Abstract
It has been proposed that one route of behavioral evolution involves novel regulation of conserved genes. Age-related division of labor in honey bee colonies, a highly derived behavioral system, involves the performance of different feeding-related tasks by different groups of individuals. Older bees acquire the colony's food by foraging for nectar and pollen, and the younger “nurse” bees feed larvae processed foods. The transition from hive work to foraging has been shown to be socially regulated and associated both with decreases in abdominal lipid stores and with increases in brain expression of genes implicated in feeding behavior in Drosophila melanogaster. Here we show that division of labor is influenced by a canonical regulator of food intake and energy balance in solitary species, the insulin/insulin-like growth factor signaling (IIS) pathway. Foragers had higher levels of IIS gene expression in the brain and abdomen than did nurses, despite their low lipid stores. These differences are likely nutritionally mediated because manipulations that induced low lipid stores in young bees also up-regulated these genes. Changes in IIS also causally influenced the timing of behavioral maturation: inhibition of the insulin-related target of rapamycin pathway delayed the onset of foraging in a seasonally dependent manner. In addition, pathway analyses of microarray data revealed that nurses and foragers differ in brain energy metabolism gene expression, but the differences are opposite predictions based on their insulin-signaling status. These results suggest that changes in the regulation of the IIS pathway are associated with social behavior.
Keywords: Apis mellifera, behavioral maturation, social insect, nutrition, foraging
An important problem in biology is to understand the molecular basis for complex behavior. It has been proposed that one route of behavioral evolution involves novel regulation of conserved genes (1). It is well established that orthologous sets of genes regulate the development of body plans across taxa (2), but this idea has only recently begun to be tested for behavior (3, 4).
Age-related division of labor in honey bee colonies involves the performance of different food-related tasks by different groups of individuals. Nurse bees feed brood for the first 1–2 weeks of adult life, process and store food for another week, and then shift to foraging for nectar and pollen at ≈2–3 weeks of age (5). This division of labor is socially regulated; bees speed up, slow down, or reverse their maturation in response to colony needs (6). Although the mechanics of foraging in honey bees are similar to food-gathering in solitary bees, there are fundamental differences. Honey bees forage to improve the fitness of the colony rather than their own; they collect food when their colony needs it. Honey bees feed on honey before exiting the hive to fuel their foraging flights, and most of the food obtained on a foraging trip is not for their own sustenance.
We hypothesized that the regulation of honey bee behavioral maturation involves previously undescribed roles for widely conserved nutrient-sensing or metabolic pathways, for the following reasons. (i) Nutrition has an important role in honey bee age-related division of labor (5). The onset age of foraging is affected by experimentally induced changes in nutritional status (7, 8) and the expression of a nutritionally related gene (9). (ii) Onset age of foraging also is affected by experimentally induced changes in the expression of genes related to feeding behavior in Drosophila (10, 11). (iii) Nurses have much larger lipid and protein nutrient stores than foragers (12). Large lipid stores may be functionally associated with nursing behavior because bees that are forced to revert from foraging to brood care do not regain large lipid reserves and are not as good at rearing brood as typical nurses (13). The striking loss of abdominal lipid that occurs before the onset of foraging (12) is thought to increase individual foraging performance (14). (iv) Nutritional differences between nurses and foragers occur even though all colony members are exposed to the same food stores inside the hive, further suggesting close coupling of nutritional status and behavior.
Insulin/insulin-like growth factor signaling (IIS) is a key regulator of both metabolism (15) and feeding-related behavior (16). Food intake or high levels of nutrient stores leads to enhanced synthesis of insulin (17) or (in insects) insulin-like peptides (ILPs) (18) and represses the synthesis of glucagon or (the insect equivalent) adipokinetic hormone (AKH) (19). IIS also up-regulates both the intracellular target of rapamycin (TOR) pathway (20) and juvenile hormone (JH) (21, 22). JH is known to be involved in the regulation of honey bee behavioral maturation (23, 24).
We tested the hypothesis that behavioral maturation in honey bees entails a previously undescribed regulation of IIS with gene expression analyses, behavioral analyses of foraging ontogeny after pharmacological manipulations, and pathway analyses of microarray data.
Results
Characterization of Insulin-Related Neuropeptide and Receptor Genes.
The honey bee genome contains genes encoding two ILPs (AmIlp1 and AmIlp2), AKH (AmAkh), and the putative receptors for these peptides (AmInR1, AmInR2, and AmAkhR) (25–27). Several lines of evidence indicate a role for AmIlp1 as a functional insulin propeptide gene. AmIlp1 is positively regulated in larvae by good larval nutrition and is much more highly expressed in larvae than AmIlp2 (27). Phylogenetic analysis also indicates that AmIlp1 is more closely related to other ILPs than is AmIlp2 [supporting information (SI) Fig. 4].
Experiment 1: Insulin-Signaling Gene Expression Is Higher in Foragers than Nurses.
Experiments in Drosophila have shown that misregulation of ILPs and AKH in the brain is sufficient to alter many IIS functions (19, 28), whereas insulin receptor signaling has distinct roles in central and peripheral tissues (29, 30). Therefore, we focused on the expression of ILPs and AKH in the brain and expression of receptors in both brain and abdominal tissues. Abdominal gene expression represents a composite of several target tissues for these peptidergic systems (including gut and fat body).
