Abstract
Endocytosis and subsequent lysosomal degradation serve as a well characterized mechanism to fine-tune and down-regulate EGFR signaling. However, other members of the EGFR/ErbB receptor family have been reported to be endocytosis-impaired. Here we demonstrate that endocytosis of ErbB4 is regulated in an isoform-specific manner: CYT-1 isoforms were efficiently endocytosed whereas CYT-2 isoforms were endocytosis-impaired. CYT-1 isoforms in endocytic vesicles colocalized with Rab5 and Rab7 indicating trafficking via early endosomes to late endosomal/lysosomal structures. A PPXY motif within the CYT-1-specific sequence that lacks from CYT-2 was necessary both for ubiquitination and endocytosis of CYT-1 isoforms and provided a binding site for a WW domain-containing ubiquitin ligase Itch. Itch catalyzed ubiquitination of ErbB4 CYT-1, promoted its localization into intracellular vesicles, and stimulated degradation of ErbB4 CYT-1. Dominant negative Itch suppressed ErbB4 CYT-1 endocytosis and degradation. These data indicate that ErbB4 isoforms differ in endocytosis and degradation by a mechanism mediated by CYT-1-specific PPXY motif interacting with a WW domain-containing E3 ubiquitin ligase.
Keywords: AIP4, Itch, ubiquitination, WW domain
EGFR (ErbB1), ErbB2 (c-Neu), ErbB3, and ErbB4 form the epidermal growth factor receptor (EGFR/ErbB) subfamily of receptor tyrosine kinases (RTKs). ErbB receptors bind EGF-like growth factors, such as EGF and neuregulins (NRGs), and mediate multiple cellular responses, such as proliferation, differentiation, migration, and survival. Aberrations in ErbB signaling have a role in carcinogenesis, and cancer drugs targeting EGFR and ErbB2 are currently in clinical use (1).
Endocytosis and subsequent lysosomal degradation of cell surface receptors serve as a powerful mechanism to attenuate ligand-induced signaling (2). Inefficient endocytosis of RTKs can lead to sustained signaling and to enhanced mitogenesis and transformation, as in the case of endocytosis-impaired mutant of EGFR (3). Endocytosis may also modify the RTK signaling output, and different patterns of downstream signal transduction cascades may be activated by RTKs residing in endosomes as compared with RTKs at the cell surface (4, 5).
EGFR serves as a well characterized example of a cell surface receptor that undergoes efficient endocytosis. Upon EGF stimulation, EGFR dimerizes and is rapidly internalized via clathrin-coated pits. Subsequently, EGFR traffics via early endosomes and recycling vesicles back to cell surface or, alternatively, via late endosomes to lysosomal degradation (2). Studies with chimeric ErbB constructs composed of domains of EGFR fused to other ErbBs indicate that EGFR, unlike ErbB2, ErbB3, or ErbB4, has intracellular motifs that facilitate endocytosis and targeting to degradation (6, 7). Thus, in contrast to EGFR, endocytosis of other ErbBs has been regarded inefficient.
In contrast to EGFR and ErbB2, which are well characterized human oncogenes and cancer drug targets, the biological responses regulated by ErbB4 are poorly understood (8, 9). This may be partly explained by the fact that alternative splicing generates four functionally unique isoforms of ErbB4 (9). For example, the ratio of expressed ErbB4 isoforms seems to change in diseased tissues, such as those affected by schizophrenia (10) or medulloblastoma (11), although the outcomes of these shifts at the molecular and cellular levels remain to be fully elucidated. Structurally, the ErbB4 isoforms differ at extracellular juxtamembrane domain (JM-a and JM-b isoforms) or at the intracellular cytoplasmic domain (CYT-1 and CYT-2 isoforms) (12, 13). The JM-a isoforms, but not the JM-b isoforms, are susceptible to two-step proteolysis by tumor necrosis factor-α converting enzyme (TACE) and γ-secretase activity, generating soluble intracellular domains (ICD) that can translocate into nucleus and regulate transcription (14–16). The CYT-1 isoforms, but not the CYT-2 isoforms, contain a direct binding site for phosphoinositide 3-kinase (PI3-K) that is necessary for NRG-induced chemotaxis and survival mediated by noncleavable ErbB4 CYT-1 in NIH 3T3 cells (12, 17). On the other hand, the cleavable JM-a CYT-2 isoform seems to be unique among the isoforms in being capable of ligand-independent signaling by a mechanism involving proteolytic generation of a stable phosphorylated ICD (14, 18).
Here we provide evidence that endocytosis and targeting of ErbB4 to late endosomal compartments is regulated in an isoform-specific manner. PPXY motif within the CYT-1 isoform-specific sequence of 16 aa that lack from CYT-2 isoforms is shown to mediate ErbB4 ubiquitination and endocytosis by providing a binding site for the WW domain-containing HECT-type E3 ubiquitin ligase Itch. Itch E3 ligase activity is demonstrated to regulate ErbB4 CYT-1 ubiquitination, subcellular localization, and degradation.
Results and Discussion
ErbB4 CYT-1 Isoforms Are Localized to Endosomal Vesicles to a Greater Extent than ErbB4 CYT-2 Isoforms.
