Abstract
Much is known about the composition and function of the postsynaptic density (PSD), but less is known about its molecular organization. We use EM tomography to delineate the organization of PSDs at glutamatergic synapses in rat hippocampal cultures. The core of the PSD is dominated by vertically oriented filaments, and ImmunoGold labeling shows that PSD-95 is a component of these filaments. Vertical filaments contact two types of transmembrane structures whose sizes and positions match those of glutamate receptors and intermesh with two types of horizontally oriented filaments lying 10–20 nm from the postsynaptic membrane. The longer horizontal filaments link adjacent NMDAR-type structures, whereas the smaller filaments link both NMDA- and AMPAR-type structures. The orthogonal, interlinked scaffold of filaments at the core of the PSD provides a structural basis for understanding dynamic aspects of postsynaptic function.
Keywords: EM tomography, high-pressure freezing, hippocampal neuron, PSD-95
The postsynaptic density (PSD), a macromolecular signaling assembly embedded in the postsynaptic membrane (PSM) of neurons, contains receptors, scaffold molecules, and cytoskeletal elements and is the primary postsynaptic site for signal transduction and signal processing (1–3). PSDs are identified by EM as a band of electron-dense material (4) 20–30 nm thick and 300 nm long. The PSDs at excitatory synapses contain glutamate receptors of the NMDA and AMPA type. Recycling of AMPA receptors at the PSD accounts for dynamic changes in synaptic transmission (5).
Prompted by success with EM tomography on presynaptic structures at the frog neuromuscular junction (6), we adapted methods to determine the superamolecular structure of the PSD. The PSD, however, is a much larger molecular machine that may include several hundred proteins (7–9).
PSDs in dendritic spines from unstimulated hippocampal cultures were prepared for tomography by high-pressure freezing and freeze substitution. We started by identifying structures containing PSD-95, a member of the family of membrane-associated guanylate kinase (MAGUK) proteins composed of PSD-95, PSD-93, SAP97, and SAP102 (10), which have many known binding partners and large numbers of copies in PSDs (11). Next, we determined the relationships of major transmembrane structures and transverse elements inside the PSD to structures containing PSD-95. A picture emerges, in which vertically oriented filaments containing PSD-95 family members link glutamate receptors with other scaffolding molecules to establish an orthogonal matrix at the core of the PSD, by which we mean the organization of the scaffolding proteins concentrated near the postsynaptic membrane. This picture provides a structural basis for further understanding many aspects of PSD function.
Results
Tomography of PSDs.
Eight dendritic spines free from ice damage [supporting information (SI) Fig. 6 and SI Methods] and with cross-sectioned PSDs were selected for EM tomography. One PSD from the reconstruction of a mushroom-shaped spine (SI Methods) was selected for extensive segmentation, rendering, and structural analysis (Fig. 1 A and B). Structures as small as 3–4 nm in diameter within 1- to 1.5-nm-thick virtual sections can be segmented. Dimensions of structures such as actin filaments (7 nm wide) (data not shown), synaptic vesicles (40 nm in diameter), and synaptic cleft (25 nm wide) are in close agreement with dimensions determined by cryo-EM and other methods (12–14).
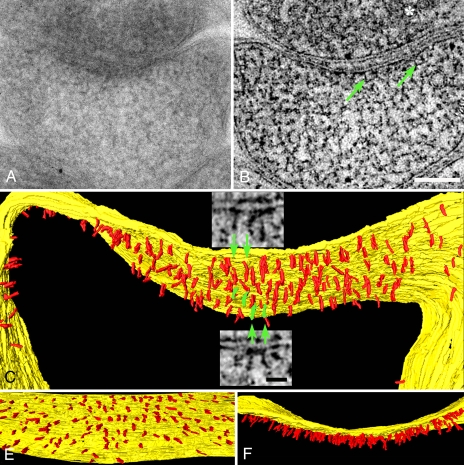
Fig. 1.
