Abstract
The Sin3-histone deacetylase (HDAC) corepressor complex is conserved from yeast to humans. Mammals possess two highly related Sin3 proteins, mSin3A and mSin3B, which serve as scaffolds tethering HDAC enzymatic activity, and numerous sequence-specific transcription factors to enable local chromatin regulation at specific gene targets. Despite broad overlapping expression of mSin3A and mSin3B, mSin3A is cell-essential and vital for early embryonic development. Here, genetic disruption of mSin3B reveals a very different phenotype characterized by the survival of cultured cells and lethality at late stages of embryonic development with defective differentiation of multiple lineages—phenotypes that are strikingly reminiscent of those associated with loss of retinoblastoma family members or E2F transcriptional repressors. Additionally, we observe that, whereas mSin3B−/− cells cycle normally under standard growth conditions, they show an impaired ability to exit the cell cycle with limiting growth factors. Correspondingly, mSin3B interacts physically with the promoters of known E2F target genes, and its deficiency is associated with derepression of these gene targets in vivo. Together, these results reveal a critical role for mSin3B in the control of cell cycle exit and terminal differentiation in mammals and establish contrasting roles for the mSin3 proteins in the growth and development of specific lineages.
Keywords: E2F, histone deacetylase, knockout, quiescence
The Sin3-histone deacetylase (HDAC) corepressor has been physically and functionally linked to diverse transcriptional complexes governing many physiological processes (1, 2). The two highly related mammalian Sin3 proteins, mSin3A and mSin3B, use their multiple interaction domains to direct chromatin-modifying activities to specific sites in the genome, most typically via sequence-specific transcription factors and their cognate binding elements. Class I HDACs, HDAC1 and HDAC2, are the principal enzymatic activities of the mSin3 complex. In addition, there are several other mSin3-associated proteins, including mSds3, p33ING1, and SAP30 (3–6).
mSin3A has been shown to be essential for early embryonic development and for the growth and survival of cultured cells that may relate to its requirement for the regulation of multiple transcriptional programs (7). Of relevance to the current study, mSin3B is expressed in cells deleted for mSin3A, suggesting that, despite their structural relatedness, mSin3B is not functionally equivalent to mSin3A. Both yeast and mammalian Sds3 are required for the maintenance of Sin3-associated HDAC enzymatic activity (3, 8). Nullizygosity for mammalian Sds3 results in early embryonic lethality and engenders marked chromosome segregation defects due to a failure in pericentric heterochromatin formation (9). mSin3A-null fibroblasts exhibited normal karyotypes (7).
In mammalian cell cycle, the G0/G1-to-S transition is a highly regulated event whose disruption represents a prerequisite for essentially all human cancers. This critical phase of the cell cycle is controlled by the retinoblastoma (Rb) family of proteins, pRb, p107, and p130, by virtue of their binding to and repression of the E2F transcription factors (10, 11). In G0 (quiescence) and G1 (resting) phases of the cell cycle, the Rb proteins associate with E2F and tether chromatin modifiers to actively repress E2F target genes encoding key cell cycle progression factors. Progression into the S (DNA synthesis) phase is brought about by CDK4/6-mediated phosphorylation of the Rb proteins, resulting in their dissociation from E2F transcription factors. The molecular basis of E2F-directed transcriptional repression in G0/G1 remains an area of active investigation, with current models supporting a physical link between an E2F/p107/p130 complex and mSin3B-HDAC1 that, upon docking to the promoters of E2F target genes, leads to local histone deacetylation and repression of transcription (12, 13). In this study, we generated mSin3B deficiency mice and cells to understand its roles in mammalian growth and development and to validate its specific capacity to regulate E2F gene targets and their linked processes in cell cycle and differentiation control.
Results
mSin3B Is Essential for Embryonic Development.