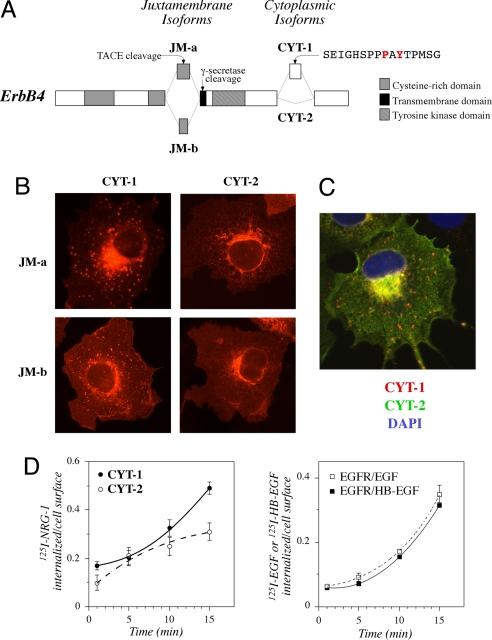
Constructs encoding all four full-length ErbB4 isoforms, i.e., the four possible combinations of alternative JM and CYT domains (Fig. 1A), with C-terminal HA tags were transiently expressed in COS-7 cells. Immunofluorescence microscopy demonstrated that isoforms with the CYT-1 type of cytoplasmic tail were more frequently localized into cytoplasmic vesicles than CYT-2 isoforms, irrespective of the JM domain (Fig. 1B). Because both the cleavable JM-a CYT-1 and the noncleavable JM-b CYT-1 localized to the vesicles, these data indicate the presence of the intact full-length ErbB4 within the vesicular structures. Similar results were obtained with ErbB4 constructs without tags or with C-terminal EGFP tags, with variable expression levels, and in the backgrounds of HeLa, NIH 3T3, and MCF-7 cells (14) (data not shown). In addition, differential subcellular localization of ErbB4 CYT isoforms was evident in confocal analysis of single cells simultaneously expressing differentially tagged CYT-1 or CYT-2 isoforms (Fig. 1C).
Fig. 1.
Subcellular localization and internalization of ErbB4 isoforms. (A) Alternative splicing generates four ErbB4 isoforms that differ at juxtamembrane (JM) or cytoplasmic (CYT) domains. JM-a isoforms, unlike JM-b isoforms, include a proteolytic cleavage site for TACE and γ-secretase. CYT-1 isoforms, unlike CYT-2 isoforms, contain a 16-aa sequence that includes a PI3-K binding site (YTPM) as well as a proline-rich protein interaction motif (PPAY). P1054 and Y1056 are shown in red. (B) COS-7 cells expressing each of the HA-tagged ErbB4 isoforms were stained with anti-HA antibody (red) and photographed under a fluorescence microscope. (C) COS-7 cells simultaneously expressing HA-tagged ErbB4 JM-a CYT-1 and Myc-tagged ErbB4 JM-a CYT-2 were stained with anti-HA (red) and anti-Myc (green) antibodies. The nuclei were stained with DAPI (blue). (D) NIH 3T3-7d transfectants stably expressing ErbB4 JM-b CYT-1, ErbB4 JM-b CYT-2, or EGFR were incubated for 1, 5, 10, or 15 min with 125I-NRG-1, 125I -EGF, or 125I-HB-EGF at 37°C. Cells were washed once with PBS and twice with an acidic buffer and lysed with NaOH. Radioactivity removed in acid washes containing surface-bound growth factors was compared with radioactivity in cell lysates containing internalized growth factors.
To determine the identity of CYT-1-positive vesicles, HA-tagged ErbB4 CYT-1 or CYT-2 was expressed in COS-7 cells together with GFP-tagged Rab5 or Rab7, markers of early and late endosomal vesicles, respectively (19). Confocal microscopy demonstrated that ErbB4 CYT-1 isoforms, but not CYT-2 isoforms, clearly colocalized with Rab5 and Rab7 in intracellular vesicular structures [supporting information (SI) Fig. 5 in SI Appendix]. The CYT-1/Rab colocalization was further reinforced by stimulation with NRG-1 (SI Fig. 5 in SI Appendix). Taken together these findings demonstrate that full-length ErbB4 CYT isoforms differ in their subcellular localization to early and late endosomes.
ErbB4 CYT-1 Is Internalized and Degraded More Efficiently than ErbB4 CYT-2.
To address whether the more efficient localization into endocytic vesicles was associated with enhanced receptor internalization, the uptake of 125I-NRG-1 was measured by using NIH 3T3-7d transfectants stably expressing the noncleavable ErbB4 JM-b CYT-1 or JM-b CYT-2. As expected, internalization of 125I-NRG-1 was significantly faster in cells expressing CYT-1 than in cells expressing CYT-2 (Fig. 1D Left). Interestingly, the kinetics of ligand-stimulated internalization by CYT-1 was similar to that of EGFR known to undergo efficient internalization after ligand stimulation (Fig. 1D Right). Consistent with differential internalization, NRG-1 stimulation induced faster down-regulation of biotinylated cell surface-associated pool of ErbB4 CYT-1 when compared with ErbB4 CYT-2 (SI Fig. 6A in SI Appendix).
