Abstract
Caspase-12 is a dominant-negative regulator of caspase-1 (IL-1β-converting enzyme) and an attenuator of cytokine responsiveness to septic infections. This molecular role for caspase-12 appears to be akin to the role of cFLIP in regulating caspase-8 in the extrinsic cell death pathway; however, unlike cFLIP/Usurpin, we demonstrate here that caspase-12 is catalytically competent. To examine these catalytic properties, rat caspase-12 was cloned, and the recombinant enzyme was used to examine the cleavage of macromolecular and synthetic fluorogenic substrates. Although caspase-12 could mediate autoproteolytic maturation of its own proenzyme, in both cis and trans, it was not able to cleave any other polypeptide substrate, including other caspase proenzymes, apoptotic substrates, cytokine precursors, or proteins in the endoplasmic reticulum that normally undergo caspase-mediated proteolysis. The dearth of potential substrates for caspase-12 also was confirmed by whole-cell diagonal-gel analysis. Autolytic cleavage within the caspase-12 proenzyme was mapped to a single site at the large–small subunit junction, ATAD319, and this motif was recognized by caspase-12 when incorporated into synthetic fluorogenic substrates. The specific activity of caspase-12 with these substates was several orders of magnitude lower than caspases-1 and -3, highlighting its relative catalytic paucity. In intact cells, caspase-12 autoproteolysis occurred in the inhibitory complex containing caspase-1. We propose that the proteolytic activity of caspase-12 is confined to its own proenzyme and that autocleavage within the caspase-1 complex may be a means for temporal limitation of the inhibitory effects of caspase-12 on proinflammatory cytokine maturation.
Keywords: apoptosis, inflammation, sepsis
The caspases are a family of cysteinyl proteases that play a critical role in apoptotic cell death as well as in inflammatory cytokine processing (1). The last and most recently discovered caspase is caspase-12, which is highly related to the cytokine processing group-I caspases that include caspase-1 (ICE, IL-1β-converting enzyme) (Fig. 1). Early studies suggested a potential role for caspase-12 in endoplasmic reticulum (ER) stress-induced apoptosis, including the cell death response to amyloid-β deposition and, therefore, the pathogenesis of Alzheimer's disease (2). Recent studies from our laboratories, however, are not consistent with these earlier findings (3, 4). Instead, caspase-12 was shown to play a role in modulating the inflammatory caspase activation pathway by a direct dominant-negative effect on caspase-1 and to have no detectable effect on apoptotic pathways. In humans, for example, an SNP results in the synthesis of either a full-length caspase-12 proenzyme or a substantially truncated protein containing only the prodomain, but not the large and small subunits of the active mature enzyme. The presence of the long polymorph, which is rare in the human population and largely confined to individuals of African descent, confers hyporesponsiveness to endotoxin-stimulated cytokine production in whole blood and is overrepresented in individuals with severe sepsis and septic mortality (3). In mice, caspase-12 gene deletion confers resistance to peritonitis and septic shock in surgical models of human sepsis, and the resulting survival advantage is conferred by enhanced bacterial clearance in caspase-12-deficient animals (4). Collectively, these data in humans and rodents demonstrate that the presence of caspase-12 attenuates responsiveness to bacterial invasion and confers a survival disadvantage. In neither case was an effect of apoptosis observed (including ER stress). Mechanistically, it was found that caspase-12 directly associated with caspase-1 and precluded normal caspase-1 activation. This interaction results in reduced cytokine processing, especially those processed by caspase-1, including IL-1β and IL-18 (IGIF, IFN-γ-inducing factor), and enhanced susceptibility to bacterial invasion as a consequence. Surprisingly, the catalytic activity of caspase-12 was not necessary for this inhibitory effect on the caspase-1 cytokine pathway (4), suggesting that, in the same way that cFLIP is a dominant-negative regulator of caspase-8 and the extrinsic cell death pathway (5, 6), caspase-12 is its counterpart for regulating the inflammatory caspase cascade. Unlike cFLIP, however, caspase-12 does not contain critical lesions in the catalytic machinery that would preclude it from proteolytic activity. Although several laboratories have struggled to detect proteolytic activity from caspase-12, it is possible that its catalytic capabilities are important for its role in caspase-1 modulation or other cellular functions. To this end, we have characterized the proteolytic properties of caspase-12.
Fig. 1.
