Abstract
We show that the times separating the birth of benign, invasive, and metastatic tumor cells can be determined by analysis of the mutations they have in common. When combined with prior clinical observations, these analyses suggest the following general conclusions about colorectal tumorigenesis: (i) It takes ≈17 years for a large benign tumor to evolve into an advanced cancer but <2 years for cells within that cancer to acquire the ability to metastasize; (ii) it requires few, if any, selective events to transform a highly invasive cancer cell into one with the capacity to metastasize; (iii) the process of cell culture ex vivo does not introduce new clonal mutations into colorectal tumor cell populations; and (iv) the rates at which point mutations develop in advanced cancers are similar to those of normal cells. These results have important implications for understanding human tumor pathogenesis, particularly those associated with metastasis.
Keywords: cancer genetics, colorectal cancer, metastasis, stem cells
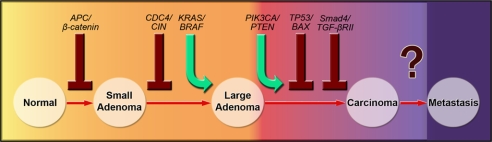
Colorectal tumorigenesis proceeds through well defined clinical stages associated with characteristic mutations (1, 2) (Fig. 1). The process is initiated when a single colorectal epithelial cell acquires a mutation in a gene inactivating the APC/β-catenin pathway (1). Mutations that constitutively activate the KRAS/BRAF pathway are associated with the growth of a small adenoma to clinically significant size (>1 cm in diameter) (3). Subsequent waves of clonal expansion driven by mutations in genes controlling the TGF-β (4, 5), PIK3CA (6), TP53 (7), and other pathways are responsible for the transition from a benign tumor (adenoma) to a malignant tumor (carcinoma). The only difference between a carcinoma and an adenoma is the ability of the former to invade the tissues underlying the colorectal epithelium. Some tumors eventually acquire the ability to migrate and seed other organs (metastasis) (8). Colorectal tumors can usually be cured by surgical excision at any stage before this last one, i.e., before metastasis to distant sites such as the liver (9).
Fig. 1.
Major genetic alterations associated with colorectal tumorigenesis. See SI Methods for further explanation.
Understanding the basic features of this evolutionary process has obvious and important implications for both scientific and medical research. But many questions remain. For example, how long does it take for a particular neoplastic cell to acquire the genetic events required for each sequential step in this progression? This question has heretofore been impossible to address in individual patients, although relevant information about bulk tumors, rather than cells, has been obtained through clinical and radiographic studies (10–12). We here describe an approach that can answer this and related questions.
Large-scale sequencing of the vast majority of protein-coding genes in human tumors has recently become possible and was applied to study the genomes of breast and colorectal cancers (13, 14). In the current study, we investigated whether the mutations discovered in the colorectal cancers evaluated in Wood et al. (14) were found in other neoplastic lesions from the same patients, an approach we call “comparative lesion sequencing.” We show that the sequencing data, when analyzed quantitatively, can be used to determine the time intervals required for development of the cells responsible for any two sequential clonal expansions. We were particularly interested in the expansion associated with metastasis. This final expansion is the least well understood at the biochemical and physiologic levels, even though it is responsible for virtually all deaths from the disease.
Results
Point Mutation Rates and Growth Kinetics of Colorectal Cancers.
Although knowledge of the precise mutation rate and tumor growth rates of these lesions are not required to make conclusions from comparative lesion sequencing, estimates of these parameters can inform their interpretation. An estimate of the point mutation rate in these tumors can be made on the basis of the results reported in ref. 14, wherein 847 nonsynonymous mutations were detected among 304 million bp sequenced at high quality. All of these mutations were somatic, i.e., not present in the germ line. Most of the lesions evaluated in ref. 14 were liver metastases, and all were mismatch-repair proficient. To convert the mutation prevalence data in ref. 14 to a mutation rate, it is necessary to know the number of divisions that the cancer cell had undergone. The most reliable way to measure cell-division time in human tumors is through the administration of DNA precursors such as BrdU to patients, followed by evaluation of BrdU incorporation plus DNA content by means of flow cytometry (15). This approach yields Tpot, defined as the time between cell divisions in the absence of cell death. Several hundred colorectal cancers have been evaluated by this method, with Tpot measured as ≈4 days (16–21). By using this figure for the cell division rate and the mutational data reported in ref. 14, the point mutation rate in colorectal cancers is estimated to be 4.6 × 10−10 mutations per base pair per generation. This rate is slightly less than that measured in various normal cell types [≈10 × 10−10 mutations per base pair per generation (22–25)]. Additional details about this estimate are provided in supporting information (SI) Methods.