To address the functional outcome of differential internalization, degradation of [35S]methionine-labeled noncleavable ErbB4 JM-b CYT-1 and JM-b CYT-2 was determined in COS-7 cells. Degradation of CYT-1 was significantly more effective than degradation of CYT-2 (SI Fig. 6B in SI Appendix). Again, the basal degradation rate of ErbB4 CYT-1, but not of ErbB4 CYT-2, was similar to that of EGFR (SI Fig. 6B in SI Appendix). CYT-1 was degraded more efficiently than CYT-2 also when stability of cleavable JM-a isoforms was analyzed by blocking protein synthesis with cycloheximide (Fig. 4C, lanes 1–4). These data indicate that CYT-1 isoforms are more efficiently internalized than CYT-2 isoforms and that the difference in internalization is associated with a difference in receptor degradation rate.
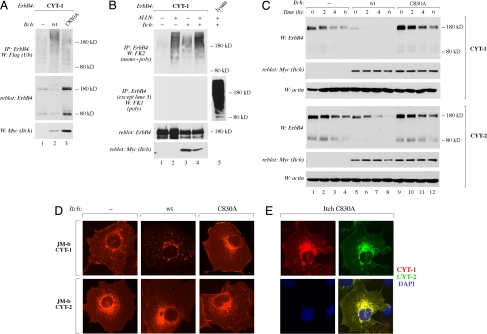
Fig. 4.
Function of Itch in ErbB4 ubiquitination, endocytosis, and degradation. (A) COS-7 cells expressing ErbB4 JM-a CYT-1 and Flag-tagged ubiquitin in the presence or absence of Myc-tagged Itch or Itch C830A were lysed, and the lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Flag antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283). Expression of Itch was analyzed by Western with anti-Myc antibody. (B) COS-7 cells expressing ErbB4 JM-a CYT-1 and Flag-tagged ubiquitin in the presence or absence of Myc-tagged Itch were treated for 2 h with or without 150 μM ALLN. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-FK1 or anti-FK2 antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283). Expression of Itch was analyzed by Western blotting with anti-Myc antibody. Total lysate from cells expressing Itch-Myc and treated with ALLN was also analyzed by Western blotting with FK1 antibody without prior immunoprecipitation (lane 5). (C) COS-7 cells expressing ErbB4 JM-a CYT-1 or ErbB4 JM-a CYT-2 in the presence or absence of Myc-tagged Itch or Itch C830A were treated with 100 μM cycloheximide for 0, 2, 4, or 6 h and lysed. ErbB4 expression was analyzed by Western blotting with anti-ErbB4 antibody (Abcam). Membranes were reblotted with anti-Myc antibody. Protein loading was controlled by blotting with anti-actin. (D) COS-7 cells expressing HA-tagged ErbB4 JM-b CYT-1 or HA-tagged ErbB4 JM-b CYT-2 in the presence or absence of Myc-tagged Itch or Itch C830A were stained with anti-HA antibody (red) and photographed under a fluorescence microscope. Itch expression was confirmed by staining with anti-Myc antibody (data not shown). (E) COS-7 cells expressing HA-tagged ErbB4 JM-a CYT-1, Myc-tagged ErbB4 JM-a CYT-2, and Flag-tagged Itch C830A. Cells were stained with anti-HA (red), anti-Myc (green), and DAPI (nuclear stain; blue).
PPXY Motif Within the CYT-1-Specific Sequence Is Necessary for Endocytosis of ErbB4 CYT-1.
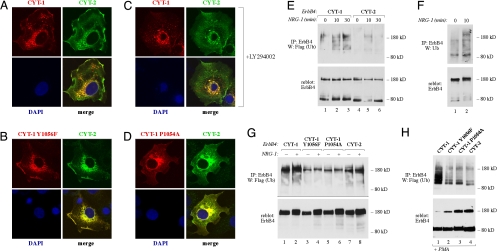
CYT-1 isoforms differ from CYT-2 isoforms structurally by containing an insert of 16 amino acids (see Fig. 1A) that are not present in the CYT-2 sequence. These 16 amino acids include one tyrosine residue, Y1056, that has previously been shown to function as the only direct docking site of ErbB4 for PI3-K (12). The Y1056 residue is localized within a YXXM (X is any amino acid) consensus binding motif for PI3-K (20) that spans the amino acids 1056–1059 as YTPM sequence in CYT-1 (Fig. 1A). Modification of phosphatidyl inositols by PI3-K may regulate endocytosis (21). To address the molecular mechanism underlying the observed differences in the subcellular targeting of the two CYT isoforms, a confocal analysis of COS-7 cells expressing a CYT-1 construct with an engineered Y1056F mutation was carried out. When CYT-1 Y1056F was expressed simultaneously with wild-type CYT-2, the difference in the intracellular targeting of the CYT isoforms was abolished (compare Fig. 2 A and B). However, inhibition of PI3-K activity with the chemical inhibitor LY294002 did not prevent the localization of CYT-1 into the cytoplasmic vesicles (Fig. 2C) whereas it efficiently blocked NRG-1-stimulated phosphorylation of Akt (data not shown). These data demonstrate that the Y1056 residue, but not coupling of PI3-K activity to Y1056, was involved in regulating differential targeting of the CYT isoforms.
Fig. 2.