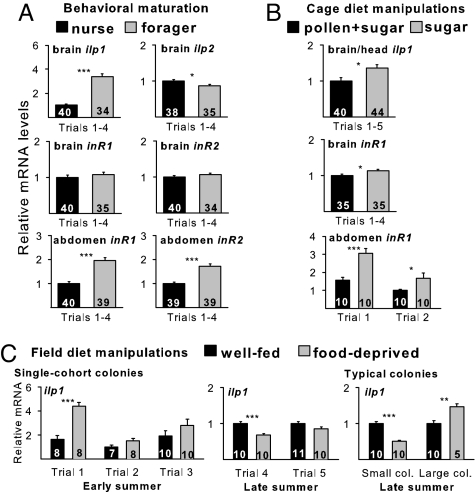
Brain AmIlp1 and abdomen AmInR1 and AmInR2 were significantly more highly expressed in foragers than nurses (Fig. 1A and SI Fig. 5). AmAkh and AmAkhR expression did not differ consistently between nurses and foragers (SI Fig. 5). These results indicate that despite low adiposity relative to nurses, foragers have enhanced insulin production and responsiveness.
Fig. 1.
Up-regulation of insulin-signaling genes in the brain and abdomen of worker honey bees during behavioral maturation and in response to poor nutrition [quantitative PCR (qPCR)]. (A) IIS gene expression in brains and abdomens of nurses and foragers. Data were pooled from four independent trials. ANOVA; brain ilp1, Pgroup (df = 1,68) < 0.0001, Pgroup×trial(df = 3,68) < 0.0001; brain ilp2, Pgroup(df=1,67) < 0.05, Pgroup×trial(df=3,67) < 0.0001; brain inR1, Pgroup (df=1,67) > 0.05, Pgroup×trial(df=3,67) > 0.05; brain inR2, Pgroup(df=1,67) > 0.05, Pgroup×trial(df=3,67) > 0.05; abdominal inR1, Pgroup(df=1,71) < 0.0001, Pgroup×trial df=3,71) = 0.05; abdominal inR2, Pgroup(df=1,71) < 0.0001, Pgroup×trial(df=3,71) > 0.05. Results from individual trials are given in SI Fig. 5. (B) IIS gene expression in brains/heads and abdomens of 4- and 6-day-old bees caged and fed pollen and sugar or a sugar-only diet. Data for brain and head ilp1 were pooled from five independent trials (ANOVA: Pdiet(df=1,74) < 0.05, Ptrial×diet(df=4,74) > 0.05). Data for brain inR1 were pooled from four independent trials (Pdiet(df=1,62) < 0.05, Pdiet×trial(df=3,62) > 0.05). Two independent trials for abdomen inR1 are shown. Data from all individual trials are shown in SI Fig. 6. (C) ilp1 expression in brains/heads after field diet manipulations. Data are shown for brain ilp1 from three early-summer trials that used single-cohort colonies (ANOVA: Pdiet(df=1,45) < 0.0001, Ptrial(df=2,45) < 0.001, Pdiet×trial(df=2,45) < 0.05). Data are shown for head ilp1 from two individual late-summer trials that used single-cohort colonies (combined analysis: Pdiet(df=1,37) < 0.01, Ptrial(df=1,37) < 0.0001, Pdiet×trial(df=1,37) > 0.05). Single trials were performed for head ilp1 with small typical colonies and large typical colonies in late summer. Main effect of group or diet for pooled trials and Student's t tests for individual trials: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Experiment 2: Foraging-Related Up-Regulation of ilp1 and inR1 Is Attributable to Nutritional Status.
Results from dietary manipulations support the hypothesis that differences between nurses and foragers in IIS gene expression are caused by the decline in nutritional status associated with behavioral maturation. Caged 4- and 6-day-old bees fed only sugar had significantly higher expression of brain (or head) ilp1 (Fig. 1B) and both brain and abdomen inR1 (Fig. 1B) than did bees fed sugar and (lipid- and protein-rich) pollen. Nutritional differences between the two diets are reflected by findings that the sugar-only diet resulted in significantly lower (forager-like) lipid stores and mRNA levels for vitellogenin (vg) (SI Fig. 6). VG is a principal storage protein in bees that is typically more abundant in nurses than foragers (31).
Poor nutrition also increased brain ilp1 expression in the field (Fig. 1C), but results varied with season and colony size. In trials performed in early summer with small colonies, chronic food deprivation increased brain ilp1 in 5-day-old bees relative to bees of the same age from well fed control colonies. This effect was significant in 1 of 3 individual trials, but an overall analysis showed a significant effect of diet on brain ilp1. The same effect was seen in late summer in (typical) large-sized colonies (one trial), but in small colonies (both single-cohort colonies and typical colonies), food deprivation decreased brain ilp1 in late summer. These results indicate that poor nutrition can increase brain ilp1 in the field as in the laboratory, but this effect depends on seasonal factors that are less potent in larger colonies. Given that foragers are less adipose than nurses, these results suggest that up-regulation of ilp1 and inR1 in foragers is attributable to their decreased nutritional status.