Comparison of Mutations Before and After Cell Culture or Xenografting.
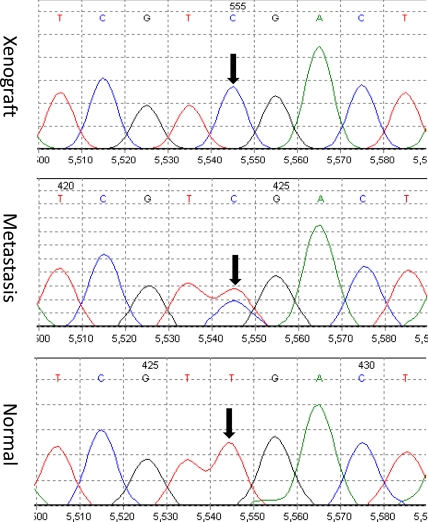
The samples used in ref. 14 were all derived from colorectal cancer cells that had been passaged for 6–12 months in vitro as cell lines or in nude mice as xenografts. Before initiating the current study of other lesions from the same patients, it was important to determine whether the mutations identified in the cultured or xenografted cells were actually present in the naturally occurring lesions before cellular expansion ex vivo. For this purpose, we analyzed 289 different mutations found in 18 cell lines or xenografts, each derived from a different patient. Five of these had been initiated and passaged in vitro, whereas the remaining 13 had been passaged as xenografts in nude mice. Two hundred eighty-seven of the 289 mutations (99.3%) found in the cell lines or xenografts were also found in the original tumors. The direct Sanger sequencing method we used in the experiments reported herein had a sensitivity of ≈25%, so that a heterozygous mutation present in <50% of the cells would not be observed (Fig. 2). These data therefore indicate that the point mutations found in colorectal cancer cell lines or xenografts only rarely arise during in vitro or in vivo experimental growth of cells after the tumors are excised from patients.
Fig. 2.
Representative examples of sequencing chromatograms of DNA from a xenograft, from the metastatic lesion from which the xenograft was derived, and from the patient's normal cells. Note that the ratio of the mutant to wild-type allele in the xenograft is higher than that in the metastatic lesion because the latter represented a mixture of neoplastic and nonneoplastic cells (stroma, white blood cells, etc.). The arrow points to the mutated base.
Comparison of Metastases with Primary Colorectal Cancers.
Paired samples of primary colorectal cancers and metastatic lesions from 10 patients were available for this study. Of the index lesions (Table 1), seven had been excised from the liver and three from mesenteric lymph nodes. We were able to evaluate an average of 28 mutations per lesion in the patients evaluated in the Discovery screen of ref. 14. The remaining index lesions had been studied only in the Validation screen, so only approximately five mutations per patient could be studied in these cases. In all, 233 somatic mutations identified in the index metastases were evaluated in the 10 cases. Of these, only seven [3.0%, 90% confidence interval (C.I.) 1.5–5.7%] were not found in the colorectal cancers from which the metastases arose (SI Table 2).
Table 1.
Summary of patient information
| Patient no. | Wood et al. (14) ID no. | Age at diagnosis | Sex | Location of colorectal tumor | TNM stage* | Site of index lesion | No. of mutations in colorectal adenoma/no. in carcinoma† | No. of mutations in colorecal carcinoma/no. in index metastasis† | No. of other metastases | No. of mutations in other metastases/no. in index metatasis† |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mx27 | 73 | F | Ascending | T3N1M0 | Liver | NA | 47/47 | 3 | 24/24 |
| 2 | Mx29 | 50 | M | Descending | T4N1M1 | Liver | NA | 7/7 | 3 | 17/17 |
| 3 | Mx34 | 83 | F | Cecum | T4N2M1 | Lymph node | 17/22 | 24/25 | 4 | 31/31 |
| 4 | Mx40 | 75 | F | Cecum | T4N1M0 | Lymph node | NA | 5/5 | 3 | 9/9 |
| 5 | Mx43 | 72 | M | Sigmoid | T3N2M1 | Liver | NA | 48/50 | 5 | 98/98 |
| 6 | Co92 | 47 | F | Cecum | T3N2M0 | Liver | NA | 8/8 | 0 | NA |
| 7 | Mx32 | 55 | F | Ascending | T3N1M0 | Liver | NA | 28/32 | 3 | 39/45 |
| 8 | Co84 | 41 | M | Cecum | T4N2M1 | Lymph node | NA | 4/4 | 0 | NA |
| 9 | Mx38 | 65 | M | Rectum | yT3N1M0 | Liver | NA | 6/6 | 3 | 17/17 |
| 10 | Co82 | 80 | F | Cecum | T3N1M0 | Colon | 6/11 | NA | 1 | 5/5 |
| 11 | Mx26 | 46 | F | Cecum | T2N2M1 | Liver | NA | NA | 1 | 3/3 |
| 12 | Co108 | 76 | F | Ascending | T4N0M1 | Liver | NA | NA | 1 | 6/6 |
| 13 | Mx41 | 55 | M | Ascending | T3N1M1 | Liver | NA | 49/49‡ | 2 | 6/6‡ |
| Average or total | 63 | 23/33 | 226/233 | 29 | 255/261 | |||||
NA, not applicable because indicated comparison could not be performed.