Standard gene-targeting technology was used to generate a germ-line conditional knockout allele of mSin3B in which Cre-mediated recombination of intronic LoxP sequences results in deletion of exon 2 and generation of a null allele [supporting information (SI) Fig. 5]. Two of the six independently generated mouse embryonic stem cell clones harboring the desired mSin3B recombination event were used to generate germ-line-transmitting chimeras as documented by Southern blot and PCR assays (Fig. 1a). FLPe-mediated deletion of the Frt-flanked Neomycin-resistance cassette generated an mSin3B-floxed allele (mSin3BL) that was judged to be functionally wild type by virtue of normal mSin3B expression and lack of gross phenotypes in mSin3BL/L mice and derivative cells (data not shown; see below). Upon Cre expression, the mSin3B floxed allele was recombined, giving rise to a functionally null mSin3B allele (Fig. 1b).
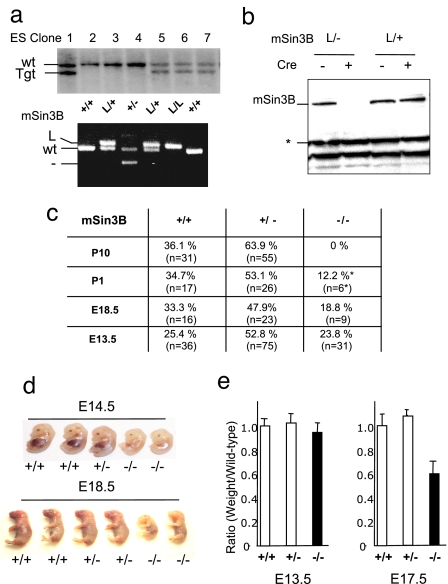
Fig. 1.
mSin3B is essential for late embryonic development. (a) (Upper) mSin3B targeted ES cell clones were identified by Southern blot analysis with the probe shown in SI Fig. 5. (Lower) PCR genotyping of embryos harboring the indicated alleles. (b) Western blot on total protein extracts from mSin3BL/+ or mSin3BL/− fibroblasts before (−) or after (+) retroviral-mediated Cre expression using an antibody against mSin3B. Equal amounts of proteins were loaded. *, Cross-reactive band, used as a loading control. (c) Genotype distribution of embryos and offspring from mSin3B heterozygotes intercrosses in a mixed background (E, embryonic day; P, day after birth). *, Found dead. The number of animals analyzed per genotype and time point are indicated in parentheses. (d) Representative litters from mSin3B heterozygote intercrosses, at E14.5 (Upper) or E18.5 (Lower). For each litter, the two embryos on the right are null for mSin3B. (e) Relative weight of mSin3B-null embryos (black bars) compared with their wild-type or heterozygotes littermates (white bars) at E13.5 (Left) or E17.5 (Right). Shown is the average from three litters for each time point.
mSin3BL/+ mice were mated to E2A-Cre transgenic mice to generate mSin3B+/− mice, which were born at the expected ratio and phenotypically indistinguishable from mSin3B+/+ through 16 months of age (data not shown). mSin3B+/− intercrosses failed to produce mSin3B−/− offspring that survived beyond postnatal day 1 (Fig. 1c). Serial developmental analysis of these mSin3B+/− intercrosses revealed all three genotypes in the expected ratios through embryonic day 15.5 (E15.5) (Fig. 1c and data not shown), pointing to essential functions for mSin3B in late gestation.
mSin3B Is Required for Differentiation of Multiple Lineages.