PPXY motif in regulation of differential endocytosis and ubiquitination of ErbB4 CYT-1 and CYT-2. COS-7 cells were transfected with constructs encoding wild-type Myc-tagged ErbB4 JM-a CYT-2 and wild-type HA-tagged ErbB4 JM-a CYT-1 (A and C), HA-tagged ErbB4 JM-a CYT-1 Y1056F (B), or HA-tagged ErbB4 JM-a CYT-1 P1054A (D). Cells expressing Myc-tagged ErbB4 JM-a CYT-2 and HA-tagged ErbB4 JM-a CYT-1 were treated for 4 h without (A) or with (C) 20 μM LY294002 to inhibit PI3-K activity. Cells were stained with anti-HA (red) and anti-Myc (green) antibodies. The nuclei were stained with DAPI (blue). (E) COS-7 cells expressing Myc-tagged ErbB4 JM-a CYT-1 or ErbB4 JM-a CYT-2 and Flag-tagged ubiquitin were stimulated for 0, 10, or 30 min with 50 ng/ml NRG-1. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Flag antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283). (F) OVCAR-3 cells were stimulated for 0 or 10 min with 50 ng/ml NRG-1. Lysates were immunoprecipitated with anti-ErbB4 (HFR-1) followed by Western blotting with anti-ubiquitin (P4D1). Membrane was reblotted with anti-ErbB4 antibody (Abcam). (G) COS-7 cells expressing HA-tagged ErbB4 JM-b CYT-1, ErbB4 JM-b CYT-1 Y1056F, ErbB4 JM-b CYT-1 P1054A, or ErbB4 JM-b CYT-2 and Flag-tagged ubiquitin were stimulated for 0 or 30 min with 50 ng/ml NRG-1. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Flag antibody. Membrane was reblotted with anti-ErbB4 antibody (Abcam). (H) COS-7 cells expressing ErbB4 JM-a CYT-1, ErbB4 JM-a CYT-1 Y1056F, ErbB4 JM-a CYT-1 P1054A, or ErbB4 JM-a CYT-2 and Flag-tagged ubiquitin were stimulated for 0 or 30 min with 100 ng/ml PMA. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Flag antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283).
The CYT-1-specific stretch of 16 aa also includes a PPXY motif that may in principle serve as a binding site for WW domain-containing proteins (22–24). The PPXY motif is located between amino acids 1053 and 1056 as a PPAY sequence in CYT-1 and thus shares the Y1056 with the YTPM binding sequence for PI3-K (Fig. 1A). To assess whether the results obtained with the Y1056F mutation (Fig. 2B) were a consequence of disrupting a functional PPXY motif, rather than a YXXM motif for PI3-K, the proline-1054 was mutated to alanine in the CYT-1-specific sequence. Indeed, P1054A mutation abolished the appearance of CYT-1-positive vesicles (Fig. 2D). These data suggest that the PPXY motif present in ErbB4 CYT-1 isoforms functions as an endocytic sorting motif.
PPXY Motif Within the CYT-1-Specific Sequence Is Necessary for Efficient Ubiquitination of ErbB4 CYT-1.
Ubiquitination is one of the key regulators of endocytosis and subsequent lysosomal degradation of cell surface receptors in both yeast and mammalian cells (25). We have previously demonstrated that membrane-anchored 80-kDa cleavage product (m80) of JM-a CYT-1 is significantly more ubiquitinated than m80 of JM-a CYT-2, when ErbB4 cleavage is induced by PMA (18). Here we demonstrate that also the full-length JM-a CYT-1 was more efficiently ubiquitinated than full-length JM-a CYT-2 (Fig. 2E). Moreover, ubiquitination of endogenously expressed full-length ErbB4 was evident in OVCAR-3 ovarian carcinoma cells naturally expressing both JM-a-type of ErbB4 isoforms (Fig. 2F and data not shown). The CYT-1-specific sequence of 16 aa does not contain lysine residues (Fig. 1A), ruling out a difference in availability of direct ubiquitination sites (12, 25). To address the significance of the CYT-1-specific PPXY motif, ubiquitination of the CYT-1 Y1056F and P1054A mutants was tested. Consistent with the effect on targeting to cytoplasmic vesicles, both mutants were ubiquitinated to a lesser extent when compared with wild-type CYT-1. This was observed both in the context of full-length noncleavable JM-b isoforms (Fig. 2G) and in the context of m80 fragments generated from full-length JM-a isoforms by PMA stimulation (Fig. 2H). These data demonstrate that, in addition to endocytosis, the CYT-1-specific PPXY motif regulates ErbB4 CYT-1 ubiquitination.
PPXY Motif Within the CYT-1-Specific Sequence Serves as a Binding Site for WW Domains of the E3 Ubiquitin Ligase Itch.