Experiment 3: TOR Nutrient-Sensing Pathway Affects Behavioral Maturation.
We tested whether insulin-signaling pathways influence honey bee behavioral maturation by determining the effect of oral treatment of rapamycin (a TOR inhibitor) on the age at onset of foraging. We hypothesized that rapamycin delays the onset of foraging because increased IIS up-regulates the TOR pathway (20).
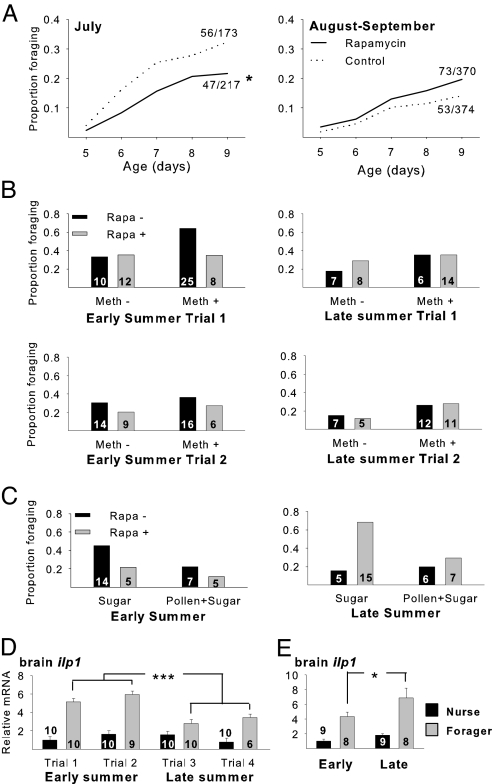
Rapamycin delayed the onset age of foraging in a seasonally dependent manner (Fig. 2A). Rapamycin caused a significant delay in foraging ontogeny in a combined analysis of five trials conducted during early summer. By contrast, rapamycin did not influence the age at first foraging in an analysis of four late-summer trials. There was a significant interaction between the effects of rapamycin treatment and season.
Fig. 2.
Delayed behavioral maturation caused by rapamycin. (A) Proportion of bees that initiated foraging at 5–9 days of age after rapamycin or control treatments. Data for early summer (July 2006) and late summer (August–September 2006) are pooled from five and four trials, respectively. Cox Proportional Hazards: all trials in 2006 (meth, methoprene; rapa, rapamycin; trt, treatment) (foragers/total), nrapa = 120/587, ncontrol = 109/547, Ptrt < 0.05, Pdateoftrial < 0.05, Ptrt×date < 0.01, Ptrial < 0.05, Ptrt×trial < 0.05; early summer 2006, nrapa = 47/217, ncontrol = 56/173, Ptrt < 0.05, Ptrial < 0.001 (Ptrt×trial > 0.05, removed from model); late summer 2006, nrapa = 73/370, ncontrol = 53/374, Ptrt > 0.05, Ptrial < 0.05, Ptrt×trial < 0.01. (B and C) Similar results were obtained in a second year. Early summer 2007, Prapa < 0.01, Ptrial < 0.001, Pmeth×diet < 0.05 (interactions P > 0.05 removed); late summer 2007, Prapa > 0.05, Ptrial < 0.05, Pmeth×diet < 0.05 (interactions P > 0.05 removed). (B) Proportion of bees that foraged before 10 days of age after combinatorial treatments with rapamycin and the JH analog methoprene. Data from individual trials are shown. Statistical analyses on data pooled from two early-summer trials and two late-season trials: Pmeth < 0.01, Prapa > 0.05, Pseason < 0.01, Prapa×season = 0.05 (interactions P > 0.05 removed). (C) Proportion of bees that foraged before 10 days of age after combinatorial treatments with rapamycin and adult diet manipulations (sugar only or pollen and honey). Data are from single early-summer and late-summer trials: Pdiet = 0.08, Prapa < 0.05, Pseason > 0.05, Prapa×season < 0.01 (interactions P > 0.05 removed). (D) Expression of ilp1 in brains of nurses and foragers collected from small colonies in early and late summer. ANOVA, followed by paired contrasts: PearlyFvs.lateF < 0.0001, PearlyNvs.lateN > 0.05 (early F, early foraging; late F, late foraging; early N, early nursing; late N, late nursing). (E) Expression of ilp1 in brains of nurses and foragers collected from large colonies in early and late summer. *, P < 0.05, ***, P < 0.001.
The seasonal effect of rapamycin is consistent with other seasonal changes related to division of labor. Late-summer bees initiate foraging later in life than early-summer bees in temperate climates (32), as in our experiments (Fig. 2A); this difference is associated with maintenance of larger lipid stores later in life (33, 34) and lower blood titers of JH (35). These changes enable late-summer bees to overwinter inside the hive and survive several months longer than do bees emerging earlier in the summer (5).