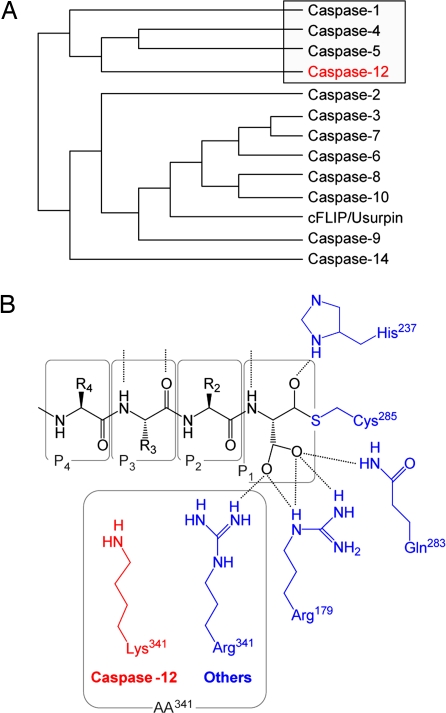
Caspase-12 features. (A) Human protein sequence alignment using the Merck multiclustal algorithm (14) places caspase-12 in the caspase-1 (ICE) group-I gene family (boxed). Mouse and rat phylogenic trees differ from human by the consolidation of human caspase-4 and -5 into rodent caspase-11. (B) An alignment of all known active mammalian caspases shows that all residues within the caspase enzymes that form the catalytic dyad and tether the P1 carboxylate side chain of substrates (blue) are absolutely conserved with the exception of Arg[ICE:341] (ICE numbering system), which is Lys in rodent caspase-12s (red).
Results
Caspase-12 Contains Key Features Necessary for Proteolysis.
For these studies, rat procaspase-12 was cloned and expressed in bacteria to produce active enzyme (National Center for Biotechnology Information accession no. AF317633.1). The deduced protein sequence was found to be 86% identical (95% similar) to mouse procaspase-12 and 48% identical (61% similar) to human caspase-12 (a 79-residue segment missing in the human sequence accounts for much of the rodent/human sequence nonidentity). A phylogenetic analysis clearly placed caspase-12 from all three species in the group-I inflammatory caspase cluster, which includes caspase-1, -4, -5, -12, and -14 (caspase-11 in the mouse and rat is the orthologue to human caspase-4 and -5) (Fig. 1A). Detailed analysis of the key residues contained within caspases that are known to be essential for proteolytic activity (based on the 3D x-ray structures of caspase-1 and -3) (7, 8) showed absolute conservation in rat caspase-12, with one exception (Fig. 1B). These conserved elements include the catalytic diad [Cys(ICE:285) and His(ICE:237)], the conserved QxCxG pentapeptide motif that harbors the catalytic cysteine residue (QACRG for rat caspase-12, like the majority of all other caspases), and three of the four residues that stabilize the Asp carboxylate side chain within the S1 subsite [Arg(ICE:179), Gln(ICE:283), and Ser(ICE:347)]. The lone exception is a conservative substitution of Arg(ICE:341) for Lys in rat caspase-12. This same substitution is present in mouse, but not human, caspase-12. With the exception that the effect of a conservative substitution in one of the four residues that stabilize the P1 Asp of caspase substrates is unknown, rodent caspase-12 appears to have the needed elements for catalytic activity. This finding is in contrast to cFLIP, where these critical residues are substantially changed, resulting in no catalytic ability (6).
Autolytic Processing of Caspase-12 in Cis and Trans.
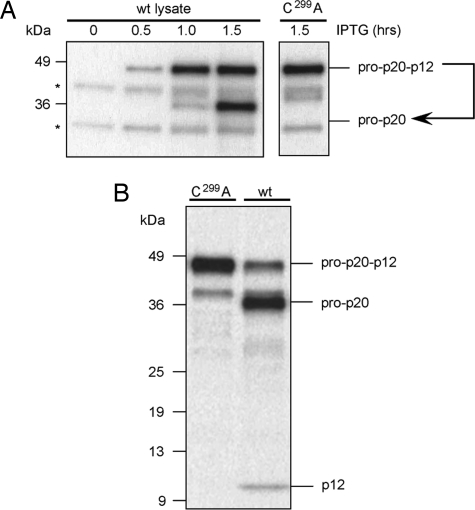
All caspases with catalytic activity are capable of autolytic processing between their large and small subunits when generated in heterologous expression systems (cis activation). Inducible bacterial expression systems are ideal for studying these events in vitro because processing does not occur during the early stages of induction, but then rapidly occurs as media acidifies (9). Rat caspase-12 expression in Escherichia coli was induced, and samples of the culture were harvested at varying time points and analyzed by SDS/PAGE and Western blotting using antisera raised against the large subunit of recombinant rat caspase-12. As occurs with other caspases in this system, procaspase-12 accumulated as its full-length proenzyme form for the first hour and then was rapidly converted to its mature form (Fig. 2A). That the maturation of caspase-12 was autolytic and not mediated by a bacterial protease was confirmed in parallel samples in which the essential catalytic Cys was mutated to Ala (C299A). To test whether caspase-12 could also autoprocess in trans, mature active caspase-12 was generated in bacteria and then was used to test for cleavage of [35S]procaspase-12. Mature active caspase-12 from bacterial expression was able to efficiently process [35S]procaspase-12 (Fig. 2B), and this activity also was ablated if the bacterial-expressed caspase-12 was inactivated through the C299A mutation. Together these data demonstrate that rat caspase-12 is proteolytically competent and is able to mediate its own maturation through autoproteolysis in both cis and trans.