* T2, carcinoma invaded muscularis propria; T3, carcinoma invaded through muscularis propria into submucosa; T4, carcinoma invaded through wall of colon into nearby tissues or organs; N0, no lymph node involvement; N1, cancer cells found in one to three nearby lymph nodes; N2, cancer found in more than three nearby lymph nodes; M0, no distant metasases identified; M1, distant metastasis identified; a “y” before the TNM stage means that the patient was treated with chemoradiotherapy prior to surgery to reduce the size of the lession.
†The numbers refer to the mutations that could be successfully sequenced. Not all mutations in an index metastatic lesion could be sequenced in other lesions of the same patient because of limitations in available material.
‡There were 49 mutations detected in the liver and two lymph node metastases that were removed at the time of surgery. A new metastasis developed 29 months later, after chemotherapy. This late metastasis contained 19 new mutations that were not present in the original metastases or carcinoma and are not included in this table (see text).
Comparisons of Different Metastatic Lesions.
In 11 of the 13 patients, we were able to examine at least one metastatic lesion different from the index lesion of the same patient. Of a total of 261 mutations evaluated, 255 (97.7%) were found in the 29 additional metastases studied (Table 1). This was the expected result, because the great majority of the mutations present in metastases were also present in the precursor advanced colorectal carcinoma, as noted above. What was more informative was the study of patients in whom mutations were identified in the metastasis but not in the precursor advanced carcinoma (henceforth denoted “metastasis-specific mutations”). Although there were only seven of these mutations identified, five of them were particularly informative because they could be assessed in other metastatic lesions from the same patients. In patient 5, both of the metastasis-specific mutations originally identified in the index liver metastasis were also identified in a mesenteric lymph node metastasis. In patient 7, three liver metastasis-specific mutations identified in the index metastasis were identified in a second, independent liver metastasis concurrently excised from the patient.
Comparison of Advanced Colorectal Carcinomas with Large Adenomas.
It is believed that colorectal carcinomas arise in preexisting, benign adenomas through the acquisition of additional genetic alterations (Fig. 1). In most cases, the adenomatous tissue is destroyed during carcinoma growth. However, in two of the cases studied here, large residual adenomas at the edges of the carcinomas were still present (Fig. 3). Paraffin-embedded sections of these lesions were carefully microdissected to separate adenomatous from carcinomatous elements and evaluated for 33 mutations known to be present in the carcinoma. Ten of the 33 mutations (30%) were not found in the adenomatous components (see SI Table 2). The differences between the fraction of mutations found in metastases but not their precursor advanced carcinomas and the fraction of mutations found in the advanced carcinomas but not in their large precursor adenomas were statistically significant (P < 0.001, two-group binomial test for equality of proportion).
Fig. 3.
Histopathology of representative lesions. (A) Primary invasive moderately differentiated adenocarcinoma (enclosed by black boundary) arising in a tubular adenoma (enclosed by red boundary) from patient 10. (B) Primary invasive moderately differentiated adenocarcinoma (enclosed by black boundary) with adjacent nonneoplastic colonic mucosa (enclosed by red boundary) from patient 2. (C) Metastatic adenocarcinoma (enclosed by black boundary) to liver (enclosed by red boundary) derived from primary colon adenocarcinoma of patient 2. All sections were stained with H&E, and the tissues within each boundary were separately microdissected.
Of the 10 mutations identified in advanced carcinomas but not large adenomas, 7 were in candidate cancer genes (CAN-genes) as defined in Wood et al. (14). This proportion is significantly different from the proportion of metastasis-specific mutations (14%) that were in CAN-genes (P < 0.001, two-group binomial test) or the proportion of mutations that were in CAN-genes among all genes with mutations (16%, P < 0.01, two-group binomial test). Two of the 10 mutations identified in carcinomas but not in their precursor adenomas were in TP53, consistent with prior data on the timing of TP53 mutations (26).