Vertical filaments at the PSD. (A) EM of a PSD in a mushroom-shaped dendritic spine. Structural details are obscured by overlap within this 120-nm-thick section. (B) Vertical filaments (green arrows) are apparent in a 1.5-nm-thick virtual section derived from the tomographic reconstruction of the section in A. The synaptic vesicle is indicated by an asterisk. (Scale bar: 100 nm.) (C) Rendering of vertical filaments (red) from the tomographic reconstruction. Vertical filaments, 5 nm in diameter and 20 nm long, contact the postsynaptic membrane (yellow). (Insets) Virtual sections from which particular vertical filaments (green) are segmented. (Scale bar: 20 nm.) (D) En face view showing uniform distribution of vertical filaments at the PSD. (E) Overlap of vertical filaments contributes to the typical thickened appearance of a PSD viewed in cross-section.
Vertical Filaments.
Filaments segmented in a series of virtual sections were classified on the basis of their location, shape, and dimensions. A large class of membrane-associated filaments at the PSD is nearly straight and vertically oriented with respect to the postsynaptic membrane (Fig. 1 B and C and SI Movie 1). We refer to filaments of this type as vertical filaments. Vertical filaments are typically 5 nm in diameter (4.9 ± 0.2 nm, n = 22) and 20 nm long (21 ± 3 nm, range 16–25 nm, n = 38). Vertical filaments within the PSD are uniformly spaced, with a nearest neighbor distance of 13.4 ± 2.8 nm (n = 31) (Fig. 1D). The thicket of vertical filaments gives rise to the typical dense appearance that is characteristic of PSDs in standard EM cross-sectional views (Fig. 1E). Vertical filaments are ubiquitous in reconstructions of PSDs, even in places where other structural elements are absent. Based on their density, there would be ≈400 vertical filaments in a 400-nm-diameter (0.16-μm2) PSD.
The dimensions of vertical filaments, and their associations with the postsynaptic membrane, suggest that they belong to the PSD-95 family of MAGUK proteins (10). These family members share many structural similarities, permitting the dimensions of a generic family member to be estimated by combining the known sizes of their domains (20 nm long, 4 nm in diameter) (SI Methods). Dimensions of fully extended PSD-95 and SAP97 molecules (16–22 nm long and 6 nm in diameter) by single-particle EM (15) also match to those of vertical filaments.
Vertical Filaments Label for PSD-95.
Conventional immuno-EM shows that the antibodies to PSD-95 specifically label PSDs (Fig. 2 A and B). When parallel labeling experiments are followed by freeze substitution and tomography, electron-dense particles corresponding to silver-enhanced Nanogold are located near vertical filaments (n = 9) (Fig. 2D Lower). We expected that the secondary antibody on the Nanogold would be enveloped in silver; in the four instances that were rendered, structures embedded in the silver grain contacted filaments up to 15 nm long that, in turn, contacted the vertical filaments (Fig. 2D Upper). Because the size of the filaments linking gold particles and vertical filaments corresponds to that expected for the primary IgG and does not match other filaments in the PSD, we tentatively conclude that we are observing antibodies bound to PSD-95 in the vertical filaments.
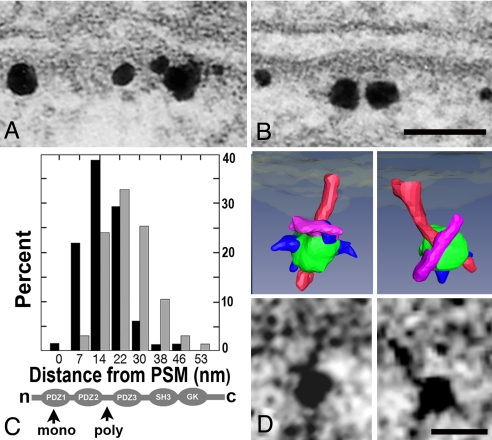
Fig. 2.