By E14.5, mSin3B−/− embryos displayed overt growth retardation and pale color (Fig. 1d) and, by E17.5, mSin3B−/− embryos weighed 40% less than mSin3B+/+ and mSin3B+/− littermates (Fig. 1e). Histological analysis of serial sections of E12.5, E14.5, E16.5, and E18.5 mSin3B−/− embryos revealed no significant dysmorphologies at the earlier stages, whereas at later embryonic stages, there was global retardation in size and notable abnormality (data not shown). Specifically, the liver appeared much paler in mSin3B−/− embryos compared with mSin3B+/+ or mSin3B+/− embryos after E14 (Fig. 1d; data not shown). With regard to generalized impairment, the abnormal hematopoietic picture in the liver and much paler appearance of mSin3B−/− embryos prompted in-depth analysis of the hematopoietic compartment. Circulating blood smears from E18.5 mSin3B−/− embryos revealed the presence of immature nucleated erythrocytes and numerous erythrocytes with nuclear fragments, so-called Howell–Jolly bodies (Fig. 2a), a profile consistent with impaired erythroid differentiation. Correspondingly, the number of circulating red blood cells (RBCs) and hemoglobin levels were significantly lower in E18.5 Sin3B−/− embryos than mSin3B+/+ and mSin3B+/− littermates (Fig. 2b; 43% decrease in RBC numbers, P < 0.01; 37% decrease in hemoglobin levels, P < 0.01). Furthermore, FACS-based analysis of lineage-specific surface markers showed a strong decrease of mature granulocytes (Gr-1+) in E18.5 mSin3B−/− bone marrows (SI Fig. 6). These defects in erythrocytes and granulocytes of mSin3B−/− embryos are reminiscent of those in E2F4 and Mad mutant mice (14–16) and therefore establish a genetic link to these transcriptional repressors in vivo. In addition, colony-forming assays on fetal liver cells from Sin3B−/− embryos revealed a marked increase in the abundance of erythroid progenitors (>2-fold increase), and to a lesser extent, of granulocyte progenitors (1.4-fold increase) (Fig. 2b). Thus, Sin3B enables the terminal differentiation of multiple hematopoietic lineages.
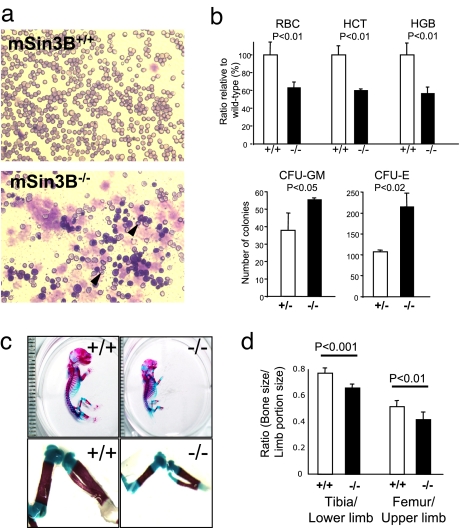
Fig. 2.
mSin3B is required for cellular differentiation during development. (a) Representative smears from peripheral blood of E18.5 mSin3B+/+ (Upper) or mSin3B−/− (Lower) embryos, stained with Wright–Giemsa. Note the high proportion of poorly differentiated erythrocytes in Sin3B-null blood, as evidenced by Howell–Jolly bodies (arrows) and nucleated cells. (b) (Upper) Analysis of peripheral blood defects in Sin3B-null mice. HCT, hematocrit; HGB, hemoglobin. Shown is the average from at least three E18.5 embryos for each genotype. The P values (P) calculated using a t test are indicated. (Lower) In vitro differentiation of E14.5 mSin3B+/− or mSin3B−/− fetal liver cells in CFU-GM or CFU-M. At least three animals were analyzed per genotype. The P values (P) are indicated. (c) Skeletal preparations of E18.5. Sin3B+/+ (Left) and Sin3B−/− (Right) embryos stained with Alcian blue (cartilage) and Alizarin red (bone). Note the diminished ossification of the long bones of the hind limb in the mSin3B−/− embryo. Each graduation corresponds to 1 mm. (d) Quantification of the ratio between bone and limb size in wild-type (white; n = 5) or null (black; n = 6) embryos for Sin3B. The P values are indicated.