Although the seasonal effect of rapamycin is consistent with the nutritional and endocrine seasonal changes, results from two 2 × 2 factorial experiments revealed that they are not causally related. These experiments were conducted independently of the original rapamycin experiments, in a second field season. As in the first field season, rapamycin delayed the onset of foraging in early summer but not late summer (Fig. 2 B and C). Methoprene (a JH analog) caused precocious foraging as expected (24) but did not significantly interact with rapamycin to regulate the onset age of foraging and did not alter the seasonal change in response to rapamycin (Fig. 2B). Similarly, bees fed a richer diet (honey and pollen) showed a trend toward a later onset of foraging relative to those fed sugar alone (a weaker effect than expected from ref. 7 but in the same direction), but diet did not significantly interact with rapamycin to regulate the onset of foraging and did not alter the seasonal change in response to rapamycin (Fig. 2C). These results indicate that factors other than JH and nutrition mediate the seasonal differences in response to rapamycin.
We did detect a seasonal change in insulin signaling itself that might explain the rapamycin results. We performed additional experiments to examine trial-by-trial variation in brain gene expression between collections of nurses and foragers made early and late in the summer. In trials that used small colonies (the same colonies as in Experiment 1), there was a late-summer decline in ilp1 expression in forager brains (Fig. 2D) but not in large colonies (Fig. 2E). Although these data come from only one field season, they suggest that insulin signaling is sensitive to seasonal factors, but, as was true for food deprivation, large colonies are buffered from these seasonal changes. Because our experiments with rapamycin were performed with small colonies, the effect on foraging ontogeny may have disappeared late in the summer because IIS was already low in these colonies.
Experiment 4: Up-Regulation of Brain Energy Metabolism Pathways in Nurses Despite Low Insulin Signaling.
We obtained additional evidence for atypical regulation of insulin signaling by annotating energy metabolism pathways with results from three previously published microarray experiments (36, 37). These experiments produced three lists of genes that were differentially expressed: (i) in the brains of nurse bees and foragers; (ii) in response to methoprene, which accelerates the onset age of foraging (23); and (iii) in response to queen mandibular pheromone (QMP), which delays the onset age of foraging (38). Insulin signaling both regulates and is regulated by changes in energy metabolism, and generally there is a positive correlation between insulin signaling and energy metabolism gene expression in a variety of species and tissues (39–41). To test whether this positive relationship is present in the honey bee brain, we mapped genes to conserved energy metabolism Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (42) and performed statistical tests for enrichment and directional bias on each list of genes generated by the microarray experiments (37).
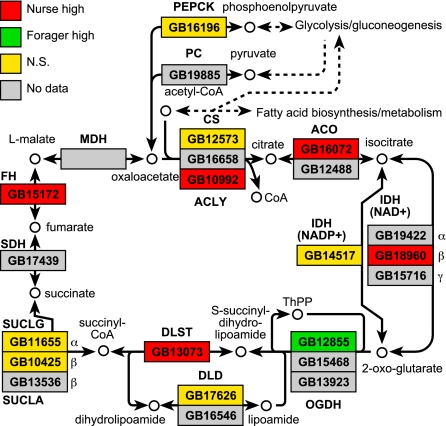
The list of genes differentially expressed in the brains of nurses and foragers was not significantly enriched for energy metabolism genes (37). However, differentially expressed energy metabolism genes were predominantly up-regulated in nurse bees (Table 1), including five of six in the citrate cycle (Fig. 3). These results suggest that the brains of nurses have higher capacity for energy metabolism than forager brains, in contrast to their lower IIS gene expression. By contrast, foragers have a higher overall metabolic rate (43) and expression of oxidative phosphorylation genes in the thorax and abdomen (44), and proteomic analyses show higher protein expression for several classes of energy metabolism enzymes in whole-body samples of foragers compared with nurses (45). These differences likely reflect the activity of tissues such as flight muscle that are more directly involved in increased forager metabolism.
Table 1.
Tests for enrichment and directional biases of energy metabolism genes on cDNA microarrays
| Gene set | Nurse vs. forager | Methoprene vs. control | QMP vs. control |
|---|---|---|---|
| Enrichment: (up + down)/all genes | |||
| All genes | 1,086/2,094 | 659/2,128 | 322/2,044 |
| Oxidative phosphorylation | 15/23 | 24/32*** | 2/22 |
| Citrate cycle | 6/8 | 5/10 | 2/7 |
| Glycolysis | 6/11 | 5/14 | 2/11 |
| Fatty acid metabolism | 4/11 | 3/10 | 3/11 |
| Combined energy metabolism | 30/51 | 37/63*** | 8/49 |
| Directional bias | Nurse-high:forager-high | Down:up | Up:down |
| All differentially expressed genes | 546:540 | 269:390 | 123:200 |
| Oxidative phosphorylation | 13:2** | 0:24*** | 1:1 |
| Citrate cycle | 5:01 | 0:05 | 1:1 |
| Glycolysis | 5:01 | 0:05 | 2:0 |
| Fatty acid metabolism | 3:01 | 0:03 | 2:1 |
| Combined energy metabolism | 25:5*** | 0:37*** | 5:3 |
Published expression data (36, 37) were mapped to KEGG energy metabolism pathways. Enrichment (over- and under-representation) of differentially expressed genes in each pathway was determined using a hypergeometric test, and directional biases of differentially expressed genes were determined using a χ2 test. Ratios in bold differed significantly from null hypothesis. **, P < 0.01; ***, P < 0.001.