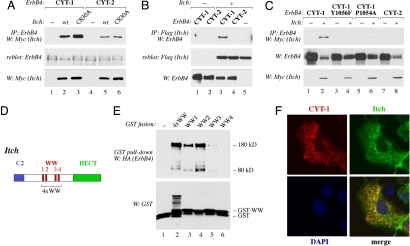
We have previously demonstrated that the membrane-anchored m80 fragments of CYT-1 and CYT-2 type are ubiquitinated to different extents whereas there is no difference between the isoforms when ubiquitination of the soluble ICDs (s80s) is analyzed (18). Here we demonstrated that the efficient ubiquitination of membrane-anchored CYT-1 isoforms depended on an intact PPXY motif (Fig. 2 G and H). Together these observations imply that the ubiquitin ligase responsible for CYT-1-specific modification contains a membrane anchor, as well as a WW domain capable of interacting with the PPXY motif. One family of ubiquitin ligases that contain both C2 domains for coupling to membrane phospholipids and two or more WW domains is the Nedd4 family of HECT-type E3 ubiquitin ligases (26). To test for an interaction between ErbB4 and two candidate members of the Nedd4 family, coimmunoprecipitation of ErbB4 together with either Nedd4 or Itch was analyzed. Whereas Nedd4 failed to demonstrate association (data not shown), Itch associated with ErbB4 in reciprocal coimmunoprecipitation experiments (Fig. 3 A and B). This association between Itch and ErbB4 was also evident when proteins endogenously expressed in OVCAR-3 cells were analyzed (SI Fig. 7 in SI Appendix), and this was confirmed by others while this manuscript was under preparation (27). Although some association was also observed between Itch and CYT-2 isoforms (Fig. 3A, lane 5), consistently more CYT-1 than CYT-2 coprecipitated with Itch (Fig. 3 A–C).
Fig. 3.
Physical interaction and colocalization of ErbB4 with Itch E3 ubiquitin ligase. (A) COS-7 cells were transfected with constructs encoding ErbB4 JM-a CYT-1 or ErbB4 JM-a CYT-2 and Myc-tagged Itch or Itch C830A. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Myc antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283). Expression of Itch was analyzed by Western blotting with anti-Myc antibody. (B) COS-7 cells were transfected with constructs encoding HA-tagged ErbB4 JM-a CYT-1 or ErbB4 JM-a CYT-2 and Flag-tagged Itch. Lysates were immunoprecipitated with anti-Flag antibody followed by Western blotting with anti-ErbB4 antibody (Abcam). Membrane was reblotted with anti-Flag antibody. Expression of ErbB4 was analyzed by Western blotting with anti-ErbB4 antibody (Abcam). (C) COS-7 cells were transfected with constructs encoding ErbB4 JM-a CYT-1, ErbB4 JM-a CYT-1 Y1056F, ErbB4 JM-a CYT-1 P1054A, or ErbB4 JM-a CYT-2 with or without construct for Myc-tagged Itch. Lysates were immunoprecipitated with anti-ErbB4 antibody (HFR-1) followed by Western blotting with anti-Myc antibody. Membrane was reblotted with anti-ErbB4 antibody (sc-283). Expression of Itch was analyzed by Western blotting with anti-Myc antibody. (D) Schematic structure of Itch including an N-terminal C2 domain for membrane anchoring, four WW domains, and a C-terminal catalytic HECT domain. 4xWW indicates a protein region spanning all four WW domains. (E) GST fusion proteins including one of the WW domains of Itch (WW1, WW2, WW3, or WW4) or all four of them in one fusion protein (4xWW) were incubated with lysates of COS-7 cells transiently expressing HA-tagged ErbB4 JM-a CYT-1. Material precipitating with glutathione Sepharose beads was analyzed by Western blotting with anti-HA antibody. Membranes were reblotted with anti-GST antibody. (F) COS-7 cells simultaneously expressing HA-tagged ErbB4 JM-b CYT-1 and Myc-tagged Itch were stained with anti-HA (red) and anti-Myc (green) antibodies. The nuclei were stained with DAPI (blue).
The interaction between Itch and either ErbB4 isoform was not dependent on the catalytic activity of the Itch HECT domain (Fig. 3D), because the catalytically inactive Itch mutant C830A demonstrated equal binding when compared with wild-type Itch (Fig. 3A). However, the avid interaction between Itch and CYT-1 depended on an intact CYT-1-specific PPXY motif as mutating either Y1056 or P1054 attenuated binding (Fig. 3C). GST pull-down experiments with single or all four Itch WW domains (Fig. 3D) fused to GST demonstrated that ErbB4 JM-a CYT-1 predominantly interacted with Itch WW domains WW2 and WW1 (Fig. 3E). Interaction of ErbB4 with WW3 and WW4 was significantly less efficient. Interestingly, other Itch interaction partners such as p73 and Nedd4-binding partner 1 have also been reported to interact mainly with the WW2 domain of Itch (28). Confocal microscopy analysis also demonstrated colocalization of Itch with CYT-1 isoforms within cytoplasmic vesicular structures (Fig. 3F). These data show that ErbB4 interacts with the WW domain-containing E3 ubiquitin ligase Itch and that ErbB4 CYT-1 associates with Itch more avidly than CYT-2 because of an interaction via the CYT-1-specific PPXY motif.
Itch Monoubiquitinates ErbB4.