Mapping and labeling of PSD-95. (A) ImmunoGold label for PSD-95 with monoclonal antibody to PDZ1 domain. (B) ImmunoGold label for PSD-95 with polyclonal antibody to the region between PDZ2 and PDZ3. Both labels are confined to the vicinity of the PSD. (Scale bar: 50 nm.) (C) Label with monoclonal antibody (black bars) is closer to the PSM than label with polyclonal antibody (gray bars). Percentage refers to the percentage of total gold label. Locations of epitopes on PSD-95 are indicated below (arrows). (D) Label associated with filaments in single virtual sections (below) and surface renderings (above). Vertical filaments are rendered in red, and silver-enhanced gold particles are rendered in green. Structures rendered in purple are the correct size to represent the primary antibody contacting the vertical filaments. Smaller structures rendered in blue may represent the secondary Fab fragment partially buried in the silver encrusted on the gold particle. (Scale bar: 10 nm.)
Vertical Orientation and Polarity of PSD-95 Molecules.
We next tested whether a population of PSD-95 molecules is vertically oriented with their N termini at the membrane. Different PSD-95 antibodies targeting different positions on vertically oriented molecules should lie at different distances from the postsynaptic membrane. A monoclonal antibody targeting residues 64–121 within the PDZ1 domain (SI Fig. 7 and SI Methods) and a polyclonal antibody to residues 291–302 in the loop between PDZ2 and PDZ3 were used. The two epitopes should be separated by 6 ± 2 nm (Fig. 2C Lower). Their actual separation in a direction perpendicular to the membrane was determined by comparing the distances from the membrane to the centers of the silver particles corresponding to an antibody label (Fig. 2 A and B). The label for the PDZ 2/3 domain was separated from the postsynaptic membrane along an axis perpendicular to its tangent by 25 ± 9 nm (n = 67), whereas that for the PDZ1 was separated by 17 ± 8 nm (n = 66), significantly closer by 8 nm (P < 0.001) (Fig. 2C Upper). Thus, many of the PSD-95 molecules in the PSD are vertically oriented with their N termini at the membrane to which they are attached (16).
Transmembrane Structures.
Structures of various sizes and shapes line the cytoplasmic side of the spine membrane. Many of them appear to line up with large structures on the external side of the membrane, suggesting that they are transmembrane structures, such as glutamate receptors. The extracellular domains of NMDA and AMPA receptors share high sequence homology (17) and are predicted to have almost identical shapes and sizes (11). The extracellular domain of intact AMPA receptor is an elongated shape that is 16 nm long, 8 nm wide, and 10 nm high (18). For any extracellular structures that closely fit these dimensions, the corresponding domain on the cytoplasmic side of the membrane was precisely segmented in three orthogonal planes. Precise alignment with a cytoplasmic domain signified a transmembrane structure and, thus, a potential glutamate receptor. We found 46 extracellular structures 15 ± 3 nm long, 8 ± 1 nm wide, and 9 ± 1 nm high in our sampled area that fit the dimensions of the extracellular domain of the glutamate receptor (Fig. 3A and SI Table 1). Transmembrane structures have two types of cytoplasmic domains. One type is flat (18 ± 3 nm long, 10 ± 1 nm wide, and 5 nm high). The other type is larger and more globular (20 ± 2 nm long, 14 ± 2 nm wide, and 16 ± 4 nm high) (SI Table 1). The dimensions of the cytoplasmic aspects of the two types of transmembrane structures fit those of AMPA and NMDA receptors as estimated by structural informatics analysis (6–7 in diameter and 3 nm high for the AMPA receptor and globular ≈20 nm in diameter for the NMDA receptor) (SI Tables 1 and 2) (19, 20). Therefore, the transmembrane structures that are slender and flat on their cytoplasmic aspects are tentatively designated as “AMPAR-type structures,” whereas transmembrane structures manifesting larger, globular cytoplasmic aspects are designated as “NMDAR-type structures.”