Fig. 3.
Citrate cycle genes are up-regulated in the brains of nurse bees. Gene expression in whole brains of nurse bees and foragers was measured on cDNA microarrays (37). Expression data were mapped to pathway diagrams compiled by KEGG. P values are based on ANOVA described in ref. 37. Abbreviations for gene names are based on SwissProt naming conventions. N.S., Not significant.
Although circulating titers of JH are higher in foragers than nurses, treatment with the JH analog methoprene caused a nurse-like shift in brain energy metabolism gene expression. Energy metabolism genes were enriched among genes regulated by methoprene in the brain, but they were up-regulated (Table 1). This result is in contrast to the finding that methoprene causes forager-like changes in overall brain gene expression (36, 37). QMP causes nurse-like overall changes (36) but did not affect brain energy metabolism gene expression (Table 1).
Discussion
Molecular pathways that regulate hunger and food-gathering behavior in solitary species influence the age at which worker honey bees shift from working in the hive to collecting food for their colony. Therefore, the regulation of honey bee division of labor, a highly derived trait, involves widely conserved nutrient-sensing or metabolic pathways, in addition to previously implicated feeding-related (10, 11) and nutritionally related genes (9).
The finding that IIS gene expression is up-regulated in the brain by low nutrient stores and in foragers (previously reported in ref. 22) differs from commonly observed patterns of expression in other species in two ways. First, the direction of the response is reversed; high levels of nutrient stores typically lead to enhanced insulin signaling (17, 18). Second, whereas we found that AmIlp1 and AmInR1 expression were positively correlated, insulin-signaling activity down-regulates insulin receptor gene expression in Drosophila and in vertebrate cell lines by inhibiting FoxO (46, 47). This feedback results in a homeostatic mechanism that ensures a rapid but brief response to nutritional changes.
Our results suggest roles for insulin signaling in the brain and fat body. Increased ilp1 production in the brain may influence behavior through local action on neuronal circuits that control foraging and also may affect non-brain targets, such as the fat bodies in the abdomen. High levels of inR1 and inR2 in the abdomen should maximize the responsiveness of abdominal tissues to circulating ILPs. However, we cannot discern whether the increase in insulin signaling during behavioral maturation is a cause or consequence of lipid loss. A few studies in other insect species suggest that ILPs can have catabolic functions in insects (48), so a causal relationship is possible. The nature of this speculative brain−abdomen communication system in bees is unknown, but similar systems are well studied in vertebrates (49).
It is possible that the combination of high brain ilp1 and high abdominal inR1 in foragers reflects a change in the adipostatic set point relative to nurses, rather than the traditional homeostatic mechanism associated with insulin signaling. In this view, the combination of high insulin synthesis and high insulin sensitivity maintains, or perhaps causes, a shift from high to low adiposity during behavioral maturation (and in response to experimental nutritional manipulations). Similar reasoning has been used to explain relationships between nutrient-sensing pathways and variation in nutrient stores in the contexts of mammalian torpor (50, 51) and insect diapause (52).
“Reversed” IIS gene expression and the suggested set point regulation do not occur in all contexts in honey bees. More typical homeostatic regulation is seen during larval development; ilp1 in honey bee larvae is up-regulated by good nutrition (27). It is not known why these differences in IIS in honey bees appear to be limited to behavioral maturation. Perhaps this is because the system of social foraging in honey bees requires that they forage when they are not personally hungry.
There were seasonal changes in IIS brain gene expression and the effects of IIS on behavioral maturation, but these changes were limited to small, not large, colonies. We speculate that this might have been because large colonies are able to maintain more stable levels of food stores (5) and that the seasonal effects we detected in late summer in small colonies would have been detected in large colonies sampled later in the fall than we did. It is possible that our use of small colonies made it easier to expose the seasonal effects of IIS in honey bee colonies.
A surprising result was that the transition from in-hive tasks to foraging was associated with a decrease in whole-brain energy metabolism gene expression that does not appear to be caused either by insulin or by JH, two hormones that have causal effects on behavioral maturation. Alternatively, insulin might regulate these changes, but in the opposite direction to other tissues and species. Perhaps high levels of brain energy metabolism are required in nurses for energy-intensive processes such as brain plasticity that are not necessarily correlated with metabolism in other tissues. Changes in brain structure occur throughout the lifespan of worker honey bees but are more intense in young bees (53).
Another explanation for the high levels of brain energy metabolism in nurse bees is that whole-brain analyses of energy metabolism pathways do not adequately reflect what is going on in specific brain regions. In most insect brains, ILPs are produced primarily in a small cluster of neurosecretory cells (48), but the distribution of insulin receptors in the bee brain is not known.
Insulin signaling influences diverse aspects of phenotypic plasticity in honey bees. Insulin signaling has been implicated in the regulation of caste (queen vs. worker) determination in honey bees (27, 54), and insulin-signaling genes are among the more promising candidate genes located in quantitative trait loci associated with genetic variation for honey bee foraging behavior (55). Several models have been proposed to explain how insulin signaling can influence diverse aspects of phenotypic plasticity in honey bees (22, 55, 56). Our experiments confirm a specific prediction of Corona et al. (22) by showing that low nutrient stores can increase insulin signaling. However, the context specificity of this effect implies that interactions among insulin signaling, nutrition, JH, Vg, and the environment are more complicated than had previously been imagined.