Fig. 2.
Caspase-12 is capable of both cis and trans autocatalytic processing. (A) Bacterial expression of either wild-type procaspase-12 or the catalytically inactivated C299A mutant were induced by IPTG for the indicated time. Anticaspase-12 immunoblotting of total bacterial lysates showed cis autoprocessing occurred in the samples containing wild-type, but not C299A, procaspase-12. (B) Bacterial lysates containing either the inactive form of caspase-12 (C299A) or the autoprocessed wild-type form were used as enzymes for in vitro processing of [35S]procaspase-12 C299A. Trans cleavage with wild-type caspase-12 yielded 36- and 12-kDa fragments.
Caspase-12 Mediates Discrete Autoproteolysis at Asp319.
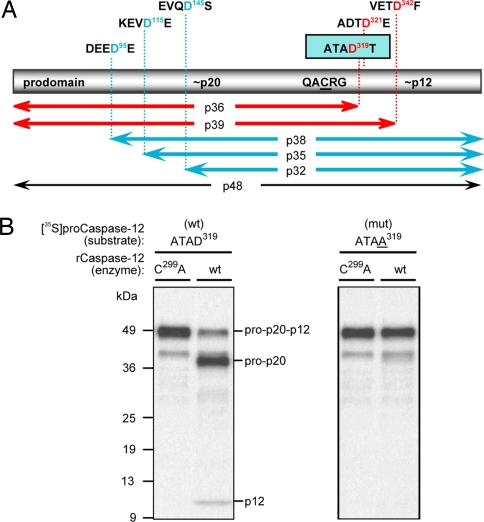
In vitro, most proteolytically active caspases are able to segregate their large subunits from their small subunits through autolytic proteolysis and to remove their N-terminal prodomains as well. If both events were occurring for rat caspase-12, then the accumulated product would be ≈20 kDa. Instead, the accumulating species after expression in bacteria (or trans cleavage of [35S]procaspase-12) was 36 kDa (Fig. 2), indicating that one event was occurring, but not the other. This finding is indicative of either a highly selective substrate requirement for caspase-12 or the unavailability of suitable automaturation sites within the caspase-12 proenzyme. To map the cleavage site to resolve this issue and to determine what the preferred motif for caspase-12 recognition might be, Asp residues across the proenzyme were systematically changed to Ala by site-directed mutagenesis (Fig. 3A) (data not shown). The resulting [35S]procaspase-12 polypeptides were used as substrate for cleavage by active caspase-12 that was generated in bacteria. A single mutation, D319A, prevented all processing activity (Fig. 3B), demonstrating that caspase-12 mediates automaturation by a single cleavage event between the large and small subunits at D319, but does not further process the large subunit from the prodomain. The sequence at that site (ATAD319T) also could shed light on the preferred sequence motif for caspase-12. Surprisingly, this motif is relatively featureless, so it is possible that the active site cleft of caspase-12 may not tolerate more elaborate side chains on the residues upstream of the P1 Asp. However, predictions made by combinatorial positional scanning would suggest that this site in caspase-12 would be amenable to efficient cleavage by both group-I (inflammatory) and group-III (apoptotic initiator) caspases (10). In limited in vitro testing, we detected such cleavages (data not shown).
Fig. 3.
Identification of procaspase-12 autoprocessing site. (A) The rat caspase-12 proenzyme has a predicted molecular mass of 48 kDa and is represented by the bar with its tripartite constituents: the N-terminal prodomain, the 20-kDa large subunit that harbors the catalytic Cys residue within the QACRG pentapeptide motif, and the 12-kDa small subunit. Potential caspase cleavage motifs are identified above the bar, with the corresponding fragment sizes indicated below the bar. (B) [35S]procaspase-12 mutants were used as substrates for in vitro cleavage assays with bacterial lysates containing either the inactive form of caspase-12 (C299A; left lanes) or the wild-type form (right lanes). Only the conversion of the ATAD319 (Left) to ATAA319 (Right) resulted in the loss of autoprocessing activity.
