Abstract
Protein acetylation and deacetylation play key roles in multiple physiological functions. Histone deacetylase 3 (HDAC3) is a highly conserved, ubiquitously expressed protein that forms multiprotein corepressor complexes to repress gene transcription. Recent studies show that HDAC3 may play a role in cell proliferation. Altered HDAC3 level increases G2/M cells, but the mechanism remains unknown. Here we show for the first time, to our knowledge, that the HDAC3 complex, including nuclear receptor corepressor (N-CoR), transducin-β-like protein 1 (TBL1), and TBL1-related protein 1 (TBLR1), is localized on the mitotic spindle. Knockdown of HDAC3 or N-CoR resulted in a collapsed mitotic spindle that was surrounded by chromosomes arranged in a dome-like configuration. Treatment of mitotic cells with Trichostatin A, an HDAC inhibitor, resulted in similar spindle defects independent of transcriptional regulation. In addition, wild-type HDAC3 but not a deacetylase-dead mutant HDAC3 rescued the phenotypes of HDAC3-depleted cells, suggesting that the enzymatic activity of HDAC3 is important for proper spindle function. Whereas the kinetochores and the spindle assembly checkpoint appeared intact in HDAC3-deficient cells, kinetochore–microtubule attachments were impaired because spindle microtubules were unstable in response to cold treatment. These data suggest that the HDAC3 complex is involved in the formation of functional mitotic spindles and proper kinetochore–microtubule attachment. The level or distribution of acetylated α-tubulin was not altered in HDAC3-deficient cells. Taken together, our studies raise the interesting possibility that acetylation–deacetylation of mitotic spindle components may be essential for mitotic spindle function.
Keywords: HDAC3, corepressor, acetylation, tubulin, mitosis
Histone acetylation status plays a critical role in epigenetic regulation of gene transcription. In addition to histones, recent studies revealed that acetylation of non-histone proteins contributes to multiple protein functions including DNA binding, protein stability, and protein–protein interaction (1). Reversible protein acetylation is also involved in the regulation of cell cycle and proliferation. Inhibitors of histone deacetylases (HDACs) are antiproliferative agents as well as potential therapeutic drugs for cancers. These drugs arrest the cell cycle at G1 and G2/M phase and induce apoptosis, but the precise mechanism remains unknown (2).
Among numerous HDACs, HDAC3 is ubiquitously expressed and conserved in a wide range of species (3–6). In mammalian cells, HDAC3 has been shown to form large corepressor complexes containing N-CoR/SMRT and additional proteins (7–11). Besides its activity to repress transcription, HDAC3 also regulates signal transduction (10, 12), apoptosis (13, 14), and cell viability (15). HDAC3 also plays important roles in the cell cycle. Both overexpression and depletion of HDAC3 lead to the accumulation of cells in G2/M phase (16, 17). A recent study reported that histone deacetylation by HDAC3 is important for phosphorylation of histone H3 on serine-10 during mitosis (18).
Here we report a specific association of the HDAC3 complex with the mitotic spindle. Knockdown of HDAC3 resulted in a unique phenotype with a collapsed spindle surrounded by chromosomes arranged in a dome shape configuration. A cold-treatment experiment demonstrated that spindle microtubules in HDAC3-depleted cells were unstable. Treatment with inhibitors of HDACs or RNA polymerase II revealed that the phenotypes were dependent on defective HDAC activity and not achieved through transcriptional regulation. Furthermore, HDAC3 knockdown and rescue confirmed that the loss of HDAC3 deacetylase activity resulted in the spindle phenotype in HDAC3-depleted cells. Overall protein assembly at kinetochores was intact, but kinetochore–microtubule attachments were impaired and spindle assembly checkpoint was activated in HDAC3-depleted cells. These data reveal the association of a HDAC3 complex with the mitotic spindle and suggest a novel role for the HDAC3 complex in regulating spindle function and kinetochore–microtubule attachments.