The functional association of the Rb family and mSin3B-HDAC1 (ref. 13 and this study) and the established role of p107 and p130 in endochondral bone development (17) also prompted analysis of the developing skeletal system. The staining of E18.5 embryonic skeletons with dyes specific for cartilage (Alcian blue) and bone (Alizarin red) revealed significant reduction in bone deposition as reflected by decreased bone/cartilage ratios in the hindlimbs of mSin3B−/− embryos relative to mSin3B+/+ controls (Fig. 2 c and d). Specifically, lower and upper hind-limb ratios of mSin3B−/− embryos were 80% (P < 0.001) and 85% (P < 0.01) of mSin3B+/+ controls, establishing an essential role for mSin3B in skeletal development. These skeletal and hematopoietic defects associated with mSin3B deficiency, and the overlap with reported phenotypes of Rb/E2F mutant strains (17) provides genetic support of a biological interaction between mSin3B and the Rb-E2F network (see Discussion).
mSin3B Regulates Cell Cycle Exit.
The intimate interrelationship of cell cycle arrest and terminal differentiation in many cell lineages and the importance of Rb-E2F in cell cycle control prompted investigation of cell cycle kinetics and cell cycle exit control in multiple independent preparations of mSin3B+/+ and mSin3B−/− mouse embryonic fibroblasts (MEFs). In contrast to profound cell cycle arrest and death associated with deficiencies of either mSds3 or mSin3A (7, 9, 18), loss of mSin3B function exerted no significant impact on the cellular proliferation or the BrdU-positive fraction upon passage under standard culture conditions (Fig. 3 a and b, 10% serum). Western blot analysis documented no change in mSin3A expression in relation to mSin3B status (Fig. 3a). Upon passage in low serum conditions (0.1% serum for 72 h), mSin3B+/+ cells ceased to proliferate and entered quiescence, whereas mSin3B−/− cells incorporated ≈3-fold more BrdU relative to mSin3B+/+ cells (Fig. 3b; P < 0.01). Consistent with impaired G0 arrest upon serum deprivation in the absence of mSin3B, mSin3B-null cells exhibit altered cell cycle distribution under these conditions, as detected by BrdU–propidium iodide incorporation. Specifically, a lower proportion of mSin3B-null cells was in the G0/G1 phase, compared with their heterozygote counterparts (SI Fig. 7a). However, mSin3B-null cells are unable to proliferate in these conditions (SI Fig. 7c), in agreement with the observed accumulation in the S and G2/M phase of the cell cycle (SI Fig. 7a). Consistent with the lack of cell cycle progression, mSin3B+/+ and mSin3B−/− cells showed a comparable inability to form colonies upon low-density seeding (data not shown). Of note, upon serum stimulation, mSin3B+/+ and mSin3B−/− cells showed resumption of normal cell cycle kinetics (SI Fig. 7b). These observations suggest that mSin3B plays a role in controlling cell cycle exit under limiting growth factor conditions, yet preserves additional cell cycle checkpoints that prevent cellular proliferation under these conditions. Together, these findings point to a specific role for of mSin3B in G0/G1 control and agree well with accompanying defects in cellular differentiation in mSin3B-null embryos.
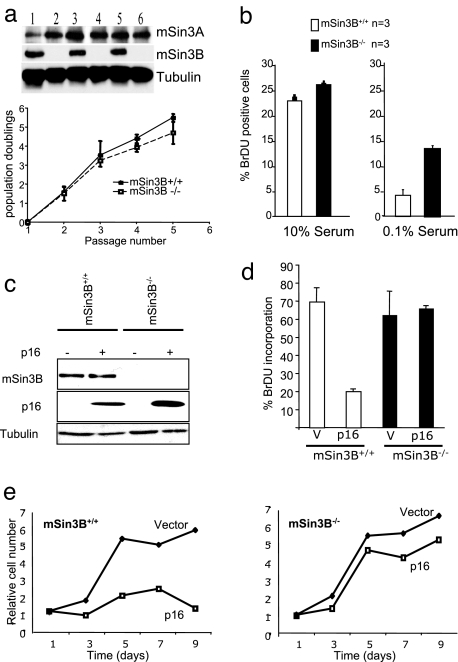
Fig. 3.