To analyze the functional significance of the interaction between ErbB4 and Itch, the role of Itch in ubiquitination of ErbB4 was analyzed. Expression of Itch increased ubiquitination of ErbB4 unlike the catalytically inactive C830A mutant of Itch (Fig. 4A), which was still capable of binding ErbB4 (Fig. 3A). These observations suggest that Itch functions as an E3 ubiquitin ligase for ErbB4 and Itch C830A as a dominant negative inhibitor of ErbB4 ubiquitination. Because the functional outcome of ubiquitination may depend on the type of ubiquitin chains attached to the substrate protein (29), we assessed whether ErbB4 CYT-1 was mono- or polyubiquitinated by Itch. An antibody, FK1, that mostly recognizes polyubiquitin chains (30) failed to recognize Itch-induced ubiquitination of ErbB4 (Fig. 4B, lanes 1–4) although it efficiently recognized polyubiquitinated proteins from total cell lysate (Fig. 4B, lane 5). In contrast, FK2, an antibody recognizing both mono- and polyubiquitin (31), readily recognized ubiquitinated ErbB4 CYT-1 both in the absence and in the presence of exogenous Itch (Fig. 4B, top blot). Consistently, Itch induced ubiquitination of ErbB4 also when transiently expressed wild-type ubiquitin was replaced by ubiquitin mutants K48R or K63R unable to form lysine-48- or lysine-63-linked polyubiquitin chains, respectively (SI Fig. 8 in SI Appendix). Moreover, Itch-induced degradation of ErbB4 was similar in the presence of wild-type ubiquitin or a ubiquitin mutant unable to form any polyubiquitin chains (Ubi-K0; data not shown). These findings indicate that Itch catalyzes attachment of mono- rather than polyubiquitin moieties to ErbB4 CYT-1.
Itch-Like E3 Ligases Are Necessary for Efficient Endocytosis and Degradation of ErbB4 CYT-1.
One well established consequence of monoubiquitination of cell surface receptors is internalization and subsequent lysosomal degradation (25). The Itch ubiquitin ligase of ErbB4 was shown to partially colocalize with ErbB4 CYT-1 in cytoplasmic vesicles (Fig. 3F), implying a role in endosomal/lysosomal targeting. To analyze the effect of Itch on ErbB4 endocytosis and degradation, CYT-1 and CYT-2 isoforms were expressed in the presence or absence of wild-type or catalytically inactive dominant negative Itch mutant. Overexpression of wild-type Itch enhanced the localization of CYT-1, but not CYT-2, isoforms into cytoplasmic vesicles (Fig. 4D) regardless of the type of the JM domain (SI Fig. 9A in SI Appendix). The specifically CYT-1-positive vesicles induced by Itch overexpression were for the most part also positive for Rab7 expression (SI Fig. 9B in SI Appendix), supporting a role for Itch in targeting CYT-1 into late endosomal/lysosomal pathway. Indeed, wild-type Itch dramatically accelerated the degradation of CYT-1 after protein synthesis was blocked with cycloheximide (Fig. 4C). In contrast, overexpression of the catalytically inactive Itch C830A totally prevented the localization of CYT-1 into vesicles (Fig. 4D) and attenuated CYT-1 degradation (Fig. 4C). CYT-2 isoforms were much less susceptible to Itch-induced changes in both localization and degradation (Fig. 4 C and D), in accordance with weaker physical association with Itch when compared with CYT-1 isoforms (Fig. 3).
To more directly analyze the significance of Itch in the differential vesicular targeting of CYT-1 and CYT-2 isoforms, full-length CYT-1 and CYT-2 coupled to different epitope tags were simultaneously overexpressed together with the dominant negative Itch C830A. Under these conditions neither CYT-1 nor CYT-2 localized to cytoplasmic vesicles (compare Fig. 4E with Fig. 1C). Moreover, when the degradation rate of CYT-1 in the presence of dominant negative Itch C830A was plotted together with the degradation rate of CYT-2 in the absence of exogenous Itch constructs, the two curves were superimposable (SI Fig. 9C in SI Appendix). These data demonstrate that Itch is sufficient for stimulating ErbB4 CYT-1 degradation. In addition, Itch-like proteins interacting with the CYT-1-specific PPXY motif are necessary for the differential degradation of ErbB4 CYT-1 and CYT-2.
Taken together, we have demonstrated that the ErbB4 isoforms were different in their susceptibility to internalization, monoubiquitination, endocytic targeting, and degradation. The molecular mechanism of this variation was differential association of a PPXY motif within the CYT-1 isoform-specific sequence with a Nedd4 family E3 ubiquitin ligase. These findings suggest that the different ErbB4 CYT isoforms have both quantitative and qualitative signaling differences. This may explain some of the controversy that currently prevails about cellular responses stimulated by ErbB4.
Materials and Methods
Expression Plasmids.
Plasmids encoding the four full-length ErbB4 isoforms with or without C-terminal HA or Myc tags have been described (14). P1054A and Y1056F mutations were generated to plasmids encoding full-length ErbB4 isoforms with HA tags by using the QuikChange site-directed mutagenesis kit (Stratagene) to generate pcDNA3.1ErbBJM-aCYT-1P1054A-HA, pcDNA3.1ErbB4JM-aCYT-1Y1056F-HA, pcDNA3.1ErbB4JM-bCYT-1P1054A-HA, and pcDNA3.1ErbB4JM-bCYT-1Y1056F-HA. To generate pCDNA3.1EGFR, a 4-kb XbaI/NotI fragment encoding full-length EGFR was digested from pEBS7EGFR (32) and inserted into pcDNA3.1(-)neo (Invitrogen). Plasmids encoding the following proteins have been described: GFP-Rab5a (33), GFP-Rab7 (34), GFP-Rab11 (35), Nedd4-Flag (kindly provided by S. Kumar, Hanson Institute, Adelaide, Australia), Itch-Myc, Itch-C830A-Myc, Itch-Flag, Itch-C830A-Flag (28), ubiquitin-Flag (36), ubiquitin-K48R-HA, and ubiquitin-K63R-HA (both kindly provided by D. Bohmann, University of Rochester Medical Center, Rochester, NY).
Cell Culture and Transfection.