Results and Discussion
HDAC3 Core Complex Is Localized on the Mitotic Spindle.
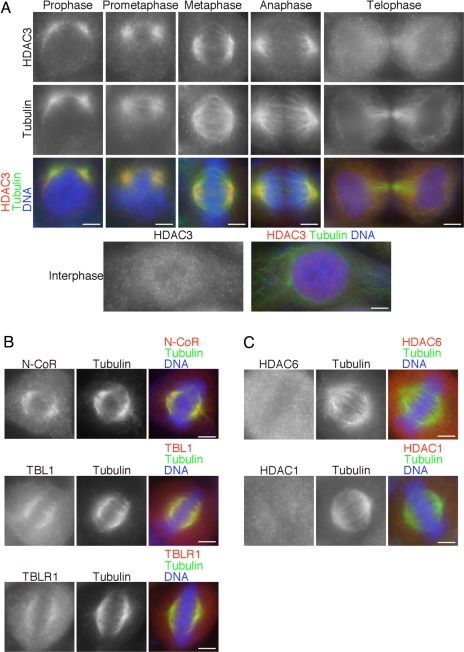
To analyze the role of HDAC3 in mitosis, we examined HDAC3 localization during mitotic progression in HeLa cells. Surprisingly, HDAC3 was localized on the mitotic spindle throughout mitosis [Fig. 1A and supporting information (SI) Fig. 5A]. HDAC3 staining was more concentrated on the spindle microtubules nearest to the poles during prophase and spread over the entire spindle in prometaphase and metaphase. HDAC3 is absent from the poles in metaphase (Fig. 1A and SI Fig. 5B). When cells were treated with the microtubule deploymerizing drug nocodazole, HDAC3 did not associate with the centrosomes, as determined by γ-tubulin staining (SI Fig. 5B), but strongly bound to the remaining fragments of microtubules (SI Fig. 5C). At telophase, HDAC3 staining became more diffused throughout the cytosol and appeared in the reforming nucleus (Fig. 1A). In interphase cells, HDAC3 was predominantly localized in the nucleus and not accumulated on microtubules. HDAC3 staining on DNA during mitosis was much weaker in comparison to that in interphase cells. In addition, HDAC3 staining over the spindle was also observed in HEK293T cells and mouse 3T3 fibroblasts (SI Fig. 5D), suggesting that HDAC3 accumulation on the mitotic spindle is a general phenomenon.
Fig. 1.
HDAC3 core complex is specifically associated with the mitotic spindle. (A) HeLa cells in each phase of mitosis and interphase were stained for HDAC3 (red), tubulin (green), and DNA (blue). (B) Mitotic HeLa cells were stained for N-CoR (Top, red), TBL1 (Middle, red), or TBLR1 (Bottom, red) with tubulin (green) and DNA (blue). (C) Mitotic HeLa cells were stained for HDAC6 (Upper, red) or HDAC1 (Lower, red) with tubulin (green) and DNA (blue). (Scale bars: 5 μm.)
HDAC3 forms a stable core complex with corepressor proteins N-CoR/SMRT and the WD40 repeat proteins TBL1/TBLR1 (7–11). Because the corepressor complex is important for HDAC3 function in interphase, we wondered whether HDAC3 is associated with these proteins on the spindle during mitosis. When cells in metaphase were stained for N-CoR, TBL1, and TBLR1, each component of the corepressor complex was also localized on the mitotic spindle (Fig. 1B and SI Fig. 6A). As observed for HDAC3, these molecules were already associated with spindle microtubules in prophase (SI Fig. 6B), suggesting that the HDAC3 complex associates with spindle microtubules before nuclear envelope breakdown. These data suggest that HDAC3 is localized on spindle microtubules as part of a core complex containing N-CoR, TBL1, and TBLR1.
