3F1 females. Transient transgenic embryos were harvested at E10.5–E13.5 and were stained to visualize β-galactosidase activity (41).
Genotyping Analysis.
We extracted total genomic DNA from yolk sacs of all transient transgenic embryos. Positive transgenic animals were confirmed by PCR.
In Situ Hybridization.
The surface ectoderm of E10.5 and E11.5 mouse embryos and HH23 chick embryos were removed from the head region with forceps in diethylpyrocarbonate (DEPC)-treated PBS. The embryos were fixed overnight in 4% paraformaldehyde dissolved in DEPC-treated PBS at 4°C. Whole-mount in situ hybridization in mouse and chick embryos was performed as described (42). After in situ hybridization, mouse embryos were processed for paraffin sectioning by 8-μm sections.
In Utero Electroporation.
Electroporation was performed as described (24) with modifications to electroporate the diencephalon. DNA solution was injected into the third ventricle, followed by negative electrode insertion into the lumen of the third ventricle. The positive electrode was placed outside, near the head of the embryo. A series of three square-wave current pulses (7 V, 100 ms) was delivered, resulting in gene transfection into one side of the diencephalic wall.
Additional Details.
Full methods and associated references are available in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Dr. Gail Martin (University of California, San Francisco) for the Fgf8 in situ probe and Dr. Yumiko Saga for help in pursuing this work by using the facility of the National Institute of Genetics. This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.O.) and from the National Institute of Genetics Cooperative Research Program (Grants 2006-A75 and 2007-B3 to N.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709398105/DC1.
References
- 1.Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: Genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 2.Hedges DJ, Batzer MA. From the margins of the genome: Mobile elements shape primate evolution. BioEssay. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 3.Ohshima K, Okada N. SINEs and LINE: Symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould SJ, Vrba ES. Exaptation: A missing term in the science of form. Paleobiology. 1982;8:4–15. [Google Scholar]
- 8.Brosius J, Gould SJ. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.”. Proc Natl Acad Sci USA. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 10.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 11.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 13.Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britten RJ, Davidson EH. Gene regulation for higher cells: A theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 15.Britten RJ. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- 16.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 18.Xie X, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci USA. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Striedter G. Principles of Brain Evolution. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 20.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 21.Echevarria D, Vieira C, Gimeno L, Martinez S. Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res Brain Res Rev. 2003;43:179–191. doi: 10.1016/j.brainresrev.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Krubitzer L, Kaas J. The evolution of the neocortex in mammals: How is phenotypic diversity generated? Curr Opin Neurobiol. 2005;15:444–453. doi: 10.1016/j.conb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Depew MJ, Simpson CA. 21st Century neontology and the comparative development of the vertebrate skull. Dev Dyn. 2006;235:1256–1291. doi: 10.1002/dvdy.20796. [DOI] [PubMed] [Google Scholar]
- 24.Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 25.Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- 26.Storm EE, et al. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 27.Grove EA, Fukuchi-Shimogori T. Generating the cerebral cortical area map. Annu Rev Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- 28.Crossley PH, Martinez S, Ohkubo Y, Rubenstein JLR. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi D, et al. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Kimura-Yoshida C, et al. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- 31.Dobreva G, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- 33.Bielle F, et al. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 34.Assimacopoulos S, Grove EA, Ragsdale CW. Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J Neurosci. 2003;23:6399–6403. doi: 10.1523/JNEUROSCI.23-16-06399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahituv N, et al. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:1906–1911. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogiwara I, Miya M, Ohshima K, Okada N. V-SINEs: A new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Res. 2002;12:316–324. doi: 10.1101/gr.212302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit AF, Riggs AD. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995;23:98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert N, Labuda D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santangelo AM, et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumiyama K, et al. Genomic structure and functional control of the Dlx3–7 bigene cluster. Proc Natl Acad Sci USA. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.