Quantification of the Level of Mutations in DNA.
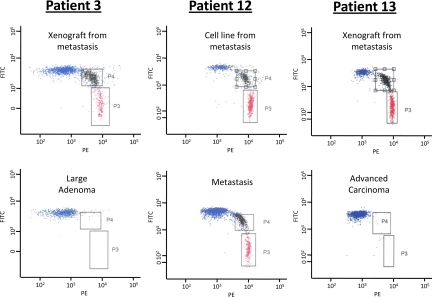
The absence of a somatic mutation in a given DNA sample, as assessed by Sanger sequencing, simply means that the mutation was not present in >25% of the analyzed DNA template molecules (i.e., >50% of the cells in the case of heterozygous mutations). Mutations present in a smaller fraction can generally not be distinguished from the background in sequencing chromatograms. To determine whether the mutations were present in a smaller but still sizable fraction of the tumor cell population, we evaluated a subset of the DNA samples via BEAMing (beads, emulsions, amplifications, magnetics) assay (see SI Methods) (27, 28). We performed 20 BEAMing assays in seven patients, focusing on those mutations that appeared to be present in late-stage lesions but not in an earlier-stage lesion of the same patient (e.g., present in metastasis but not in the advanced colorectal carcinoma). In 19 of these assays, no mutations were observed (examples in Fig. 4). Because the sensitivity of the BEAMing assays was ≈0.01%, we conclude that <1 in 2,500 cells in the precursor lesion contained any of these 19 mutations, thus suggesting that at least one major clonal expansion occurred between the two stages analyzed in each case.
Fig. 4.
Representative examples of BEAMing assays from the indicated patients and lesions. In patient 13, the mutation shown represents one that was present in a new metastasis that occurred 29 months after chemotherapy (see Application to Individual Patients). The red dots correspond to beads attached to mutant DNA fragments [labeled with phycoerythrin (PE)], the blue dots correspond to beads attached to WT DNA fragments [labeled with fluorescein (FITC)], and the black dots correspond to beads attached to both WT and mutant DNA fragments.
Colorectal Cancer Evolution: Mathematical Assessment.
The data in Table 1 can be used to determine the relative timing of the birth of the founder cells (Fcells) that gave rise to the various tumor cell populations described above (Fig. 5). The basis for this analysis is that all somatic mutations present in clonal fashion in an adenoma (i.e., present in all cells of the tumor) must have been present in its cell of origin (its founder cell). These mutations accumulated during the life span of this founder cell and include those that occurred during the turnover of normal stem cells before the onset of tumorigenesis. As tumors progress, they accumulate additional mutations that become fixed in the founder cells of subsequent neoplastic states. The founder cell of the advanced carcinoma, for example, will harbor all of the mutations present in the precursor adenoma plus additional mutations that occurred in the interim. The length of this interim period can be estimated by measuring the number of additional mutations in the progressed lesion.
Fig. 5.
Evolution of a lethal cancer. Each cell-filled cone represents one or more clonal expansions (see SI Methods for details). The times required for the evolution of the large adenoma founder cell to an advanced carcinoma founder cell (ΔLAd,ACa) and evolution of the advanced carcinoma founder cell to metastatic founder cell (ΔACa,Met) were determined by comparative lesion sequencing. Other intervals, such as the time (Texp) required for the expansion of the metastasis founder cell FCellMet to the size detected in our patients, were estimated as described in SI Methods. The model posits that there are at least two clonal expansions, denoted by question marks, that are not associated with any known genetic alterations.
The founder cells of interest are (i) the one (FcellMet) that gave rise to the final clonal expansion resulting in the index metastasis; (ii) the last common ancestor (FcellACa) of the advanced carcinoma and FcellMet; and (iii) the last common ancestor (FcellLAd) of the large adenoma and FcellACa. The birth date (T) of a founder cell is defined as the age of the patient when the founder cell underwent its first division. As shown in the SI Methods, the interval (ΔTACa,Met) between the birth date of founder cells FcellMet and FcellACa can be approximated as
where FACa,Met is the fraction of the mutations in the metastasis that were not found in the advanced carcinoma (i.e., 1 − [number of mutations in advanced carcinoma/number of mutations in metastasis]). Similarly, the interval (ΔTLAd,ACa) between the birth dates TLAd and TACa of founder cells FcellACa and FcellLAd, respectively, can be approximated as
where FLAd,ACa is the fraction of mutations in the advanced carcinoma that were not found in the large adenoma. Similar equations can be applied to any two lesions that represent the clonal expansions of two founder cells as long as one of the two founder cells is a direct descendent of the other.