We tested the possibility that other HDACs might also be localized on the mitotic spindle. Another member of the HDAC family, HDAC6, associates with α-tubulin in interphase and promotes microtubule dynamicity and cell motility by deacetylating Lys-40 of α-tubulin (19, 20). Immunostaining of mitotic cells showed that HDAC6 was diffusely localized throughout the cytosol and not found on the spindle (Fig. 1C). HDAC1 was also localized in the cytosol and did not associate with the spindle (Fig. 1C) (21). Thus, HDAC3 is unique among the HDACs to be localized on the spindle at mitosis.
Knockdown of HDAC3 Results in Collapsed Mitotic Spindle with Impaired Kinetochore–Microtubule Attachments.
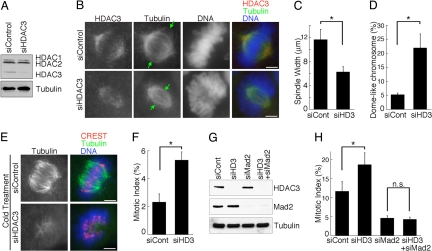
To elucidate the physiological function of HDAC3 in mitotic cells, we examined the effect of HDAC3 knockdown (Fig. 2 A and B). Reduction of HDAC3 on mitotic spindles resulted in two unique phenotypes. First, the mitotic spindle appeared to be smaller and collapsed. We measured the width of the collapsed spindles (indicated by the distance between the arrows in Fig. 2B) and found that the width of the spindles in HDAC3-depleted cells was reduced by 50% relative to that observed in control cells (Fig. 2C). Second, the chromosomes appeared to be excluded from the bipolar spindle structure in HDAC3-deficient cells (Fig. 2B). There was a significant 4-fold increase in cells containing chromosomes that were arranged in a dome-like configuration surrounding the mitotic spindle after HDAC3 knockdown (Fig. 2D and SI Movies 1 and 2). These two phenotypes suggest that HDAC3-depleted cells are unable to make proper chromosome alignment because of defects in the mitotic spindles and/or kinetochore–microtubule attachment.
Fig. 2.
HDAC3 regulates spindle morphology and microtubule–kinetochore attachment. (A) Lysates from HeLa cells transfected with control siRNA (siControl) or siRNA against HDAC3 (siHDAC3) were immunoblotted against HDAC3 and α-tubulin. An HDAC3 antibody (BD Transduction Laboratories) that also recognizes HDAC1 and 2 (14) was used to show specific reduction of HDAC3. (B) Mitotic HeLa cells transfected with control or HDAC3 siRNAs were stained for HDAC3 (red), tubulin (green), and DNA (blue). (C) The width of the spindles, shown by the distance between the arrows in B, was measured for 40 cells and is expressed as the average width (± SD). *, P < 0.01. (D) One hundred mitotic cells were counted, and cells with collapsed spindles surrounded by dome-shaped chromosomes are represented as percentages of total mitotic cells. The graph shows the average (± SD) of three independent experiments. *, P < 0.01. (E) Mitotic HeLa cells transfected with control or HDAC3 siRNAs were incubated at 4°C for 10 min and stained for CREST (red), tubulin (green), and DNA (blue). (Scale bars: 5 μm.) (F) Mitotic index was increased in unperturbed HDAC3-depleted HeLa cells. The graph shows the average (± SD) of three independent experiments. *, P < 0.01. (G) Specific knockdown of HDAC3 and/or Mad2 was confirmed by immunoblotting. (H) HeLa cells depleted of HDAC3, Mad2, or both HDAC3 and Mad2 were synchronized as described in Materials and Methods. Mitotic index (average percentages ± SD) was determined from three independent experiments. *, P < 0.05; n.s., not significant.
To further analyze the defective spindle phenotype, we tested the stability of the spindle microtubules in response to cold treatment (22). Spindle microtubules without stable kinetochore attachment are unstable and are selectively destroyed by cold temperature. Although cold treatment moderately affected the spindle morphology of control cells (Fig. 2E) (23), the overall structure of the kinetochore-attached microtubules remained robust in control cells. In contrast, cold treatment resulted in a highly unstable and much disintegrated mitotic spindle in HDAC3-deficient cells (Fig. 2E). This result suggests that the spindle microtubules are not properly attached to kinetochores in HDAC3-deficient cells.