Caspase-12 Proteolytic Activity Is Confined to Self.
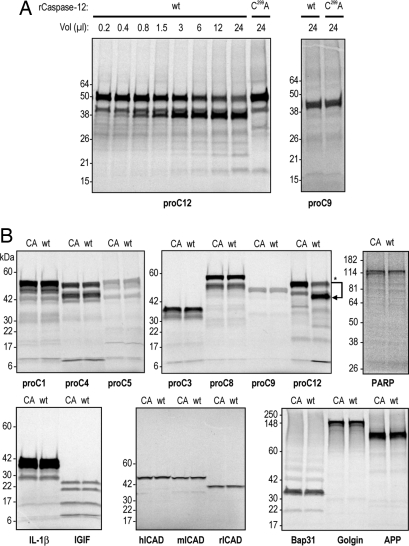
Although more recent data do not support a role for caspase-12 in mediating ER stress-induced apoptosis, earlier work suggested that caspase-12 could cleave and activate caspase-9, the principle initiator caspase of the intrinsic cell death pathway (11). We examined caspase-9 (Fig. 4A) among multiple known caspase substrates (Fig. 4B), including other caspase proenzymes (caspase-1, -4, and -5 from the group-I inflammatory subfamily, caspase-3 from the group-II effector subfamily, and caspase-8 and -9 from the group-III intrinsic and extrinsic pathways). We also examined caspase-1 substrates (IL-1β and IL-18/IGIF), representative apoptotic substrates (PARP and ICAD), proteins cleaved in the ER during apoptosis (Bap31 and Golgin), and the amyloid-β precursor protein (APP). Each cleavage reaction was performed on the respective [35S]polypeptide and included a parallel incubation with catalytically inactivated C299A caspase-12. Even at high concentrations for extended incubation times (up to 6 h), none of these proteins was cleaved by caspase-12 except the [35S]procaspase-12 positive control (Fig. 4B). To comprehensively extend the number of potential substrates examined, recombinant caspase-12 was used in a whole-proteome cleavage analysis using diagonal-gel PAGE coupled with MALDI-TOF mass spectroscopy (data not shown). This procedure has been applied successfully to the identification of mitochondrial caspase-3 substrates (12). Although this procedure yielded hundreds of putative substrates for caspase-1 and -3, only 11 were identified for caspase-12. All 11 of these potential substrates turned out to be false positives. Validation experiments included digestion of cell lysates with caspase-12 and detection of cleavage by Western analysis, in addition to in vitro transcription/translation of the putative substrate and cleavage of the resulting product with caspase-12. Collectively, these data indicate that, although caspase-12 is very effective at macromolecular recognition and cleavage of itself after D319, it does not effectively recognize any other cellular polypeptide that was examined. There are many caveats to these studies (our inability to examine potential substrates of low abundance, the potential of cofactor requirements, and the possibility that assay conditions are not appropriate), yet it is clear that caspase-12, under in vitro conditions that are highly permissive for other caspases, does not recognize polypeptide substrates other than itself, which it cleaves with high efficiency. This finding suggests the possibility that self-cleavage, as a potential mechanism for self-regulation, may be the primary function of caspase-12's catalytic abilities.
Fig. 4.
Macromolecular cleavage by caspase-12 does not extend to other known caspase substrates. Bacterial lysates containing either the inactive form of caspase-12 (C299A or CA) or the autoprocessed wild-type form of caspase-12 (wt) were used as enzyme for in vitro processing of the indicated [35S]precursor proteins. (A) Recombinant caspase-12 (rCaspase-12) titration shows concentration responsiveness in the reaction mixtures (Left), and the maximum concentration was used to test for rCaspase-12 processing of procaspase-9 (proC9) (Right). (B) Multiple caspase substrates were tested by the same procedure as described for A, with incubation times of 90 min. Only procaspase-12, used as a positive control, was cleaved.
Caspase-12 Recognition of Synthetic Substrates and Inhibitors.