mSin3B is dispensable for cellular proliferation but required for cell cycle exit in vitro. (a) (Upper) Western blot analysis of mSin3B wild-type (lanes 1, 3, 5) and null (lanes 2, 4, 6) primary fibroblasts with the indicated antibodies. (Lower) Growth curves from mSin3B wild-type (n = 3) and null (n = 3) primary fibroblasts at passage 4. (b) BrdU incorporation after a 2-h pulse of 20 μM BrdU by mSin3B wild-type (white bars) or null (black bars) early-passage primary MEFs in 10% serum (Left) or 0.1% serum (Right). At least 200 cells were counted per point. Error bars indicate standard deviations. (c) Western blot of extracts from mSin3B wild-type or null MEFs infected or not with p16 using the indicated antibodies. (d) Proliferation of mSin3B wild-type (n = 3) or null (n = 3) MEFs after p16 or vector (V) infection, assayed by BrdU incorporation. At least 100 cells were counted for each point. (e) Growth curve of mSin3B wild-type (Left) or null (Right) MEFs after p16 or vector infection.
The role of mSin3B in cell cycle control was also examined in a setting other than growth factor regulation: p16INK4a induces growth arrest in wild-type cells through its ability to inactivate G1 cyclin-dependent kinases 4 and 6 (19). As shown in Fig. 3 c–e, retroviral transduction of p16INK4a into early passage mSin3B+/+ and mSin3B−/− MEFs resulted in decreased BrdU incorporation and cell proliferation in mSin3B+/+ cells, whereas mSin3B−/− cells were refractory to p16INK4a-induced growth arrest. Because E2F4 and E2F5, Rb, and at least one Rb-related protein are required for p16INK4a-mediated growth arrest (20), these cell culture findings, together with the in vivo phenotypes, reinforce the functional link between mSin3B and the Rb-E2F pathway.
mSin3B Contributes to the Repression of E2F Gene Targets in Vivo.
We have demonstrated that mSin3B plays a central function in cell cycle exit upon low-serum conditions. Quiescence corresponds to a G0 state of the cell cycle, which is characterized on the molecular level by E2F/p130-driven transcriptional repression of key cell cycle regulatory genes, including E2F1 (21) by mSin3B and E2F/p130 interactions on the promoter of E2F target genes in quiescent cells (13). To substantiate the physiological relevance of these cellular and biochemical observations, we determined the requirement for mSin3B in repressing the transcription of E2F target genes in vivo. To this end, we somatically deleted mSin3B via the IFN-inducible Mx-Cre transgenic system whose expression can be strongly induced by i.p. injection of polyinositic:polycytodylic acid (pIpC) that activates an endogenous IFN response (22). Mx-directed Cre expression is particularly robust in the liver, as evidenced by complete conversion of the Sin3BL allele to a Sin3B-null allele (Fig. 4a) and by total or near-complete loss of mSin3B protein in liver extracts only in mSin3BL/− Mx-Cre mice that were treated with pIpC (Fig. 4b).
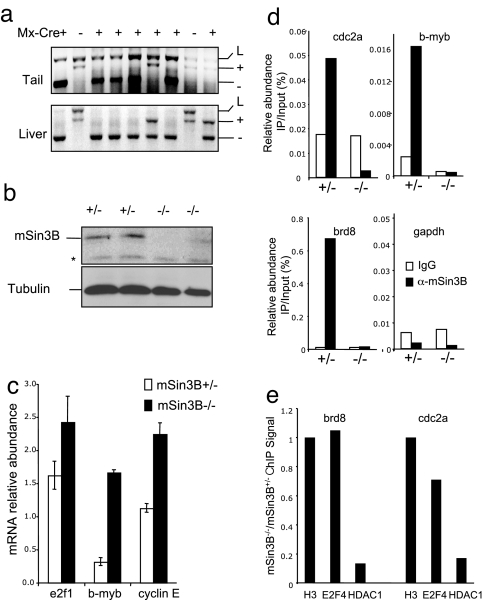
Fig. 4.