We observed a significant although not dramatic increase in the mitotic index in unperturbed HDAC3-depleted cells (Fig. 2F), suggesting that the spindle assembly checkpoint is activated to arrest cells in mitosis. However, given the spindle defects, we anticipated that the spindle checkpoint would be activated more robustly in HDAC3-deficient cells. To further examine this, we depleted HDAC3 and/or the spindle checkpoint protein Mad2 in cells synchronized by a thymidine block and release to enrich for mitotic cells (Fig. 2 G and H). Whereas the overall mitotic index in both control and HDAC3-depleted cells was further increased, HDAC3-deficient cells with spindle defects did not accumulate to high levels (Fig. 2H). One possibility is that the spindle phenotype is transient and that HDAC3-deficient cells eventually achieve proper chromosome alignment and complete cell division, thus exhibiting a delay in mitotic progression. Importantly, the mitotic accumulation of HDAC3-depleted cells was abrogated when the cells were simultaneously depleted of Mad2 (Fig. 2H), suggesting that a Mad2-dependent spindle checkpoint is activated in these cells. Taken together, these results suggest that knockdown of HDAC3 resulted in mitotic spindle defects, problems in kinetochore–microtubule attachment, and activation of the spindle assembly checkpoint.
The N-CoR/HDAC3 Complex Is Required for Regulation of Mitotic Spindle.
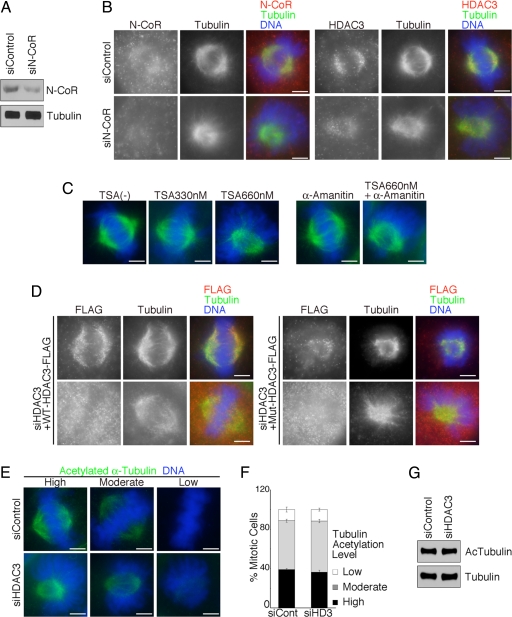
Because HDAC3 is primarily associated with N-CoR/SMRT to form a stable corepressor complex, we tested the effect of depleting N-CoR on spindle morphology. N-CoR depletion from the mitotic spindle resulted in a collapsed spindle surrounded by dome-shaped chromosomes (Fig. 3 A and B and SI Fig. 7A), a phenotype similar to that observed in HDAC3-deficient cells (Fig. 2). Interestingly, HDAC3 was also mislocalized from the mitotic spindle in N-CoR-deficient cells (Fig. 3B). These data suggest that N-CoR is important for HDAC3 localization as well as for its function at mitotic spindles. In interphase cells, N-CoR/SMRT is necessary not only for the formation of the core complex but also for the full enzymatic activity of HDAC3 (24). It remains to be determined whether N-CoR also regulates HDAC3 enzymatic activity in mitosis.
Fig. 3.