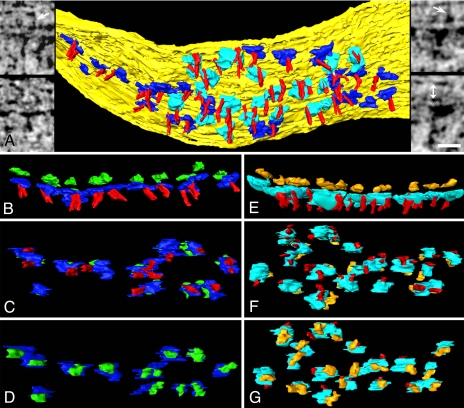
Fig. 3.
Transmembrane structures at the PSD. (A) Cytoplasmic surface of the membrane (yellow) at the PSD, showing AMPAR-type (blue) and NMDAR-type cytoplasmic domains (cyan), and vertical filaments (red). (Left Insets) AMPAR-type structures in virtual sections from the tomographic reconstruction. Arrow points to a transcleft filament. (Right Insets) NMDAR-type structures. Arrow points to a transcleft filament, and double arrow (lower right) indicates the extent of synaptic cleft. (Scale bar: 20 nm.) (B–D) AMPAR-type structures shown in cross-section (B, extracellular domain green), en face from inside the spine (C), and en face from outside the spine (D). Cytoplasmic domains of AMPAR-type structures are contacted by vertical filaments (C). (E–G) NMDAR-type structures shown in cross-section (E, extracellular domain gold), en face from inside the spine (F), and en face from outside the spine (G). Cytoplasmic domains of NMDAR-type structures are contacted by one or two vertical filaments (F).
AMPAR-Type Structures.
AMPAR-type structures are arrayed around the periphery of the PSD, with a mean nearest neighbor distance of 24 ± 6 nm (range 14–36 nm, n = 17) (Fig. 3 A–D). Extrapolation based on the 29 AMPAR-type structures in our sampled area predicts 30–100 AMPAR-type structures in a typical 400-nm-diameter PSD (SI Methods). Each AMPAR-type structure (28 of 29) is typically contacted near its center by a vertical filament (Fig. 3 B and C). Virtually every extracellular domain of the AMPAR-type structures within the PSD associates with a 3- to 4-nm-diameter filament spanning the synaptic cleft (Fig. 3A Left Inset). The transcleft filaments that contact the presynaptic membrane are 24 ± 1 nm long (range 18–30 nm, n = 9).
NMDAR-Type Structures.
The cytoplasmic domains of NMDAR-type structures are the largest structures near the membrane. The NMDAR-type structures (Fig. 3A Right Inset) cluster in a central region of the PSD, typically ≈130 nm in diameter, forming a regularly spaced (nearest neighbor distance 32 ± 4 nm, range 23–39 nm, n = 17) rhombic lattice (Fig. 3 A, F, and G). The cytoplasmic side of each NMDAR-type structure typically associates with two vertical filaments that contact their edges (Fig. 3 E and F). The extracellular domains of NMDAR-type structures also associate with fine filaments in the synaptic cleft (Fig. 3A Right Inset).
Horizontal Filaments.
Two types of filaments have a horizontal orientation parallel to the postsynaptic membrane and are largely limited to the PSD. In virtual sections, one type has a diameter of 4–5 nm and a length of ≈20 nm, and it lies 10–20 nm from the membrane at the PSD (Fig. 4A, purple, and B). The second type lies slightly further (15–20 nm) from the membrane and is 30–35 nm long and 5–6 nm in diameter (Fig. 4 A, white, and C). Each type intersects with other members of its own type while interweaving with and contacting the vertical filaments (Fig. 4A), lending them a layered appearance (Fig. 4D).
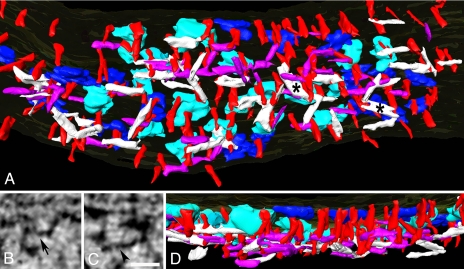
Fig. 4.