mSin3B directly represses E2F target genes transcription in vivo. (a) (Upper) PCR for mSin3B alleles on DNA from tail (before pIpC injection) or liver (10 days after pIpC injection) from Sin3B/Mx-Cre transgenic animals. (b) Western blot with the indicated antibodies from liver extracts 10 days after pIpC injection from mSin3BL/+Mx-Cre+ mice (+/−) or mSin3BL/−Mx-Cre+ mice (−/−). *, Nonspecific band. (c) qPCR analysis of mRNA abundance in mSin3B+/− or mSin3B−/− livers as generated in a. Shown is the average abundance of cDNAs after RT-PCR corresponding to the indicated transcripts, normalized to β-2-microglobulin. Three animals for each genotype were used. (d) ChIP assay on the promoter of the indicated E2F target genes and GAPDH, using liver extracts from mSin3BL/+Mx-Cre+ mice (+/−) or mSin3BL/−Mx-Cre+ mice (−/−) with control antibody (IgG, white bars) or anti-mSin3B antibody (black bars). Shown is the average of two independent experiments performed in duplicate. (e) Fold changes of the ChIP signal with E2F4 and HDAC1 antibodies on the promoter of cdc2a and brd8, in liver extracts from mSin3BL/−;Mx-Cre+ mice (−/−) compared with liver extracts from mSin3BL/+;Mx-Cre+ mice (+/−). The results are normalized to the corresponding H3 abundance. Shown is a representative experiment.
We analyzed the abundance of transcripts corresponding to several E2F4 target genes in livers taken from 4-week-old mice injected with pIpC. All E2F4 targets examined were significantly up-regulated from 1.5- to 5-fold in the mSin3B-deficient livers (Fig. 4c). To further strengthen the direct link between mSin3B and transcriptional repression of E2F targets, chromatin-immunoprecipitation assays were performed to determine whether mSin3B resides on the promoters of these E2F target genes in vivo. In mSin3B+/+ livers from 4-week-old mice, anti-mSin3B chromatin immunoprecipitates (ChIP) were significantly enriched for several E2F target promoters relative to an irrelevant antibody and GAPDH promoter controls (Fig. 4d). To ensure the mSin3B specificity of the PCR signals, we documented lack of enrichment of E2F target promoters above background in anti-mSin3B ChIP derived from mSin3B-deficient livers (Fig. 4d). Finally, to investigate the molecular mechanisms underlying mSin3B-driven repression of E2F targets, we investigated the impact of mSin3B inactivation on the enrichment in E2F4 and HDAC1 at E2F target promoters. As shown in Fig. 4e, E2F4 levels at the brd8 or cdc2a promoters are not significantly affected by loss of mSin3B. By contrast, HDAC1 recruitment at these promoters largely depends on the presence of mSin3B, because mSin3B inactivation results in a >5-fold reduction in HDAC1 levels by ChIP (Fig. 4e). Importantly, E2F4 and HDAC1 were not recruited at the GAPDH promoter, a non-E2F target (data not shown). These observations are consistent with the model that mSin3B is tethered to E2F target promoters by E2F repressors and plays a central role in the recruitment of chromatin modifiers at these loci. Together with previous genetic data, these biochemical assays establish a direct and physiologically relevant link between mSin3B and the transcriptional repression of E2F4 target genes in vivo via the capacity of mSin3B to tether histone deacetylases to E2F4-regulated promoters.
Discussion
In this study, we provide in vivo evidence of a role for mSin3B cell cycle exit control and repression of E2F gene targets and in the differentiation of erythrocytes and chondrocytes. The reported phenotypes are consistent with an essential contribution of mSin3B to E2F-mediated transcriptional repression. Finally, despite highly related structures and overlapping expression patterns of mSin3A and -B, their genetic inactivation in the mouse results in strikingly different cellular and organismal phenotypes, indicating functional dichotomy between the mSin3 proteins.