Enzymatic activity of HDAC3 is essential for proper spindle formation and chromosome alignment. (A) Specific knockdown of N-CoR was confirmed by immunoblotting. (B) Mitotic HeLa cells transfected with control or N-CoR siRNAs were stained for N-CoR (Left, red) or HDAC3 (Right, red) with tubulin (green) and DNA (blue). (C) At 2.5 h after release from 2 mM thymidine block, 330 nM or 660 nM TSA with or without 2 μg/ml α-amanitin was added to the culture media. Cells were cultured for an additional 5 h, and the enriched mitotic cells were stained for tubulin (green) and DNA (blue). (D) HDAC3-depleted HeLa cells were reconstituted with either siRNA-resistant FLAG-tagged wild-type HDAC3 (Left) or deacetylase-dead Y298H mutant HDAC3 (Right) and stained for FLAG (red), tubulin (green), and DNA (blue). (E) Mitotic HeLa cells transfected with control or HDAC3 siRNAs were stained for acetylated α-tubulin on Lys-40 (green) and DNA (blue). Cells with high, moderate, and low levels of tubulin acetylation are represented. (Scale bars: 5 μm.) (F) One hundred mitotic cells were counted, and the distribution of tubulin acetylation levels (average percentages ± SD) was determined from three independent experiments. (G) Lysates from HeLa cells transfected with control or HDAC3 siRNAs were used for immunoblotting against acetylated α-tubulin and total α-tubulin.
Enzymatic Activity of HDAC3 Is Essential for Proper Spindle Formation and Chromosome Alignment.
We next addressed whether HDAC3 enzymatic activity is involved in spindle formation. First, we used Trichostatin A (TSA), an inhibitor of HDACs, to see whether inhibition of HDAC activity would result in mitotic spindle defects. Cells synchronized at the G1/S phase were incubated with TSA for 5 h and harvested at the peak of mitosis. In the presence of 330 nM TSA, the majority of cells contained properly aligned chromosomes with a few chromosomes lying external to the spindle poles (Fig. 3C) as reported (25). However, after treatment with 660 nM TSA, the spindle was collapsed and the chromosomes were arranged in a dome-like configuration surrounding the bipolar spindle (Fig. 3C and SI Fig. 7B). Although TSA also inhibits other HDACs, the similarity in the spindle defects between the TSA-treated cells and HDAC3-depleted cells suggests that the enzymatic activity of HDAC3 is involved in mitotic spindle formation.
Next, we determined whether the spindle phenotype is a consequence of altered transcriptional activity, because HDAC3 is a well known transcriptional regulator. Cells were treated with TSA together with α-amanitin, an inhibitor of RNA polymerase II. Treatment with α-amanitin alone did not change the morphology of the mitotic spindles (Fig. 3C). When cells were incubated with 660 nM TSA in the presence of α-amanitin, they exhibited phenotypes similar to those of TSA-treated cells and HDAC3-depleted cells (Fig. 3C and SI Fig. 7B). These findings suggest that HDAC3 regulates mitotic spindle formation in a transcription-independent manner. This is consistent with a previous report that actinomycin D did not affect accumulation of cells in G2/M phase induced by HDAC inhibitors (18).
To further determine the significance of the enzymatic activity of HDAC3, we knocked down HDAC3 and rescued HDAC3-deficient cells with FLAG-tagged siRNA-resistant wild-type HDAC3 or deacetylase-dead Y298H mutant HDAC3 (26) constructs. Although exogenously expressed HDAC3 was diffusely distributed in most cells probably because of high levels of overexpression (Fig. 3D Lower), FLAG-tagged HDAC3 is clearly localized to mitotic spindles in some cells (Fig. 3D Upper). Importantly, overexpression of wild-type HDAC3 restored proper spindle morphology and chromosome alignment in HDAC3-deficient cells (Fig. 3D Left). In contrast, deacetylase-dead mutant HDAC3 did not rescue either the morphology of the spindle, which remained small and collapsed, or the dome-like configuration of chromosomes in HDAC3-depleted cells (Fig. 3D Right). These data suggest that the deacetylase activity of HDAC3 is necessary for proper spindle formation and chromosome alignment.