COS-7 cells, HeLa cells, wild-type NIH 3T3 cells, and NIH 3T3-7d transfectants stably expressing ErbB4 isoforms (12, 37) were maintained in DMEM, 10% FCS (Promocell), and 1% l-glutamine–penicillin–streptomycin solution (Sigma–Aldrich). OVCAR-3 cells were cultured in RPMI medium 1640, 10% FCS, 1% l-glutamine–penicillin–streptomycin solution, and 10 μg/ml insulin (Sigma–Aldrich). Cells were transfected by using FuGENE 6 Transfection Reagent (Roche) according to the manufacturer's instructions.
Internalization Analysis.
Recombinant NRG-1, EGF, and HB-EGF were iodinated (38), and the quantity of cell-surface-bound and internalized growth factors was determined as previously described (6, 38). Shortly, NIH 3T3-7d transfectants expressing ErbB4 JM-b CYT-1, ErbB4 JM-b CYT-2, or EGFR were incubated for 1, 5, 10, or 15 min in the presence of 20 ng/ml 125I-NRG-1, 125I-EGF, or 125I-HB-EGF at 37°C. Cells were washed with ice-cold PBS, and the cell-surface-bound ligands were removed by two washes with an acidic (pH 2.8) buffer. The washed cells were lysed with NaOH releasing internalized factors. Radioactivity removed by acid washes and in cell lysates (containing surface-bound and internalized factors, respectively) was measured with a γ-counter.
Immunofluorescence and Confocal Microscopy.
Transfected COS-7 cells were fixed with methanol. Mouse anti-Myc antibody (clone 9E10; Zymed), rat anti-HA antibody (epitope 12CA5; Zymed), Alexa Fluor 488 goat anti-mouse antibody (Molecular Probes), and Alexa Fluor 568 goat anti-rat antibody (Molecular Probes) were used. DAPI (Sigma–Aldrich) was used to visualize the nuclei. Immunostained cells were analyzed by using an Olympus BX60 microscope with the 3.00 434 soft imaging system or by using an Axiovert 200M confocal microscope with LSM 510 Meta (Zeiss). Confocal images of single 0.8-μm sections are shown.
Ubiquitination of ErbB4.
To study the ubiquitination of ErbB4, COS-7 cells were transfected with HA- or Myc-tagged ErbB4 constructs and a plasmid encoding ubiquitin-Flag. Cells were starved without serum for 6 h, stimulated with 0 or 50 ng/ml NRG-1 for 10 or 30 min or with 0 or 100 ng/ml PMA for 30 min, and lysed. Aliquots of the lysates corresponding to 200–400 μg of total protein were immunoprecipitated with anti-ErbB4 antibody (HFR-1; Neomarkers), separated in 8% SDS/PAGE gels, and analyzed by Western blotting using an anti-Flag antibody (Sigma–Aldrich). Membranes were reblotted with anti-ErbB4 (Santa Cruz Biotechnology or Abcam).
To analyze ubiquitination of endogenous ErbB4, OVCAR-3 cells were starved overnight, stimulated with 0 or 50 ng/ml NRG-1 for 10 min, and lysed. Aliquots of lysates corresponding to 2 mg of total protein were immunoprecipitated with anti-ErbB4 antibody (HFR-1) and analyzed by Western blotting using anti-ubiquitin antibody (P4D1; Santa Cruz Biotechnology). Membrane was reblotted with anti-ErbB4 (sc-283, Santa Cruz Biotechnology or Abcam).
To study the effect of Itch on ErbB4 ubiquitination, COS-7 cells were transfected with ErbB4 JM-a CYT-1-HA and ubiquitin-Flag with or without Itch-Myc or Itch-C830A-Myc. Cells were starved without serum for 6 h, treated with or without 150 μM N-acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN; Calbiochem) for 2 h, and lysed. One milligram of total protein was immunoprecipitated with anti-ErbB4 antibody (HFR-1), separated in 8% SDS/PAGE gels, and blotted with mouse monoclonal FK1-antibody (Biomol) or mouse monoclonal FK2-antibody (Biomol) recognizing either polyubiquitinated or both poly- and monoubiquitinated proteins, respectively. Membranes were reblotted with anti-HA and anti-Myc antibodies.
Coimmunoprecipitation of ErbB4 and Itch.
To study the putative interaction of ErbB4 with Itch, COS-7 cells expressing HA-tagged ErbB4 proteins JM-a CYT-1, JM-a CYT-1 Y1056F, JM-a CYT-1 P1054P, or JM-a CYT-2 with or without Itch-Myc, Itch-Flag, or Itch-C830A-Myc were used. Cells were starved without serum for 6 h and lysed. Aliquots of the lysates corresponding to 500–650 μg of total protein were immunoprecipitated with anti-ErbB4 antibody (HFR-1) or anti-Flag antibody followed by reciprocal Western blotting using an anti-Myc antibody or anti-ErbB4 antibody (Abcam), respectively. Membranes were reblotted with anti-ErbB4 (sc-283), anti-Myc, or anti-Flag antibodies.
GST Pull-Down Assay.
Constructs encoding GST fusion proteins including each or all four of the WW domains of Itch and procedures have been described (28).
ErbB4 Degradation Analysis.