Filament network at the PSD. (A) Meshwork of horizontal filaments at the core of the PSD. The shorter type (purple) is 4–5 nm in diameter and ≈20 nm long, and the longer type (white) is 5–6 nm in diameter and 30–35 nm long. The asterisks indicate sheet-like structures. (B and C) Shorter (B, arrow) and longer (C, arrowhead) types of horizontal filament in virtual sections. (Scale bar: 20 nm.) (D) Cross-sectional view of the PSD in A showing layering of horizontal filaments. The shorter filaments (B, purple) lie somewhat closer to the postsynaptic membrane than the longer filaments (C, white).
One or both ends of the shorter type of filament (Fig. 4A) contacts a vertical filament. The shorter type of filament may contact vertical filaments associated with any of the adjacent AMPAR- or NMDAR-type structures. One or both ends of the longer, larger type of filament typically contact vertical filaments that contact adjacent NMDAR-type structures and, in a few instances, adjacent AMPAR-type structures. The longer type of filament appears to be more concentrated under the NMDAR-type structures. When rendered, some of these filaments assume a sheet-like appearance (Fig. 4A).
Discussion
Tomographic reconstructions from freeze-substituted hippocampal cultures provide the first views of the molecular organization of the PSDs from dendritic spines.
Vertical Filaments Are a Major Structural Element of the PSD.
They form an array of nearly vertically oriented, membrane-associated filaments that label for PSD-95. These evenly spaced vertical filaments are the most abundant structural entity in the PSD. The number of vertical filaments also closely matches the total number of PSD-95 family members in a typical PSD (11), suggesting that most, if not all, of the vertical filaments are members of the PSD-95 family.
AMPAR-Type Structures.
Classification of the AMPAR-type structure was based on the close match of the extracellular aspects of transmembrane structures with the dimensions expected of AMPA receptors (18). Although the sizes of the cytoplasmic domains of AMPAR-type structures are close to predicted, they are two to three times longer than expected in the plane parallel to the membrane, possibly representing proteins, such as TARPs known to associate with AMPA receptors (21).
There are several reasons to believe that the AMPAR-type structures correspond to AMPA receptors. Because AMPA receptors complex with TARPs that bind to PSD-95 family members (21–23), we anticipated that vertical filaments would contact the AMPAR-type structures. Also, AMPA receptors are preferentially distributed at the periphery of PSDs (24, 25), where we find the AMPA-type structures, and the nearest neighbor distance between AMPAR-type structures (24 ± 6 nm), matches the spacing between AMPA receptors derived from replica labeling (20 ± 4 nm) (28). Finally, our estimate of 30–100 AMPAR-type structures at the PSD agrees with estimates of the numbers of AMPA receptors (26, 27).
NMDAR-Type Structures.
Although the size and shape of extracellular domains of NMDAR-type structures are indistinguishable from those belonging to AMPAR-type structures (11), their cytoplasmic domains differentiate them. Their diameter is consistent with that predicted for NMDA receptors except that they are ≈6 nm taller possibly due to other proteins associating with NMDA receptors.
Each large cytoplasmic domain of the NMDA-type structure typically associates with two PSD-95 containing vertical filaments around its rim. Indeed, it is known that PSD-95 binds to NR2 (28, 29) and that each NMDA receptor includes two copies of NR2 (17). Almost all of the NMDAR-type structures cluster at the center of the PSD, where they are arranged in a rhombic lattice, allowing for four to five NMDAR-type structures arranged per lattice line. This arrangement would allow for 16–25 NMDA receptors in the central cluster of a typical PSD, which is consistent with previous estimates of 20–30 NMDA receptors (11, 30).
Extracellular domains of NMDAR- and AMPAR-type structures associate with transcleft filaments, which are likely to be adhesion molecules (31, 32). Their molecular identities require further investigation.
Two Types of Horizontal Filament Link Vertical Filaments.