Note that these equations are entirely independent of the actual mutation rates and cell division times (Tpot), which likely vary among different patients and cancers. They only require that the mutation rate and cell division times, whatever they are, are constant throughout each patient's life. The approximations in Eqs. 1 and 2 are accurate as long as the number of mutations with positive or negative effects upon growth is small compared with the number of neutral mutations. As detailed in SI Methods, this requirement is, in general, expected to be met. Mutations thereby act as a clock, providing information similar to that obtained through the use of sequence divergence to assess the relatedness of organisms or cells during evolution or development (29, 30).
Application to Individual Patients.
One of the major results of the current study is that FLAd,ACa is much greater than FACa,Met, meaning that it takes much longer for a large adenoma to evolve into an advanced carcinoma than for such a carcinoma to metastasize. Assumptions that limit the accuracy of the times determined through these equations are given in SI Methods. Their implementation can best be illustrated through their application to five patients in the current study in whom a minimum of 25 mutations could be evaluated (Table 1).
Patient 1 was 73 years old when she developed an advanced carcinoma of the ascending colon that was 4 cm in diameter and of stage T3N1M0 (T3 refers to the stage of the carcinoma, which, in this case, had grown completely through the muscularis propria; N1 indicates that cancer was found in at least one but less than four lymph nodes; M0 indicates that no distant metastases were evident at the time of surgery). Fifteen months later, a liver metastasis of 5 cm in diameter was found to have developed. All 47 mutations found in the metastasis were also found in the advanced carcinoma in the colon (FACa,Met = 0.0). Application of Eq. 1 indicated that the metastasis originated from a cell (FcellMet) whose birth occurred very soon after the birth of the cell (FcellACa) that founded the advanced carcinoma (C.I., 0–3.4 years).
Patient 3 was 83 years old when she developed an advanced carcinoma of the ascending colon that was 9 cm in diameter and of stage T4N2M1 (N2 indicates that cancers cells were found in more than three mesenteric lymph nodes). A residual adenoma that surrounded the carcinoma was identified at the time of surgery. A small (<1-cm diameter) mesenteric lymph node metastasis was found to contain 25 mutations that were subsequently evaluated in other lesions of this patient. Of these, 24 were found in the colorectal carcinoma (FACa,Met = 0.04). Application of Eq. 1 indicated that the advanced carcinoma founder cell was born 3.2 years (C.I., 0.4–7.1 years) before the lymph node metastasis founder cell was born. In contrast, evaluation of the same mutations in the large adenoma from which the carcinoma developed revealed an FLAd,ACa of 0.23. Application of Eq. 2 indicated that the large adenoma founder cell was born 17 years (C.I., 7.9–30.9 years) before the advanced carcinoma founder cell. In the ≈17 years between the birth of FcellLad and FcellACa, the tumor underwent waves of clonal expansion driven by mutations in TP53 and the other genes (SI Table 2) presumably required for invasion and further growth of this tumor. Once it acquired these capabilities, a cell (FcellMet) capable of lymph node metastasis appeared within a relatively short period.
Patient 5 was 72 years old when he developed an advanced carcinoma of the sigmoid colon that was 1.5 cm in diameter and of stage T3N2M1, accompanied by an 8.9-cm liver metastasis. Comparative lesion sequencing indicated that the metastasis founder cell FcellMet was born 2.8 years (C.I., 0.6–4.9 years) after the birth of the advanced carcinoma founder cell FcellACa. A large (1.3-cm) mesenteric lymph node metastasis and two smaller mesenteric lymph node metastases were also evaluated from this patient. The larger lymph node contained the same 50 mutations identified in the liver metastasis, including the two mutations not found in the primary colorectal carcinoma; the two smaller lymph nodes did not contain these two mutations. Thus, the 1.3-cm mesenteric lymph node metastasis and liver metastasis founder cells may have both been derived from a small population of cells within the carcinoma that had acquired metastatic capability. Alternatively, the liver metastasis could have originated from the large mesenteric lymph node metastasis. In this case, comparative lesion sequencing indicates that the liver metastasis founder cell must have been born soon after (0 years; C.I., 0.0–2.0 years) the birth of the of lymph node metastasis founder cell.