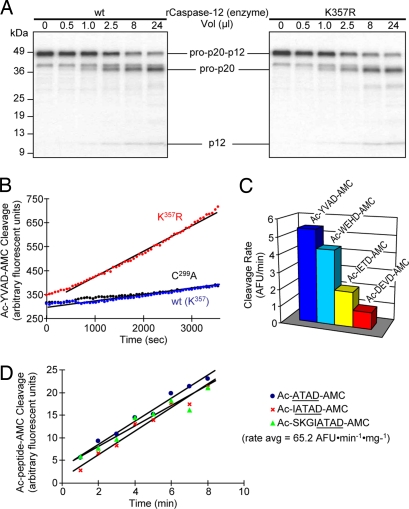
Within the spectrum of macromolecular substrates tested, caspase-12 only recognized its own proenzyme as a substrate. Therefore, we wanted to profile the caspase-12 enzyme for its ability to cleave fluorogenic ligands that are recognized by other caspase family members. We used a subset of four tetrapeptide-aminomethylcoumarins that were determined to represent the preferred substrate motifs for other caspases by positional scanning combinatorial libraries (10). These included the preferred group-I, -II, and -III substrates (Ac-WEHD-AMC, Ac-IETD-AMC, and Ac-DEVD-AMC, respectively), as well as the canonical caspase-1 (ICE) substrate (Ac-YVAD-AMC). Recombinant rat caspase-12 was unable to cleave any of these ligands despite robust cleavage by other caspases (data not shown). Owing to the conservative change in rodent caspase-12s of Arg(ICE:341) to Lys and the important role that this residue plays in forming the S1 subsite that tethers the carboxylate side chain of the P1 Asp, we mutated this residue to Arg [K357R mutant; residue 357 being the equivalent to Arg(ICE:341) in the preferred ICE nomenclature system]. This K357R-modified form of caspase-12 displayed enhanced catalytic activity toward synthetic tetrapeptide substrates (Fig. 5A) without concomitant changes in proteolytic efficiency toward its macromolecular substrate, procaspase-12 (Fig. 5B). In profiling the four canonical fluorogenic caspase substrates, K357R-modified caspase-12 retained a group-I (caspase-1/ICE-like) substrate preference (Fig. 5C). This substrate preference is consistent with the phylogenetic association of caspase-12 with other members of the group-I caspase subfamily (caspase-1, -4, and -5) (see Fig. 1A). To study the native enzyme without a corrective modification, however, we synthesized fluorogenic peptides corresponding to the self-recognition sequence within procaspase-12 at the junction of the large and small subunits (see Fig. 3A). These peptides included the tetrapeptide, Ac-ATAD-AMC, and two extended peptides, Ac-IATAD-AMC and Ac-SKGIATAD-AMC. All three peptides were cleaved by wild-type rat caspase-12 at approximately the same rate and yielded the same specific activity of 65.2 AFU·min−1·mg−1 (Fig. 5D) (data not shown). In comparing the efficiency of Ac-ATAD-AMC cleavage to that of caspase-3 on its preferred substrate (Ac-DEVD-AMC), the specific activity was approximately four to five orders of magnitude lower (3,940 AFU·min−1·mg−1). As one of the most catalytically efficient caspases, caspase-3 is a benchmark for other proteases in this gene family. For comparison, the Kcat/Km for caspase-3 (vs. preferred substrate) is comparable to caspase-1, 10-fold higher than caspase-4 and -5, 900-fold higher than caspase-2, and 3,600-fold higher than caspase-9 (13). Therefore, the approximated catalytic efficiency of caspase-12 places it as the least catalytically active caspase known. In summary, these data show that caspase-12 is catalytically competent, but that its specificity is highly confined to cleavage of its corresponding proenzyme at the macromolecular level and to the corresponding tetrapeptide motif in synthetic substrates. This high degree of specificity is further supported by the inability of canonical peptide-aldehyde caspase inhibitors to block in vitro cleavage of [35S]procaspase-12 by recombinant active caspase-12 [group-I (Ac-YVAD-CHO and Ac-WEHD-CHO), group-II (Ac-DEVD-CHO), and group-III (Ac-IETD-CHO) inhibitors were each tested up to 100 μM without effect] (data not shown).
Fig. 5.
Activity of caspase-12 and K357R-modified caspase-12 on fluorogenic synthetic substrates. (A) Recombinant caspase-12 was generated by expression in bacteria and tested for cleavage of [35S]procaspase-12 (C299A) in vitro by using the indicated volumes of either wild-type caspase-12 (Left) or the K357R-modified caspase-12 (Right). No difference in cleavage was observed. (B) Various forms of recombinant caspase-12 were tested for activity against the canonical caspase-1 substrate, Ac-YVAD-AMC. No activity was detected with wild-type or catalytically incapacitated C299A caspase-12, whereas the K357R modification restored activity against this synthetic substrate. (C) The K357R-modified caspase-12 was tested for cleavage activity against a panel of fluorogenic substrates that are able to detect all known caspases. The preferred recognition pattern is consistent with other group-I caspases. (D) Indicated fluorogenic ligands corresponding to the autocleavage motif within procaspase-12 were prepared by solid-phase synthesis. Unmodified wild-type caspase-12 was expressed in bacteria and tested for fluorogenic activity against these and other fluorogenic ligands. Caspase-12 specific activity is 0.0652 AFU·min−1·mg−1.