Our results support the notion that molecular pathways that govern nutritional state and feeding behavior in solitary animals represent one “toolkit” that can be used in the evolution of division of labor in social insects (4). Learning how and why some components of insulin-signaling pathways are more evolutionarily labile than others will help understand the molecular basis of behavior.
Methods
Behavioral Collections.
Honey bees (Apis mellifera) were collected from four small colonies (≈10,000 bees; Experiment 1, Trials 1–4) and two large colonies (≈30,000 bees; Trials 5 and 6) while performing nursing or foraging behaviors, observed and identified in typical fashion (10). Nurses were collected after they repeatedly placed their heads into honeycomb cells containing larvae (10), and foragers were collected as they returned to the hive with visible loads of pollen on their legs. In Experiment 4, nurse bees were identified by age and location in the hive, not behavior; these also are robust identification methods (57). Once captured, bees were flash-frozen in liquid nitrogen, and qPCR analyses were performed on insulin-signaling and AKH-signaling genes in brain and abdomen. Collection timing is described in Results.
Nutritional and Pharmacological Treatments.
One-day-old bees were obtained by removing frames of pupae from typical field colonies and placing them in an incubator (34°C and 80% relative humidity). For behavioral analyses, bees were marked with a dot of colored paint (Testor's PLA) on the thorax. Groups of 35 (lipid analyses) or 50 bees (behavioral and molecular analyses) were placed into Plexiglas cages (36), caged for 3–5 days in constant darkness, and fed ad libitum a sugar diet (sugar syrup, 50% sucrose/water wt/vol; or bee candy, 80% confectioner's sugar, 20% sugar syrup) or pollen paste (45% pollen/45% honey/10% water). Rapamycin (LC Laboratories) was administered chronically during the entire caging period and delivered orally, 10 mg/g in food. Methoprene was administered chronically for the entire caging period and delivered orally, 4 mg/g food (58). For analyses of foraging ontogeny, bees were placed into colonies after 3 days. For lipid and RNA analyses, bees were flash-frozen in liquid nitrogen (10). qPCR analyses were performed on the genes that showed the most consistent differences between nurses and foragers, ilp1 and inR1.
Food Deprivation.
Paired single-cohort colonies were established (7), each with a queen and 1200 1-day-old bees derived from the same source colonies. Food-deprived colonies were fed honey for 2 days, then completely deprived of food for 2 days. Well fed colonies were provided with excess honey and pollen for the entire trial. For food deprivation of colonies with typical age demographies, 1-day-old bees were marked with paint for identification and placed into a pair of size-matched colonies. Two days later, all honeycomb frames that contained food were removed from one colony and replaced with empty frames, and the paired colony was sham manipulated. After 1–2 days of food deprivation, focal bees were collected. One trial was performed with small colonies (≈10,000 bees, occupying one Langstroth hive box), and one trial was performed with large colonies (≈30,000 bees, occupying three hive boxes). Bees were collected by flash-freezing at dawn when they were 5-days-old, before the onset of foraging, and qPCR analyses were performed on ilp1 and control gene expression in brains or heads.
RNA Extraction and qPCR.
Total RNA was extracted from dissected brains (44), whole heads, or whole abdomens. cDNA was synthesized from 200 ng of total RNA. qPCR was performed by using an ABI Prism 7900 sequence detector using specific primers (SI Table 2). Results for experimental genes were normalized to a validated control gene, rp49, using a standard curve or ΔΔCt method or to an exogenous RNA spiked into a master mix before cDNA synthesis (22, 44) (see SI Table 2).
Quantification of Abdominal Lipid.
Lipid from abdominal fat bodies was extracted in chloroform/methanol and quantified by using a colorimetric assay with vanillin/phosphoric acid (8).
Behavioral Analyses of Age at First Foraging.
Methods were slightly modified from ref. 11. After 3 days of treatment in cages, all surviving bees (>90% survival) were placed into a single-cohort colony made with 1000 1-day-old (untreated) bees and a queen. Each trial included one to three cages (50–150 bees) per group. Colonies were observed for at least 3 h/day for the following 5–7 days, including the first 5 days on which bees foraged. Bees were captured briefly as they returned from their first foraging flight, identified by treatment group, and marked with an additional dot of paint on the abdomen so that they could be identified as experienced foragers. At the conclusion of the experiment, colonies were killed by flash-freezing with liquid nitrogen, and all focal bees remaining in the hive were censored. We performed nine independent trials with rapamycin-treated bees and untreated controls fed bee candy, four trials with combinatorial rapamycin and methoprene treatments fed pollen paste, and two trials with rapamycin treatments in combination with bee candy or pollen paste diet. Dates of trials are given in Results.
Statistical Analyses.