Patient 7 was 55 years old when she developed an advanced carcinoma of the ascending colon that was 3.5 cm in diameter and stage T3N1M0. Twenty months later, two metastases of 3.5- and 4-cm diameter were found in the liver. Comparative lesion sequencing of the 4-cm liver metastasis and the colorectal cancer indicated the metastasis founder cell was born 6.6 years after the carcinoma founder cell (C.I., 1.8–8.6 years). Two mesenteric lymph node metastases removed at the time of the initial surgery and the 3.5-cm liver metastasis noted above were also evaluated. Three metastasis-specific mutations were identified in both liver metastases but not in either nodal metastasis.
Patient 13 was 55 years old when he developed an advanced carcinoma of the ascending colon that was 2.5 cm in diameter and stage T3N1M1. A 7-cm metastasis in the right lobe of the liver and a metastasis in a mesenteric lymph node were removed at the time of surgery. Twenty-nine months after this resection, a new liver metastasis of 3.1-cm diameter was detected in the left lobe and completely excised. One year later, another metastasis in the liver, of diameter 3.5 cm, was identified. The metastases that were identified 29 and 41 months after the initial diagnosis both had 19 mutations that were not found in the advanced carcinoma or metastatic lesions excised at the initial surgery, with FACa,Met = 0.28. In contrast, all of the mutations identified in the metastatic lesions removed at the initial surgery were also present in the advanced carcinoma removed concurrently. We interpret this result in the following way. Chemotherapy consisting of irinotecan, leucovorin, and 5-FU administered in the 9 months after the initial surgery pruned most of the micrometatastic cells remaining in the liver. One of these cells was resistant to the chemotherapy and became the founder cell of the new metastasis and its later recurrence. The chemotherapy had induced many new mutations in this cell, consistent with the known mutagenicity of irinotecan and perhaps exacerbated by the 5-FU (31). Eq. 1 cannot be used to estimate the relative birth date of this cell because comparative lesion sequencing requires that the mutation rate be constant throughout the tumorigenic process (see SI Methods). It is notable that this patient was the only one of the patients analyzed in depth in our study who had been treated with irinotecan before the development of a new metastatic lesion.
Discussion
A Temporally Defined Model of Colorectal Cancer.
The data and approach used in the current study can be used to temporally model some of the key genetic events in colorectal tumorigenesis. As illustrated in Fig. 5, comparative lesion sequencing suggests that the average time interval between the birth of a large adenoma founder cell and the birth of an advanced carcinoma founder cell is 17 years (C.I., 10.9–26.5 years). However, the average interval between the birth of the advanced carcinoma founder cell and the liver metastasis founder cell is only 1.8 years (C.I., 0.9–3.1 years).
Information about the birth times of the founder cells giving rise to various neoplastic stages has not heretofore been available. However, our estimates of these values are consistent with clinical and radiological observations on bulk tumors. For example, the time between the appearance of small adenomas and the diagnosis of a carcinoma has been estimated at 20–25 years from studies of patients with familial adenomatous polyposis (11). Similarly, serial studies of sporadic colorectal tumor patients have indicated that the transition from large adenoma to carcinoma takes ≈15 years (11). Our estimates are also consistent with the long doubling times of tumors determined by serial radiologic studies or serial measurements of the CEA serum biomarker (10, 12, 32, 33). Such studies have indicated mean doubling times that are generally 2–4 months in metastases and much longer in adenomas and carcinomas.
Biological Implications.
Our findings suggest that virtually all of the mutations necessary for metastasis are already present in all of the cells of the antecedent carcinoma. These data are compatible with two distinct models. In model A, none of the carcinoma cells can give rise to a metastasis, but they are close to being able to do so; one or a few more genetic alterations are required. In model B, all of the carcinoma cells can give rise to metastasis; no more genetic alterations are required. Data derived from the current study, involving comparisons of different metastatic lesions from the same patients, are compatible with either model.
Model A.
If every cell in the cancer cell population were capable of giving rise to a metastasis, it is extremely unlikely that any two independent metastases would harbor an identical mutation not found in the carcinoma. However, as described in Results, we identified five metastasis-specific mutations that were each present in more than one metastasis from the same patient (patients 5 and 7). If the founder cells of one of these two metastases were not a direct descendent of the other, these data would support the idea that a small population of cells within the carcinoma had acquired additional alterations that endowed them with the capacity to metastasize. Such alterations could be the point mutations actually identified as metastasis-specific (SI Table 2) or any other heritable event [whole chromosome gains or losses, chromosome translocations or amplifications, or certain epigenetic changes (34)].