The shorter (≈20 nm long) type of horizontal filament typically associates with vertical filaments contacting either NMDAR- or AMPAR-type structures as if linking adjacent receptors via the vertical filaments. The lengths of these links would explain the ≈20-nm nearest neighbor distance among AMPAR-type structures. The longer (≈30 nm) type of horizontal filament associates with vertical filaments contacting both AMPAR- and NMDAR-type structures, but they predominately link adjacent NMDAR-type structures, which might explain their ≈30-nm spacing. The two types of horizontal filaments lie in slightly separate layers, lending the PSD a laminar organization (33, 34).
Because PSD-95 family members have many potential binding partners, the molecular identities of the horizontal filaments that they contact may be complex, even including a few multimerized PSD-95 family members (35). GKAP/SAPAP is likely to be part of the horizontal filament system because it binds to the GK domain of PSD-95. There are ≈150 copies of GKAP/SAPAP in a PSD (11), where it exists in 95- and 130-kDa forms (36). These molecular mass are in line with those expected of 5-nm filaments either the same length (20 nm) or 1.5 times the length (30 nm) of the vertical filaments. Another prevalent scaffolding molecule, Shank, binds to GKAP/SAPAP, so the sheet-like structures formed by horizontal filaments may include both GKAP and Shank (37). Indeed, Shank has a SAM domain that can polymerize into sheet-like structures (38).
Organization of the Core of the PSD.
PSD-95 has been regarded as a central player in the core organization of the PSD due to its prevalence in the PSD and its multiple binding sites for receptors and other scaffolding molecules. Our findings that a large array of vertically oriented filaments dominates the core structure of the PSD, and that the vertical filaments include PSD-95 in an extended configuration, show how PSD-95 and related family members could link other core components of the PSD into a coherent structure, as schematized in Fig. 5. The vertical orientation of PSD-95 puts its PDZ domains near the membrane in positions to bind directly, or through intermediate proteins, to glutamate receptors and deploys the SH3-GK domains further from the membrane, where they can contact a family of horizontally oriented filaments, including GKAP and, perhaps, Shank. The vertical orientation and extended shape assumed by PSD-95 family members lying in the core of the PSD thus puts them in a position to form a stable orthogonal structure in the core of the PSD-linking receptors and other scaffolding molecules.
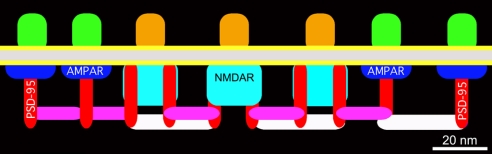
Fig. 5.
Core structure of the PSD based on tomographic reconstructions. Components and spacings between components are drawn approximately to scale. Dominant structure is an array of vertical filaments containing PSD-95 (red). NMDAR-type structures, differentiated by their large cytoplasmic extensions (cyan), concentrate in the center of the PSD. AMPAR-type structures, differentiated by their flattened cytoplasmic aspects (blue), surround the NMDAR structures. Virtually all NMDAR- and AMPAR-type structures are contacted by vertical filaments. Short horizontal filaments (purple) link vertical filaments associated with both NMDAR- and AMPAR-type structures. Longer horizontal filaments (white) concentrated under the NMDAR-type structures cross-link the vertical filament meshwork.
The lability of AMPA receptors relative to NMDA receptors is thought to make a contribution to synaptic plasticity (5). Details of the orthogonal arrangement of core components show how PSD-95 family members might stabilize the NMDA receptors arrayed in the center of the PSD while allowing AMPA receptors arrayed around the periphery to turn over more quickly. The longer type of horizontal filament is the right length to link vertical filaments from adjacent NMDA receptors and further stabilize them at their characteristic spacing of ≈30 nm in the center of the PSD. The shorter type of horizontal filaments are the right length to link vertical filaments attached to adjacent AMPA receptors and stabilize them at their spacing of ≈20 nm at the periphery of the PSD, and link AMPA to NMDA receptors at their interface. Thus, the horizontal filaments close the loop to create a framework on which to organize and stabilize NMDA and AMPA receptors beyond that realized by their individual associations with vertical filaments.