Following normalization procedures, qPCR and lipid data were analyzed by using one-, two-, or three-factor ANOVA in SAS (PROC MIXED). ANOVA was followed by pairwise comparisons with Tukey post hoc corrections for multiple comparisons. Age at first foraging results were treated as survival data and analyzed by using Cox Proportional Hazards (PROC PHREG). Unless otherwise indicated, the main effect of trial was confounded with variation between qPCR runs and thus is not shown. Main effects of group, treatment, or diet for pooled trials are shown.
Pathway Analyses of Microarray Data.
Lists of differentially expressed genes (ANOVA; nurse vs. forager, P < 0.001; methoprene vs. untreated, P < 0.05; QMP vs. solvent, P < 0.05) were obtained from three previously conducted microarray experiments (36, 37). These lists were annotated with a revised set of Apis−Drosophila melanogaster orthologs kindly provided by C. Elsik of Georgetown University (personal communication). Based on these orthologs, we mapped the gene lists to the following KEGG energy metabolism pathways: oxidative phosphorylation (map00190), citrate cycle (tricarboxylic acid cycle; map00020), glycolysis (map00010), and fatty acid metabolism (map00071) (42). We did this with online GeneMerge (59) and visualized these maps by manually annotating KEGG pathways with honey bee orthologs (www.genome.jp/KEGG). We performed two statistical tests on the results. First, in GeneMerge, we tested whether differentially expressed genes were enriched for energy metabolism pathways relative to the reference population of genes on the array (hypergeometric test). Second, we tested whether there was a directional bias among differentially expressed genes in each pathway toward higher expression in hive bees or foragers (χ2 test).
Supplementary Material
Acknowledgments.
We thank K. Pruiett and T. Newman for expert assistance in the field and laboratory, respectively; A. L. Toth for sample collection and tissue preparation; S. P. Johnson for tissue preparation and qPCR; D. Moyse for tissue preparation; S. H. Woodard for sample collection; C. Elsik for ortholog analysis; S. Aref for statistical consulting; and J. L. Beverly, D. J. Emlen, P. E. Gold, M. B. Sokolowski, and members of G.E.R.'s laboratory for reviewing the manuscript. This work was supported by grants from the National Institutes of Health (NIH) and the National Science Foundation (NSF) to G.E.R., by an NIH training fellowship and an NSF predoctoral fellowship to S.A.A., and by a National Institute on Drug Abuse Center Grant to J. V. Sweedler.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800630105/DC1.
References
- 1.Robinson GE, Ben-Shahar Y. Social behavior and comparative genomics: New genes or new gene regulation? Genes Brain Behav. 2002;1:197–203. doi: 10.1034/j.1601-183x.2002.10401.x. [DOI] [PubMed] [Google Scholar]
- 2.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Malden, MA: Blackwell Publishing; 2005. p. 234. [Google Scholar]
- 3.Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: social life in molecular terms. Nat Rev Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 4.Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends Genet. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Winston ML. The Biology of the Honey Bee. Cambridge, MA: Harvard Univ Press; 1987. p. 294. [Google Scholar]
- 6.Leoncini I, et al. Regulation of behavioral maturation by a primer pheromone produced by adult worker honey bees. Proc Natl Acad Sci USA. 2004;101:17559–17564. doi: 10.1073/pnas.0407652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz DJ, Huang ZY, Robinson GE. Effects of colony food shortage on behavioral development in honey bees. Behav Ecol Sociobiol. 1998;42:295–303. [Google Scholar]
- 8.Toth AL, Kantarovich S, Meisel AF, Robinson GE. Nutritional status influences socially regulated foraging ontogeny in honey bees. J Exp Biol. 2005;208:4641–4649. doi: 10.1242/jeb.01956. [DOI] [PubMed] [Google Scholar]
- 9.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5:673–677. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Shahar Y, Dudek NL, Robinson GE. Phenotypic deconstruction reveals involvement of manganese transporter malvolio in honey bee division of labor. J Exp Biol. 2004;207:3281–3288. doi: 10.1242/jeb.01151. [DOI] [PubMed] [Google Scholar]
- 12.Toth AL, Robinson GE. Worker nutrition and division of labour in honeybees. Anim Behav. 2005;69:427–435. [Google Scholar]
- 13.Robinson GE, Page RE, Strambi C, Strambi A. Colony integration in honey bees: mechanisms of behavioral reversion. Ethology. 1992;90:336–348. [Google Scholar]
- 14.Blanchard GB, Orledge GM, Reynolds SE, Franks NR. Division of labour and seasonality in the ant Leptothorax albipennis: Worker corpulence and its influence on behaviour. Anim Behav. 2000;59:723–738. doi: 10.1006/anbe.1999.1374. [DOI] [PubMed] [Google Scholar]
- 15.Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz MW, et al. Evidence that plasma leptin and insulin levels are associated with body adiposity via different mechanisms. Diabetes Care. 1997;20:1476–1481. doi: 10.2337/diacare.20.9.1476. [DOI] [PubMed] [Google Scholar]
- 18.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 19.Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- 20.Edgar BA. How flies get their size: Genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- 21.Tu MP, Yin CM, Tatar M. Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocrinol. 2005;142:347–356. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Corona M, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci USA. 2007;104:7128–7133. doi: 10.1073/pnas.0701909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson GE. Regulation of honey-bee age polyethism by juvenile-hormone. Behav Ecol Sociobiol. 1987;20:329–338. [Google Scholar]