Model B.
General support for this model comes from the fact that there so few additional alterations identified in the metastases compared with the advanced carcinomas. The finding that mutations not found in the carcinoma were identified in two anatomically distinct metastases could be explained if the founder cells of the two metastases had both come through a bottleneck after they migrated from the primary colorectal carcinoma. In Patient 5, this could have occurred if the liver metastasis had developed from a cell within the mesenteric lymph node metastasis that contained the same mutations. In Patient 7, this could have occurred if both liver metastases' founder cells had developed in lymph node metastases that were not detected or excised.
The reason that progression of the large adenoma to advanced carcinoma takes so much longer than the progression of the latter to metastasis is presumably because many more mutations and clonal expansions are required (some of which are indicated in Fig. 5). Moreover, some of the genes responsible for the adenoma-to-carcinoma progression have been identified (SI Table 2 and Fig. 5). One reasonable interpretation of the data is that the capacity to invade through layers of the bowel wall without dying, thereby becoming an advanced colorectal cancer, is the most challenging step in the process that eventually leads to metastasis. Once that step occurs, few additional steps are required for metastasis to take off. The advent of large-scale cancer genome sequencing provides uniquely valuable biomarkers to study tumor evolution. The study of additional mutations and lesions using the approach described in this work could definitively answer a variety of long-standing questions about the basic nature of the metastatic process in humans (35–39).
Materials and Methods
DNA samples from tumor samples or their derived xenografts or cell lines were obtained and purified by using slight modifications of those described (13). Two hundred eighty-nine exons in which a mutation had been identified in an index lesion studied in refs. 13 or 14 were PCR-amplified in all other available DNA samples from the patient. DNA samples from xenografts, cell lines, and frozen tissues were amplified by using the primers described (14). New amplicons of smaller size were designed for the DNA purified from paraffin-embedded samples. When sequencing chromatograms were difficult to interpret in the DNA purified from tumor samples, we reevaluated the mutation in question either by cloning the PCR product and sequencing individual clones or by performing a BEAMing assay (27, 28). Additional, more detailed methods are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank D. H. Nguyen for preparing Fig. 5; J. Lutterbaugh and L. Kasturi for expert technical assistance; Y. He for advice about flow cytometry; and D. Shibata, D. Williams, and J. T. Vogelstein for critical comments. This work was supported by the Virginia and D. K. Ludwig Fund for Cancer Research, Public Health Service Grants CA43460, CA57345, CA62924, CA121113, CA116867, CA120237, CA127306, CA043703, CA62924, CA105090, and GM078986, and by a gift from the National Colon Cancer Research Alliance. N.B. was partially supported by a grant from the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative.
Footnotes
Conflict of interest statement: Under separate licensing agreements between The Johns Hopkins University and Exact Sciences Corporation and Genzyme Corporation Oncology, V.E.V., K.W.K., and B.V are entitled to a share of royalty received by the University on sales of products described in this article/presentation. V.E.V., K.W.K., and B.V and the University own Genzyme Molecular Oncology stock, which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712345105/DC1.
References
- 1.Kinzler KW, Vogelstein B. Lessons from hereditary colon cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz S, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 5.Thiagalingam S. Evaluation of chromosome 18q in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 7.Baker SJ, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 8.Joyce DL, et al. Preoperative positron emission tomography to evaluate potentially resectable hepatic colorectal metastases. Arch Surg. 2006;141:1220–1226. doi: 10.1001/archsurg.141.12.1220. discussion, p 1227. [DOI] [PubMed] [Google Scholar]
- 9.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, et al. Metastatic tumor doubling time: Most important prehepatectomy predictor of survival and nonrecurrence of hepatic colorectal cancer metastasis. World J Surg. 2004;28:263–270. doi: 10.1007/s00268-003-7088-3. [DOI] [PubMed] [Google Scholar]
- 11.Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer. 1975;36:2251–2270. doi: 10.1002/cncr.2820360944. [DOI] [PubMed] [Google Scholar]