HDAC3 Does Not Alter Acetylation of α-Tubulin.
α-Tubulin is one of the most abundant non-histone proteins that are subject to acetylation. Because HDAC3 is associated with spindle microtubules throughout mitosis, it is possible that HDAC3 regulates spindle function by directly deacetylating α-tubulin. To address this question, we stained HDAC3-depleted cells with anti-acetylated α-tubulin (Lys-40) antibody that recognizes the key acetylated residue on α-tubulin (19, 20). We found that mitotic cells show a range of high, moderate, and low levels of α-tubulin acetylation (Fig. 3 E and F). Furthermore, knockdown of HDAC3 did not affect the distribution of α-tubulin acetylation levels or the global acetylation level of α-tubulin (Fig. 3 E–G). These data indicate that α-tubulin is not a target molecule of HDAC3. Considering the heterogeneous distribution and levels of acetylated α-tubulin in mitotic cells, the significance of acetylation of tubulin on spindle microtubules remains to be determined.
Knockdown of HDAC3 Does Not Affect Kinetochore Formation.
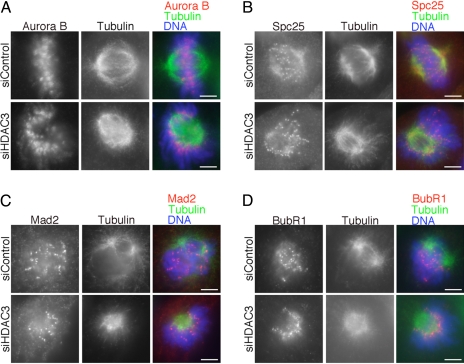
The collapsed and cold sensitive spindle with the dome-like chromosome configuration in HDAC3-deficient cells suggests that kinetochores were not properly captured by microtubules. To determine whether the lack of microtubule–kinetochore attachment is due to defective kinetochore structure, we next examined the localization of various kinetochore proteins to assess the integrity of the kinetochore structure in HDAC3-deficient cells. We found that key kinetochore proteins such as Aurora B kinase (Fig. 4A) and polo-like kinase 1 (Plk1) (SI Fig. 8A) were localized properly at the kinetochores in HDAC3-deficient cells. Others have reported that, when cells are treated with HDAC inhibitors, Aurora B assembly at centromere/kinetochore is delayed in G2 phase but that the level of Aurora B at kinetochores in these cells recovers to control levels by late prophase (25). Our finding that kinetochore Aurora B level in HDAC3-depleted cells was not affected at prometaphase/metaphase is consistent with their report. Our data showed that at least the kinetochore assembly of Aurora B remains intact even after depletion of HDAC3.
Fig. 4.
Kinetochore protein assembly is intact in HDAC3-depleted cells. Mitotic HeLa cells treated with control siRNA or siRNA against HDAC3 were stained for Aurora B (A, red), Spc25 (B, red), Mad2 (C, red), and BubR1 (D, red) with tubulin (green) and DNA (blue). (Scale bars: 5 μm.) For C and D, we examined control cells in prometaphase, at a time when the spindle checkpoint is active and the levels of checkpoint proteins Mad2 and BubR1 are high at unattached kinetochores.
We next examined the localization of Spc25, a key component of the Ndc80/Hec1 complex that regulates microtubule–kinetochore attachment (27–29). Similar to HDAC3 knockdown, depletion of the Ndc80 complex has been shown to result in a fragile spindle (23, 28). However, the staining level of Spc25 at kinetochores was similar in control and HDAC3-depleted cells (Fig. 4B). Thus, HDAC3 contributes to microtubule–kinetochore attachment in a manner that does not affect kinetochore localization of the Ndc80/Hec1 complex.