The arrangement of regular-spaced vertical filaments and glutamate receptors nevertheless leaves open the possibility that receptors could be added or deleted at the edges of the PSD. A large number of vertical filaments do not associate with glutamate receptor structures, and these filaments could bind additional AMPA receptors arriving at the edge of the PSD or accept receptors into the horizontal filament meshwork if they were already attached to a vertical filament (39). Further recruitment of AMPA receptors into the PSD may depend on the enlistment of PSD-95 family members into the vertical filament scaffold at the periphery of the PSD as the PSD increases in diameter (24, 26, 40, 41), whereas reduction in PSD-95 would lead to the loss of peripheral AMPA receptors (42–44).
Methods
Cultured Hippocampal Neurons.
Dissociated rat hippocampal neurons (E20) were plated onto glia in a Bal-Tec 3-mm gold specimen chamber and maintained for 3 weeks in 10% CO2 (45). Neurons on the specimen carrier were monitored by reflection microscopy.
Freeze Substitution.
Cultures were frozen at 2100 Bar with a Bal-Tec HPM 010 machine (Techno Trade) in 124 mM NaCl, 2 mM KCl, 1.24 mM KH2PO4, 1.3 mM MgCl2, 2.5 mM CaCl2, 30 mM glucose, 25 mM Hepes, and 0.5% ovalbumin (pH 7.4; osmolarity of 325). Samples were covered with hexadecane before freezing. Specimen carriers were left on frozen saturated uranyl acetate and 2% acrolein in HPLC-grade acetone at −160°C for 15 min in an AFS Leica unit, ramped from −160 to −90°C in 14 h, held at −90°C for 8 h, ramped to −60° in 6 h, held for 12 h, and infiltrated in Lowicryl HM20 resin in acetone. Lowicryl was polymerized by UV at −50°C. Acrolein helped stabilize membrane structure, and uranyl acetate gave a fine-grained stain that extended evenly through sections and did not overstain. Sections ≈100–200 nm thick were cut en face and mounted on Formvar/carbon-coated grids with 10-nm gold particles applied to both sides as fiducial markers.
EM Tomography.
PSDs at mature synapses in areas free of ice crystal damage were photographed in an FEI Tecnai 300-kV electron microscope with a field-emission gun at a dose of ≈300 electrons per nm2 per image. Series were acquired in two axes at tilt increments of 2° from +74° to −74°. Pixel sizes were 0.48–0.75 nm (2,048 × 2,048 image). Series were reconstructed, and the dual-axis 3D volumes merged with IMOD (46) at a typical alignment error of <0.3 pixels. The 3D volume (tomogram) was analyzed with EM3D (6), segmented semiautomatically, and surface-rendered with Amira (Mercury Computer Systems).
In Amira, the magic wand tool was used to determine the density of part of a structure, and then the whole structure was automatically outlined. Ambiguities in outlining density in one cross-sectional view were resolved by segmenting the same structure in two other views. Objects were segmented one by one, and objects were grouped into classes, allowing relationships between these objects, individually and as a class, to be defined. All measurements were performed either in EM3D or with ImageJ (http://rsb.info.nih.gov/ij) in snap shots of surface-rendered objects obtained from Amira. All data are reported as mean ± SD.
Antibodies and Immunocytochemistry.
Monoclonal antibody to PSD-95 is from Affinity BioReagents (MA1–046). A polyclonal antibody to residues 290–307 designed by Ayse Dosemeci was made by New England Peptide. For details on mapping the epitope recognized by ABR PSD-95 antibody, see SI Methods. For conventional ImmunoGold labeling, cells were fixed in 4% paraformaldehyde for 45 min, incubated with the primary antibody for 1 h, secondary antibody-conjugated to 1.4-nm Nanogold (47), and silver enhanced (HQ kit; Nanoprobes). For tomography, cultures were fixed and immunolabeled with the polyclonal PSD-95 antibody and Nanogold second antibody as described above, and silver enhanced for 6–8 min, followed by the high-pressure freezing, freeze substitution, and embedding protocol used to prepares cultures for tomography. When primary antibody was eliminated from the protocol, no specific labeling was observed.