Caspase-12 Autoproteolysis Occurs in the Caspase-1 Complex in Intact Cells.
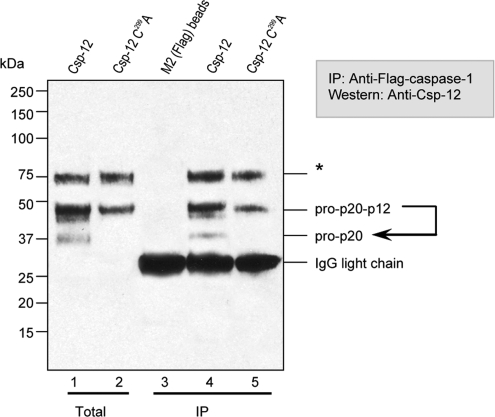
We have previously demonstrated that caspase-12 forms a complex with caspase-1 in intact cells and prevents caspase-1 activation and subsequent cytokine processing (4). We therefore examined whether caspase-12 autocleavage was occurring in these complexes (Fig. 6) by cotransfecting HEK293T cells with procaspase-1 plus procaspase-12. Caspase-1 was immunoharvested after cell lysis, and bound species of caspase-12 were detected by immunoblotting with specific anticaspase-12 antisera. The caspase-12 species that were associated with caspase-1 included both the full-length caspase-12 proenzyme and its autoproteolytic product (the 36-kDa prodomain/large-subunit fragment; the small subunit would not be detected if associated with the complex because the large subunit is the epitope for the detecting antibody). That the caspase-12 cleavage product was generated by autoproteolysis and not by other proteases, such as the associated caspase-1, was confirmed in parallel samples in which catalytically inactivated C299A caspase-12 was used. No processed caspase-12 was detectable in these complexes with caspase-1, demonstrating that processing is autolytic. Together these data demonstrate that caspase-12 undergoes autocatalyic cleavage in the caspase-1 complex and that the resulting cleavage fragment remains associated with caspase-1, although we have not explored potential differences in dissociation kinetics between cleaved versus uncleaved caspase-12.
Fig. 6.
Procaspase-12 forms a complex with caspase-1 and is partially autoprocessed in the complex. HEK293T cells were cotransfected with expression vectors harboring Flag-tagged procaspase-1 (all lanes) plus either procaspase-12 (lanes 1 and 4) or the catalytically incapacitated C299A mutant (lanes 2 and 5). After 24 h, cells were harvested and lysed. One tenth of the lysate was directly applied to SDS/PAGE (lanes 1 and 2), and the remainder was immunoharvested with antibodies directed against the caspase-1 Flag epitope tag (lanes 4 and 5; lane 3 was processed in the same way, except that only lysis buffer was used). Immunoblotting for the large subunit (p20) of caspase-12 revealed that procaspase-12 and the resulting autocleavage product were both immunoharvested with caspase-1. The asterisk indicates a band of unknown identity that is detected by prebleed control serum.