- 24.Sullivan JP, Fahrbach SE, Robinson GE. Juvenile hormone paces behavioral development in the adult worker honey bee. Horm Behav. 2000;37:1–14. doi: 10.1006/hbeh.1999.1552. [DOI] [PubMed] [Google Scholar]
- 25.Hummon AB, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 26.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJ. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol. 2006;80:1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler DE, Buck N, Evans JD. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol. 2006;15:597–602. doi: 10.1111/j.1365-2583.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 29.Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA. 2005;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 31.Amdam GV, Norberg K, Hagen A, Omholt SW. Social exploitation of vitellogenin. Proc Natl Acad Sci USA. 2003;100:1799–1802. doi: 10.1073/pnas.0333979100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuda H. Population and bioeconomic studies on the honeybee colonies: 6. The relationship between work efficiency and population size in a honeybee colony. Res Popul Ecol. 1983;25:249–263. [Google Scholar]
- 33.Maurizio A. The influence of pollen feeding and brood rearing on the length of life and physiological condition of the honeybee: Preliminary report. Bee World. 1950;316:9–12. [Google Scholar]
- 34.Snodgrass RE. Anatomy of the Honey Bee. Ithaca, NY: Comstock; 1956. p. 334. [Google Scholar]
- 35.Huang ZY, Robinson GE. Seasonal changes in juvenile hormone titers and rates of biosynthesis in honey bees. J Comp Physiol B. 1995;165:18–28. doi: 10.1007/BF00264682. [DOI] [PubMed] [Google Scholar]
- 36.Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. Pheromone-mediated gene expression in the honey bee brain. Proc Natl Acad Sci USA. 2003;100:14519–14525. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield CW, et al. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pankiw T, Huang Z, Winston ML, Robinson GE. Queen mandibular gland pheromone influences worker honey bee (Apis mellifera L.) foraging ontogeny and juvenile hormone titers. J Insect Physiol. 1998;44:685–692. doi: 10.1016/s0022-1910(98)00040-7. [DOI] [PubMed] [Google Scholar]
- 39.Webb GC, Akbar MS, Zhao C, Steiner DF. Expression profiling of pancreatic beta cells: Glucose regulation of secretory and metabolic pathway genes. Proc Natl Acad Sci USA. 2000;97:5773–5778. doi: 10.1073/pnas.100126597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girard J, Perdereau D, Foufelle F, Prip-Buus C, Ferré P. Regulation of lipogenic enzyme gene expression by nutrients and hormones. FASEB J. 1994;8:36–42. doi: 10.1096/fasebj.8.1.7905448. [DOI] [PubMed] [Google Scholar]
- 41.Jünger MA, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison JM. Caste-specific changes in honeybee flight capacity. Physiol Zool. 1986;59:175–187. [Google Scholar]
- 44.Corona M, Hughes KA, Weaver DB, Robinson GE. Gene expression patterns associated with queen honey bee longevity. Mech Ageing Dev. 2005;126:1230–1238. doi: 10.1016/j.mad.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Wolschin F, Amdam GV. Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Science. 2007;5:10. doi: 10.1186/1477-5956-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Ann Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 49.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 50.Klingenspor M, Niggemann H, Heldmaier G. Modulation of leptin sensitivity by short photoperiod acclimation in the Djungarian hamster, Phodopus sungorus. J Comp Physiol B. 2000;170:37–43. doi: 10.1007/s003600050005. [DOI] [PubMed] [Google Scholar]
- 51.Tups A, et al. Photoperiodic regulation of insulin receptor mRNA and intracellular insulin signaling in the arcuate nucleus of the Siberian hamster, Phodopus sungorus. Am J Physiol. 2006;291:R643–R650. doi: 10.1152/ajpregu.00807.2005. [DOI] [PubMed] [Google Scholar]
- 52.Williams KD, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc Natl Acad Sci USA. 2006;103:15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fahrbach SE, Farris SM, Sullivan JP, Robinson GE. Limits on volume changes in the mushroom bodies of the honey bee brain. J Neurobiol. 2003;57:141–151. doi: 10.1002/neu.10256. [DOI] [PubMed] [Google Scholar]
- 54.Patel A, et al. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS One. 2007 doi: 10.1371/journal.pone.0000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunt GJ, et al. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften. 2007;94:247–267. doi: 10.1007/s00114-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page RE, Amdam GV. The making of a social insect: Developmental architectures of social design. BioEssays. 2007;29:334–343. doi: 10.1002/bies.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seeley TD. Adaptive significance of the age polyethism schedule in honeybee colonies. Behav Ecol Sociobiol. 1982;11:287–293. [Google Scholar]
- 58.Schulz DJ, Sullivan JP, Robinson GE. Juvenile hormone and octopamine in the regulation of division of labor in honey bee colonies. Horm Behav. 2002;42:222–231. doi: 10.1006/hbeh.2002.1806. [DOI] [PubMed] [Google Scholar]
- 59.Castillo-Davis C, Hartl D. GeneMerge–post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.