Protein Structural Informatics.
Differentiation between structured and natively unstructured protein conformations was based on primary sequence (20). Estimates of the sizes of protein domains were done as described in ref. 19. For further details, see SI Methods.
Supplementary Material
Acknowledgments.
We thank Jung-Hwa Tao-Cheng (National Institute of Neurological Disorders and Stroke, Bethesda) for electron micrographs; Virginia Crocker for immunolabeling; Bechara Kachar for valuable suggestions about freeze substitution; John Lisman, Wayne Albers, Carolyn Smith, and Ayse Dosemeci for valuable comments on the manuscript; John Chludzinski for help with figures; the EM3D (Stanford University, Stanford, CA) team; and the IMOD team (University of Colorado, Boulder, CO) for software support. This work was supported by National Institute of Neurological Disorders and Stroke and National Institute of Biomedical Imaging and BioEngineering Intramural Research Programs.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800897105/DC1.
References
- 1.Siekevitz P. The postsynaptic density: A possible role in long-lasting effects in the central nervous system. Proc Natl Acad Sci USA. 1985;82:3494–3498. doi: 10.1073/pnas.82.10.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- 4.Palay SL. Synapses in the central nervous system. J Biophys Biochem Cytol. 1956;2:193–202. doi: 10.1083/jcb.2.4.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 6.Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ. The architecture of active zone material at the frog's neuromuscular junction. Nature. 2001;409:479–484. doi: 10.1038/35054000. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, et al. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci USA. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng D, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Dosemeci A, et al. Composition of the synaptic PSD-95 complex. Mol Cell Proteomics. 2007;6:1749–1760. doi: 10.1074/mcp.M700040-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 11.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: A more quantitative view. Ann Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 12.Volkmann N, et al. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 13.Zuber B, Nikonenko I, Klauser P, Muller D, Dubochet J. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc Natl Acad Sci USA. 2005;102:19192–19197. doi: 10.1073/pnas.0509527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucic V, Yang T, Schweikert G, Forster F, Baumeister W. Morphological characterization of molecular complexes present in the synaptic cleft. Structure (London) 2005;13:423–434. doi: 10.1016/j.str.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa T, et al. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44:453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Craven SE, El-Husseini AE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 17.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 19.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 20.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 21.Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 23.Schnell E, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci USA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- 25.Kharazia VN, Weinberg RJ. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci Lett. 1997;238:41–44. doi: 10.1016/s0304-3940(97)00846-x. [DOI] [PubMed] [Google Scholar]
- 26.Matsuzaki MHN, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nusser Z, et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 28.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 29.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nimchinsky EA, Yasuda R, Oertner TG, Svoboda K. The number of glutamate receptors opened by synaptic stimulation in single hippocampal spines. J Neurosci. 2004;24:2054–2064. doi: 10.1523/JNEUROSCI.5066-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saglietti L, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Wang CY, et al. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen JD, et al. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci. 2003;23:11270–11278. doi: 10.1523/JNEUROSCI.23-35-11270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valtschanoff JG, Weinberg RJ. Laminar organization of the NMDA receptor complex within the postsynaptic density. J Neurosci. 2001;21:1211–1217. doi: 10.1523/JNEUROSCI.21-04-01211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E, Cho KO, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 36.Kim E, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naisbitt S, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 38.Baron MK, et al. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 39.Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 40.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 41.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Husseini A-D, et al. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 43.Colledge M, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elias GM, et al. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Mayer ML, Vyklicky L, Jr, Westbrook GL. Modulation of excitatory amino acid receptors by group IIB metal cations in cultured mouse hippocampal neurones. J Physiol. 1989;415:329–350. doi: 10.1113/jphysiol.1989.sp017724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 47.Dosemeci A, et al. Glutamate-induced transient modification of the postsynaptic density. Proc Natl Acad Sci USA. 2001;98:10428–10432. doi: 10.1073/pnas.181336998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.