Finally, we examined the levels of checkpoint proteins Mad2 and BubR1 and a kinetochore motor protein, CENP-E, which are normally reduced at kinetochores after proper kinetochore–microtubule attachment (30). The levels of Mad2, BubR1 (Fig. 4 C and D), and CENP-E (SI Fig. 8B) were remarkably accumulated at kinetochores in HDAC3-deficient cells, to a similar extent as that found on unattached kinetochores in control cells during prometaphase when spindle checkpoint activity is robust. These results suggest that the spindle checkpoint is activated in HDAC3-deficient cells that exhibit a collapsed spindle, in agreement with the Mad2-dependent increase in mitotic index observed in these cells (Fig. 2H). Taken together, our data suggest that, in HDAC3-depleted cells, (i) the mitotic spindle is defective, (ii) kinetochore–microtubule attachments are impaired, (iii) the overall structure of the kinetochore is intact, and (iv) the spindle checkpoint is active.
Our data demonstrate for the first time, to our knowledge, the localization of HDAC3 complex at the mitotic spindle and reveal a novel role for HDAC3 in regulating spindle formation and microtubule–kinetochore attachments. Although our data do not address the possibility that HDAC3 activity on histone H3 deacetylation (31) and mitotic chromatin modification (18) may contribute to chromosome capture by spindle microtubules, our studies clearly extend a role of HDAC3 in regulating the structure and/or function of the mitotic spindles. We determined that α-tubulin is not a target of deacetylation by HDAC3. We speculate that some spindle-associated proteins might be regulated by the HDAC3 complex via deacetylation. Our studies suggest that HDAC3 is likely to be one of the major HDACs involved in the regulation of mitosis. Because HDAC inhibitors are being developed as anti-cancer agents (2), understanding how HDAC3 regulates mitotic spindle functions would be important for improving future anti-cancer therapies.
Materials and Methods
Antibodies.
Rabbit anti-HDAC3 (1:1,000), rabbit anti-N-CoR (1:1,000), rabbit anti-TBL1 (1:1,000), rabbit anti-TBLR1 (1:1,000), and rabbit anti-HDAC1 (1:1,000) were previously described (9, 11). CREST antiserum (1:20,000) was a gift from William Brinkley (Baylor College of Medicine). Rabbit anti-Spc-25 antibody (1:500) was a gift from P. Todd Stukenberg (University of Virginia, Charlottesville, VA). The following antibodies were purchased from commercial sources: mouse β-tubulin (1:2,000) and mouse acetylated α-tubulin (Lys-40) (1:2,000) (Sigma); rabbit α-tubulin (1:500) (GeneTex), mouse Aurora B kinase (1:500) (BD Transduction Laboratories), mouse HDAC6 (1:500) (Santa Cruz Biotechnology), mouse BubR1 (1:500) (Abcam), and rabbit Mad2 (1:500) (Bethyl). For HDAC3 immunoblotting, mouse HDAC3 antibody (BD Transduction Laboratories) or rabbit HDAC3 antibody (Bethyl) was used.
Cell Culture, siRNA, and Immunofluorescence.
HeLa cells were cultured on poly-lysine-coated coverslips in D-MEM supplemented with 10% FBS and antibiotics. Transfection with control siRNA or siGENOME HDAC3-specific siRNA (Dharmacon) was performed by using TransIT-siQuest (Mirus) according to the manufacturer's instruction. Two days after transfection, the cell cycle was synchronized by treatment with 2 mM thymidine for 14.5 h. Cells were released from thymidine block and cultured for 7 h to enrich for mitotic cells. Cells were rinsed in PHEM buffer (60 mM K-Pipes/25 mM Hepes/10 mM EGTA/2 mM MgSO4, pH 6.9) and fixed in 4% formaldehyde in PHEM buffer for 20 min. Coverslips were rinsed in PBS, permeabilized in 0.5% Triton X-100 in PBS for 15 min, rinsed in PBS, and blocked in Antibody Solution (100 mM K-Pipes/1 mM MgSO4/1 mM EGTA/1.83% l-lysine/1% BSA/0.1% NaN3, pH 7.2) or TBS-T with 5% nonfat dry milk for 1 h. If preextraction was necessary, cells were permeabilized in 0.5% Triton X-100 in PHEM for 1 min before fixation. Coverslips were incubated with primary antibodies in Antibody Solution or TBS-T with 5% nonfat dry milk overnight at 4°C, rinsed in PBS, incubated with Alexa Fluor 488-, Alexa Fluor 555-, or Texas-Red-conjugated secondary antibodies (Molecular Probes) in Antibody Solution or TBS-T with 5% nonfat dry milk for 1 h, rinsed in PBS, and mounted in ProLong Gold antifade reagent with DAPI (Molecular Probes).