Our genetic and biochemical studies established a key role for mSin3B in E2F-mediated gene repression and cell cycle exit. Although these observations and overlapping differentiation defects support clear links to the p107–p130 complex, it is worth noting that p107–p130 double-null MEFs exhibit normal cell cycle arrest in low serum conditions (23), suggesting that mSin3B-driven repression extends beyond p107–p130 targets. It is tempting to speculate that an mSin3B–Rb interaction underlies this difference, because MEFs triply null for all Rb family members fail to arrest under low serum conditions (24, 25), and mSin3B deficiency is associated with overexpression of p107–p130 targets (b-myb, E2F1) and at least one Rb-specific target (cyclin E) (ref. 23; this study). Together, these results suggest that mSin3B plays a central role in repression driven by the entire Rb-related family of proteins in vivo. However, although most phenotypes reported in embryos inactivated for p107–p130 are found in mSin3B−/− embryos, Rb−/− embryos die significantly earlier than mSin3B−/− embryos. Specifically, Rb−/− embryos die around E13.5 from defective erythropoiesis and massive neuronal death (reviewed in ref. 26). Along with the decreased rate of S-phase progression of Sin3B−/− fibroblasts compared with Rb−/−;p107−/−;p130−/− fibroblasts in low serum conditions (refs. 24 and 25; this study), this suggests that Rb is likely to have mSin3B-independent functions in cell cycle exit and differentiation. In connection with this hypothesis, Binne et al. (27) recently demonstrated an E2F-independent mechanism for Rb-mediated inhibition of G1-to-S progression, mediated by a direct interaction between Rb and the APC/C complex. Alternatively, mSin3A may partially compensate for the absence of Sin3B in Rb-mediated cell cycle regulation, as suggested by the transcriptional profile and pathway analysis of mSin3A-deficient cells (7).
We have demonstrated here that mSin3B inactivation results in impaired cellular differentiation in multiple lineages, namely chondrocytes, erythrocytes, and granulocytes. Although cell cycle exit appears to be required for terminal differentiation of many lineages, the molecular machinery that integrates these interlocking events is not well understood. The opposite effects of Myc and Mad family proteins on cell proliferation and cellular differentiation, the impaired differentiation of granulocytes in Mad1- or mSin3B-deficient mice, and the physical interactions between mSin3B and Mad indicate that mSin3B plays an important role in executing Mad-directed processes in vivo. Similarly, comparable embryonic and cellular phenotypes associated with deficiencies in Rb family proteins, E2F4 and mSin3B, indicate that the Rb-E2F network is a major point of mSin3B action in vivo.
Methods
Generation of an mSin3B Conditional Null Allele.
A conditional mSin3B targeting construct was generated by flanking the second exon of mSin3B gene with a 5′ LoxP-Frt-pgk-neomycin cassette and a 3′ LoxP site (7, 9). After electroporation in 129/SvOla embryonic stem cells, stable recombinant clones were selected with neomycin and amplified. Correct homologous recombination was verified by Southern blot analysis using 3′ and 5′ probes, external to the targeting constructs. Targeted ES cell clones were injected into C57BL/6 blastocysts. Excision of the pgk-neo cassette was obtained upon breeding of the germ-line-targeted mice to CAGG-Flpe transgenic mice (28). Recombination of the two LoxP sites was obtained in vivo upon breeding with EIIACre transgenic mice (29). mSin3B+/− and mSinBL/L animals were maintained in a mixed C57BL/6-FVB background.
Analysis of Hematopoietic Defects.
mSin3B+/− mice were intercrossed, and embryos were collected at different time points. For circulating blood analyses, E18.5 embryos were collected, and circulating blood smears were stained by using the Hema 3 Stain system (Fisher) and analyzed by light microscopy. Complete blood counts from circulating blood were obtained by using a Cell-Dyn 4000 counter (Abbott).