To study the effect of Itch on degradation of ErbB4, COS-7 cells transiently expressing HA-tagged ErbB4 JM-a CYT-1 or JM-a CYT-2 with or without Itch-Myc or Itch-C830A-Myc were used. Cells were starved without serum for 12 h and treated with 100 μM cycloheximide (Sigma–Aldrich) in medium without serum for 2, 4, or 6 h, as previously described (18). Cells were lysed, and 50-μg samples of total protein were separated by 8% SDS/PAGE and analyzed by Western blotting using an anti-ErbB4 antibody (Abcam). Expression of Itch or the Itch-C830A mutant was detected with anti-Myc antibody.
Supplementary Material
Acknowledgments.
We thank Dr. Graham Carpenter (Vanderbilt University, Nashville, TN) for valuable comments on the manuscript, Maria Tuominen and Minna Santanen for excellent technical assistance, and Mika Savisalo for plasmid construction. This work has been supported by the Academy of Finland, the Finnish Cancer Organizations, the Finnish Cultural Foundation, the Foundation for the Finnish Cancer Institute, the Kimmel Scholar Award, Ohio Cancer Research Associates, the Sigrid Jusélius Foundation, and Turku University Central Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708333105/DC1.
References
- 1.Hynes NE, Lane HA. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Marmor MD, Yarden Y. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 3.Wells A, Welsh JB, Lazar CS, Wiley HS, Gill GN, Rosenfeld MG. Science. 1990;247:962–964. doi: 10.1126/science.2305263. [DOI] [PubMed] [Google Scholar]
- 4.Vieira AV, Lamaze C, Schmid SL. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 5.Ye H, Kuruvilla R, Zweifel LS, Ginty DD. Neuron. 2003;39:57–68. doi: 10.1016/s0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- 6.Baulida J, Kraus MH, Alimandi M, Di Fiore PP, Carpenter G. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 7.Sorkin A, Di Fiore PP, Carpenter G. Oncogene. 1993;8:3021–3028. [PubMed] [Google Scholar]
- 8.Gullick WJ. J Pathol. 2003;200:279–281. doi: 10.1002/path.1335. [DOI] [PubMed] [Google Scholar]
- 9.Junttila TT, Sundvall M, Maatta JA, Elenius K. Trends Cardiovasc Med. 2000;10:304–310. doi: 10.1016/s1050-1738(01)00065-2. [DOI] [PubMed] [Google Scholar]
- 10.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 11.Ferretti E, Di Marcotullio L, Gessi M, Mattei T, Greco A, Po A, De Smaele E, Giangaspero F, Riccardi R, Di Rocco C, et al. Oncogene. 2006;25:7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- 12.Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M. Oncogene. 1999;18:2607–2615. doi: 10.1038/sj.onc.1202612. [DOI] [PubMed] [Google Scholar]
- 13.Elenius K, Corfas G, Paul S, Choi CJ, Rio C, Plowman GD, Klagsbrun M. J Biol Chem. 1997;272:26761–26768. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- 14.Määttä JA, Sundvall M, Junttila TT, Peri L, Laine VJ, Isola J, Egeblad M, Elenius K. Mol Biol Cell. 2006;17:67–79. doi: 10.1091/mbc.E05-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni CY, Murphy MP, Golde TE, Carpenter G. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 16.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Cell. 2006;127:185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 17.Kainulainen V, Sundvall M, Maatta JA, Santiestevan E, Klagsbrun M, Elenius K. J Biol Chem. 2000;275:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- 18.Sundvall M, Peri L, Maatta JA, Tvorogov D, Paatero I, Savisalo M, Silvennoinen O, Yarden Y, Elenius K. Oncogene. 2007;26:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- 19.Zerial M, McBride H. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 20.Engelman JA, Luo J, Cantley LC. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 21.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 22.Aqeilan RI, Donat V, Gaudio E, Nicoloso MS, Sundvall M, Korhonen A, Lundin J, Isola J, Sudol M, Joensuu H, et al. Cancer Res. 2007;67:9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 23.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 24.Komuro A, Nagai M, Navin NE, Sudol M. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 25.Hicke L, Dunn R. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 26.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Bioessays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 27.Omerovic J, Santangelo L, Puggioni EM, Marrocco J, Dall'armi C, Palumbo C, Belleudi F, Di Marcotullio L, Frati L, Torrisi MR, et al. FASEB J. 2007;21:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- 28.Oberst A, Malatesta M, Aqeilan RI, Rossi M, Salomoni P, Murillas R, Sharma P, Kuehn MR, Oren M, Croce CM, et al. Proc Natl Acad Sci USA. 2007;104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haglund K, Dikic I. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 31.Fujimuro M, Sawada H, Yokosawa H. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 32.Egeblad M, Jaattela M. Int J Cancer. 2000;86:617–625. doi: 10.1002/(sici)1097-0215(20000601)86:5<617::aid-ijc3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 33.Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. Mol Biol Cell. 2003;14:1882–1899. doi: 10.1091/mbc.E02-10-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. EMBO J. 2002;21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. J Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K, Yarden Y. Traffic. 2002;3:740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang K, Sun J, Liu N, Wen D, Chang D, Thomason A, Yoshinaga SK. J Biol Chem. 1996;271:3884–3890. [PubMed] [Google Scholar]
- 38.Elenius K, Paul S, Allison G, Sun J, Klagsbrun M. EMBO J. 1997;16:1268–1278. doi: 10.1093/emboj/16.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.