For the analysis of microtubule stability, culture media were quickly changed to ice-cold media and cells were incubated at 4°C for 10 min, followed by fixation and permeabilization at room temperature using PBS instead of PHEM buffer (22).
For the HDAC3 knockdown and rescue experiments, siRNA-resistant wild-type or Y298H mutant FLAG-tagged HDAC3 expression vectors were generated by introducing silent mutations (underlined) into the siRNA-targeted sequence AAAGCGATGTGGAGATTTA. Transfection with siRNA oligos and DNA vectors was performed by using TransIT-siQuest and TransIT-LT1 (Mirus) according to the manufacturer's instruction.
Images were acquired with a Nikon TE2000 wide-field microscope system (Nikon), with the same exposure times from specific siRNA- or drug-treated cells and control cells. The width of spindles was measured by using NIS-Elements (Nikon).
Statistical Analysis.
Data were confirmed in multiple independent experiments. Data quantification was performed with Student's t test, and data are expressed as the mean ± SD.
Supplementary Material
Acknowledgments.
We thank Dr. William (B.R.) Brinkley for CREST antiserum and Dr. P. Todd Stukenberg for Spc25 antibody. This work was supported by National Institutes of Health Grant DK53176 (to L.-y.Y.-L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710140105/DC1.
References
- 1.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 3.Yang WM, et al. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 4.Johnson CA, Barlow AL, Turner BM. Molecular cloning of Drosophila melanogaster cDNAs that encode a novel histone deacetylase dHDAC3. Gene. 1998;221:127–134. doi: 10.1016/s0378-1119(98)00435-1. [DOI] [PubMed] [Google Scholar]
- 5.Mahlknecht U, Hoelzer D, Bucala R, Verdin E. Cloning and characterization of the murine histone deacetylase (HDAC3). Biochem Biophys Res Commun. 1999;263:482–490. doi: 10.1006/bbrc.1999.1389. [DOI] [PubMed] [Google Scholar]
- 6.Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guenther MG, et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 8.Wen YD, et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 11.Yoon HG, et al. Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Lf, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 13.Xia Y, et al. c-Jun downregulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Mol Cell. 2007;25:219–232. doi: 10.1016/j.molcel.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escaffit F, et al. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol Cell Biol. 2007;27:554–567. doi: 10.1128/MCB.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takami Y, Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J Biol Chem. 2000;275:16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- 16.Dangond F, et al. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 17.Wilson AJ, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 20.Tran AD, et al. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J Cell Sci. 2007;120:1469–1479. doi: 10.1242/jcs.03431. [DOI] [PubMed] [Google Scholar]
- 21.Kruhlak MJ, et al. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 22.Brinkley BR, Cartwright J., Jr Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann NY Acad Sci. 1975;253:428–439. doi: 10.1111/j.1749-6632.1975.tb19218.x. [DOI] [PubMed] [Google Scholar]
- 23.DeLuca JG, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins AR, et al. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle. 2005;4:717–726. doi: 10.4161/cc.4.5.1690. [DOI] [PubMed] [Google Scholar]
- 26.Lahm A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCleland ML, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 28.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 29.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 31.Vermeulen M, et al. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Mol Cell Biol. 2004;24:2364–2372. doi: 10.1128/MCB.24.6.2364-2372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.