In vitro hematopoietic progenitor assays were performed by using fetal liver cells from E15.5 embryos. Single-cell suspensions were plated on the appropriate methylcellulose medium (MethoCult M3334 for CFU-E and M3534 for CFU-GM, Stem Cell Technology) according to the manufacturer's instructions. The colonies were counted 3 (CFU-E) or 5 days (CFU-GM) after plating.
Skeletal Preparations.
E18.5 embryos were collected and treated with NaCl (1 M) overnight. Skin and internal organs were removed, and embryos were then fixed in 95% EtOH overnight. Cartilage was stained with Alcian blue (Sigma) for 24 h. Soft tissues were dissolved in 2% KOH overnight, and bones were stained with Alizarin red S (Sigma). Skeletons were destained for 4 days in 20% glycerol and 1% KOH and analyzed by light microscopy.
Cellular Analysis.
MEFs were generated from E13.5 embryos and grown in DMEM plus 10% FCS and penicillin/streptomycin. Retroviral infections were performed as described in ref. 9 with the appropriate retroviral constructs. Growth curves, BrdU incorporation, cell cycle distribution, and protein analysis were conducted as described in refs. 7 and 9.
Molecular Analyses of Sin3B-Deleted Hepatocytes.
Expression of the Cre recombinase was achieved upon three i.p. injections of 100 μg of pIpC over a 4-day period in 4-week-old Mx-Cre transgenic animals. Ten days after the first injection, mice were killed and livers collected. Proteins, DNA, and RNA were isolated from these livers by using standard procedures. To investigate transcript abundance, reverse transcription was performed by using MuMLV polymerase and oligo(dT) primers, followed by real-time PCR analysis using primers specific for E2F targets (primer sequences available on request). Results were reported as relative to the abundance of β-2-microglobulin transcripts. Chromatin immunoprecipitation on liver was performed following a protocol provided by the Barton laboratory (30) and followed by real-time PCR analysis using primers specific for E2F target promoters (sequences available on request).
Supplementary Material
Acknowledgments.
We thank James Horner and Michelle Neptune of the DFCI transgenic and gene-targeting facility for generating the targeted ES cells and the germ-line mouse strains. We acknowledge Yao Yao for advice in the design of the targeting vector, Jan-Hermen Dannenberg for helpful discussions, Samuel Perry for expert histology assistance, and Lili Miao for technical help in the initial phases of this project. We are indebted to Chris Van Oevelen for help with E2F target analyses, Branka Dabovic and Poshala Aluwihare for help with skeleton stainings, and Heather Harding for helpful discussion regarding hematopoietic defects. We are grateful to Michelle Barton and Wen-Wei Tsai for invaluable help with liver ChIP experiments. We thank Brian Dynlacht, Lawrence Gardner, David Levy, Isabelle Marie, and Lili Yamasaki for critical reading of the manuscript and helpful discussions This work was funded by National Institutes of Health Grant RO1CA86379 (to R.A.D.), the Whitehead Fellowship for Junior Faculty (to G.D.), the Rett Syndrome Research Foundation (G.D.), and the March of Dimes (G.D.). K.B.G. and N.S. were supported by National Institutes of Health Training Grants CA009161 and GM066704, respectively. R.A.D. is the American Cancer Society Research Professor and an Ellison Medical Foundation Senior Scholar and is supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710285105/DC1.
References
- 1.Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 2.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2004;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 3.Alland L, et al. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol Cell Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laherty CD, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 7.Dannenberg JH, et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechner T, et al. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J Biol Chem. 2000;275:40961–40966. doi: 10.1074/jbc.M005730200. [DOI] [PubMed] [Google Scholar]
- 9.David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 12.Balciunaite E, et al. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayman JB, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley KP, et al. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humbert PO, et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell. 2000;6:281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 16.Rempel RE, et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 17.Cobrinik D, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 18.Cowley SM, et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 20.Gaubatz S, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 21.Smith EJ, Leone G, DeGregori J, Jakoi L, Nevins JR. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 23.Hurford RK, Jr, Cobrinik D, Lee MH, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 24.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 27.Binne UK, et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 29.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25:2147–2157. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.