- 12.Welin S, Youker J, Spratt JS., Jr The rates and patterns of growth of 375 tumors of the large intestine and rectum observed serially by double contrast enema study (Malmoe technique). Am J Roentgenol Radium Ther Nucl Med. 1963;90:673–687. [PubMed] [Google Scholar]
- 13.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Terry NH, White RA. Flow cytometry after bromodeoxyuridine labeling to measure S, G2+M phase durations plus doubling times in vitro and in vivo. Nat Protoc. 2006;1:859–869. doi: 10.1038/nprot.2006.113. [DOI] [PubMed] [Google Scholar]
- 16.Michel P, et al. Comparison between endoscopic and surgical sampling for the measurement of potential doubling time in colorectal cancer. Cytometry. 1997;29:273–278. doi: 10.1002/(sici)1097-0320(19971101)29:3<273::aid-cyto11>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.Schiepers C, et al. The effect of preoperative radiation therapy on glucose utilization and cell kinetics in patients with primary rectal carcinoma. Cancer. 1999;85:803–811. doi: 10.1002/(sici)1097-0142(19990215)85:4<803::aid-cncr7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 18.Terry NH, et al. Cellular kinetics in rectal cancer. Br J Cancer. 1995;72:435–441. doi: 10.1038/bjc.1995.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rew DA, Wilson GD, Taylor I, Weaver PC. Proliferation characteristics of human colorectal carcinomas measured in vivo. Br J Surg. 1991;78:60–66. doi: 10.1002/bjs.1800780120. [DOI] [PubMed] [Google Scholar]
- 20.Fuller CE, Davies RP, Williams GT, Williams ED. Crypt restricted heterogeneity of goblet cell mucus glycoprotein in histologically normal human colonic mucosa: A potential marker of somatic mutation. Br J Cancer. 1990;61:382–384. doi: 10.1038/bjc.1990.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okayasu I, et al. Significant increase of colonic mutated crypts correlates with age in sporadic cancer and diverticulosis cases, with higher frequency in the left- than right-side colorectum. Cancer Sci. 2006;97:362–367. doi: 10.1111/j.1349-7006.2006.00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araten DJ, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 24.DeMars R, Held KR. The spontaneous azaguanine-resistant mutants of diploid human fibroblasts. Humangenetik. 1972;16:87–110. doi: 10.1007/BF00393992. [DOI] [PubMed] [Google Scholar]
- 25.Elmore E, Kakunaga T, Barrett JC. Comparison of spontaneous mutation rates of normal and chemically transformed human skin fibroblasts. Cancer Res. 1983;43:1650–1655. [PubMed] [Google Scholar]
- 26.Baker SJ, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 27.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: Single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 29.Nowak MA. Evolutionary Dynamics. Cambridge, MA: Harvard Univ Press; 2006. [Google Scholar]
- 30.Shibata D, Tavare S. Counting divisions in a human somatic cell tree: How, what and why? Cell Cycle. 2006;5:610–614. doi: 10.4161/cc.5.6.2570. [DOI] [PubMed] [Google Scholar]
- 31.Baguley BC, Ferguson LR. Mutagenic properties of topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:213–222. doi: 10.1016/s0167-4781(98)00137-7. [DOI] [PubMed] [Google Scholar]
- 32.Koga H, Moriya Y, Akasu T, Fujita S. The relationship between prognosis and CEA-dt after hepatic resection in patients with colorectal carcinomas. Eur J Surg Oncol. 1999;25:292–296. doi: 10.1053/ejso.1998.0644. [DOI] [PubMed] [Google Scholar]
- 33.Staab HJ, Anderer FA, Hornung A, Stumpf E, Fischer R. Doubling time of circulating cea and its relation to survival of patients with recurrent colorectal cancer. Br J Cancer. 1982;46:773–781. doi: 10.1038/bjc.1982.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ptashne M. On the use of the word “epigenetic.”. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 35.Talmadge JE. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 2007;67:11471–11475. doi: 10.1158/0008-5472.CAN-07-2496. [DOI] [PubMed] [Google Scholar]
- 36.Fidler IJ. Origin and biology of cancer metastasis. Cytometry. 1989;10:673–680. doi: 10.1002/cyto.990100602. [DOI] [PubMed] [Google Scholar]
- 37.Waghorne C, Thomas M, Lagarde A, Kerbel RS, Breitman ML. Genetic evidence for progressive selection and overgrowth of primary tumors by metastatic cell subpopulations. Cancer Res. 1988;48:6109–6114. [PubMed] [Google Scholar]
- 38.Price JE, Bell C, Frost P. The use of a genotypic marker to demonstrate clonal dominance during the growth and metastasis of a human breast carcinoma in nude mice. Int J Cancer. 1990;45:968–971. doi: 10.1002/ijc.2910450532. [DOI] [PubMed] [Google Scholar]
- 39.Scheel C, Onder T, Karnoub A, Weinberg RA. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 2007;67:11476–11479. doi: 10.1158/0008-5472.CAN-07-1653. discussion, pp 11479–11480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.