Discussion
In both humans and mice, the presence of full-length caspase-12 (manifest by the infrequent read-through polymorphism in humans and by wild-type caspase-12 in mice) results in the attenuation of inflammatory responsiveness and enhanced vulnerability to septic infection and mortality (3, 4). It appears that the primary mechanism by which caspase-12 confers hyporesponsiveness to infection is by dampening the formation of key cytokines [IL-1β and IL-18 (and INF-γ as a consequence)] through a dominant-negative effect on caspase-1 (ICE), the protease that mediates proteolytic maturation of these cytokines. In this regard, the mechanism is similar to the dominant-negative effect of cFLIP on caspase-8 in the extrinsic cell death pathway (5). Although caspase-12 possesses catalytic activity as demonstrated here, this activity is dispensable for the dominant-negative effect on caspase-1. This finding begs the question as to whether the catalytic activity is relevant in vivo or is an inconsequential evolutionary remnant. For example, cFLIP is a member of the caspase-8/caspase-10 gene cluster and arose from tandem gene duplication. It has been catalytically inactivated by multiple incapacitating mutations in residues that are necessary for catalysis, but not for dominant-negative recognition of caspase-8. This catalytic incompetency may be manifested in caspase-12 by another route; namely, the highly restricted substrate specificity and low catalytic efficiency of this enzyme. Alternatively, the catalytic activity may be important for events in the caspase-1 cascade that are not evident within the experiments conducted here. For example, at early stages of complex formation, caspase-12 may hold caspase-1 in check via its dominant-negative effect, but eventually release its inhibition through autoproteolysis. This built-in latency may be important to prevent excessively rapid activation of the inflammatory caspase cascade under normal circumstances, but may become a liability under severe pathogenic conditions, such as in sepsis. We are currently exploring these possibilities by generating gene-modified mice harboring the C299A catalytically inactivated caspase-12 and by humanizing caspase-12-deficient mice with either the long form of human caspase-12 or its C299A counterpart. In summary, caspase-12 is highly unique in that it functions as a dominant-negative regulator of the inflammatory caspase cascade, yet it possesses proteolytic activity of unknown function or consequences. Furthermore, caspase-12 is unique in its high degree of macromolecular specificity, a feature that distinguishes it from all other caspases. An understanding of the role of this proteolysis may be important to provide an opportunity to modulate caspase-12 given the role of this caspase in conferring vulnerability to sepsis.
Methods
Cloning and Mutagenesis of Rat Caspase-12.
The 1,263-bp ORF for rat procaspase-12 was PCR-amplified from a rat lung cDNA library (Clontech) and inserted into pBluescript II SK+ (Promega). The full sequence was deposited to the National Center for Biotechnology Information (accession no. AF317633.1). The insert was subcloned into the prokaryotic expression vectors pET11d and pET20b(+) (Novagen) to generate a C-terminal His-tagged fusion protein. Where indicated, the cDNA for rat procaspase-12 in pET11d was mutagenized by using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene) to convert residues as indicated. The constructs' sequence integrity was confirmed by sequencing.
Preparation of Bacterial Lysates Expressing Rat Caspase-12.
The cDNA for wild-type or mutant procaspase-12, subcloned in pET11d, was transformed into the protease-deficient strain of E. coli BL21 (DE3) pLysS (Novagen). Protein expression was induced in cultures exponentially growing in M9 medium with 100 μg/ml carbenicillin by adding 1 mM isopropyl-d-thiogalactoside (IPTG) and persuing incubation at either 28°C or 37°C. For soluble fraction samples, 2.5-ml aliquots of bacterial culture were removed at various time points and centrifuged. Pellets were resuspended in 500 μl of cold ICE buffer III [50 mM Hepes/KOH (pH 7.0), 2 mM EDTA, 0.1% (wt/vol) CHAPS, 10% (wt/vol) sucrose, and 5 mM DTT], lysed by sonication on ice, and cleared by centrifugation at 4°C. For total fraction samples, 1-ml aliquots of bacterial culture were removed at various time points and centrifuged. Pellets were lysed by resuspension in 200 μl of 1× Laemmli SDS loading buffer and sonicated to shear DNA.
Generation of Rat Caspase-12 Antiserum.
E. coli BL21(DE3) pLysS was transformed with the cDNA for rat caspase-12 p20 subcloned into pET11d. Protein expression was induced in exponentially growing cells by a 5-h induction at 37°C in M9 medium containing 1.5 mM IPTG. Under such conditions, rat caspase-12 p20 was primarily localized in the inclusion body fraction, where it was the only protein detectable by SDS/PAGE. Inclusion bodies were purified, denatured in 6 M guanidine HCl and 25 mM Tris (pH 7.4), and used directly for rabbit immunization.
SDS/PAGE and Immunoblotting.
For Coomassie blue staining, 10 μl of bacterial lysates were fractionated by SDS/4–20% PAGE (Novex) and stained with Simply Blue Safestain (Invitrogen) according to the manufacturer's instructions. For Western blotting, 1 μl of bacterial lysate was fractionated by SDS/4–20% PAGE (Novex) and electrophoretically transferred to a nitrocellulose membrane. Blots were incubated in blocking buffer [Tris-buffered saline (pH 7.4) with 0.05% Tween-20 (0.05% TBST), 5% milk (blotting grade blocker nonfat dry milk from Bio-Rad)] for 1 h and in primary antibody diluted 1/20,000 in blocking buffer for 1 h. Blots were washed three times in 0.1% TBST for 5 min. They were then further incubated for 1 h at room temperature in secondary IgG coupled to horseradish peroxidase diluted 1/3,000 in blocking buffer. After washing three times in 0.3% TBST for 5 min, followed by three times in 0.1% TBST for 5 min, detection was performed by using ECL (Amersham).
In Vitro Cleavage of Radiolabeled Proteins.
[35S]Methionine-labeled proteins were obtained by coupled in vitro transcription/translation using the Promega TNT reticulocyte lysate system as per the manufacturer's instructions. Cleavage of the radiolabeled product was performed by incubation at 37°C in the presence of soluble bacterial lysate in a final reaction volume of 25 μl. Unless otherwise indicated, cleavage mixtures were incubated for 90 min. Cleavage reactions were terminated by the addition of 5× SDS Laemmli loading buffer, resolved by SDS/10–20% PAGE (Novex), and visualized by autoradiography (BioMax MR film; Kodak).
Caspase Fluorogenic Enzyme Enzymatic Assays.
Caspase catalytic activity was measured by using a continuous fluorometric assay based on the proteolytic cleavage of a substrate with the general structure 7-amino-4-methylcoumarin (AMC). Reactions contained 50 μl of soluble bacterial lysate and 10 μM substrate in a final volume of 200 μl of ICE buffer III. AMC release was measured in a Cytofluor 96-well plate reader by using an excitation wavelength of 380 nm and an emission wavelength of 460 nm. Synthetic inhibitors and fluorogenic substrates were synthesized in our laboratories, but are broadly available (without license) through several vendors.
Coimmunoprecipitation Experiments.
HEK293T cells were cotransfected by using Lipofectamine Plus reagent (Invitrogen) with Flag-tagged caspase-1 C[ICE:285]A and caspase-12 in either the wild-type form or C[ICE:285]A (C299A) catalytically inactivated mutant. At 24 h after transfection, cell lysates were prepared in buffer B [20 mM Tris·HCl (pH 8.0), 150 mM KCl, 10% glycerol, 5 mM MgCl2, 0.1% Nonidet P-40, and a protease inhibitor mixture] and caspase-1, -5, and -9 were immunoprecipitated by using Flag M2 agarose beads (Sigma–Aldrich) at 4°C for 2 h, followed by three washes of the beads with lysis buffer. Immunoconjugates were eluted from the beads by using Flag peptides (Sigma–Aldrich) and were processed for Western blot analysis by using anticaspase-12 antiserum.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Creagh EM, Conroy H, Martin SJ. Immunol Rev. 2003;193:10–21. doi: 10.1034/j.1600-065x.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 2.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 3.Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS, et al. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 4.Saleh M, Mathison JC, Wolinski MK, Bensinger SJ, Fitzgerald P, Droin N, Ulevitch RJ, Green DR, Nicholson DW. Nature. 2006;440:1064–1068. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- 5.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, et al. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 6.Rasper DM, Vaillancourt JP, Hadano S, Houtzager VM, Seiden I, Keen SL, Tawa P, Xanthoudakis S, Nasir J, Martindale D, et al. Cell Death Differ. 1998;5:271–288. doi: 10.1038/sj.cdd.4400370. [DOI] [PubMed] [Google Scholar]
- 7.Rotonda J, Nicholson DW, Fazil KM, Gallant M, Gareau Y, Labelle M, Peterson EP, Rasper DM, Ruel R, Vaillancourt JP, et al. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 8.Walker NP, Talanian RV, Brady KD, Dang LC, Bump NJ, Ferenz CR, Franklin S, Ghayur T, Hackett MC, Hammill LD, et al. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 9.Roy S, Bayly CI, Gareau Y, Houtzager VM, Kargman S, Keen SL, Rowland K, Seiden IM, Thornberry NA, Nicholson DW. Proc Natl Acad Sci USA. 2001;98:6132–6137. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 11.Morishima N, Nakanishi K, Takenouchi H, Shibata T, Yasuhiko Y. J Biol Chem. 2002;277:34287–34294. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]
- 12.Ricci JE, Munoz-Pinedo C, Fitzgerald P, Bailly-Maitre B, Perkins GA, Yadava N, Scheffler IE, Ellisman MH, Green DR. Cell. 2004;117:773–786. doi: 10.1016/j.cell.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Calvo M, Peterson EP, Rasper DM, Vaillancourt JP, Zamboni R, Nicholson DW, Thornberry NA. Cell Death Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 14.Yuan J, Amend A, Borkowski J, DeMarco R, Bailey W, Liu Y, Xie G, Blevins R. Bioinformatics. 1999;15:862–863. doi: 10.1093/bioinformatics/15.10.862. [DOI] [PubMed] [Google Scholar]