Abstract
A loxP-transposon retrofitting strategy for generating large nested deletions from one end of the insert DNA in bacterial artificial chromosomes and P1 artificial chromosomes was described recently [Chatterjee, P. K. & Coren, J. S. (1997) Nucleic Acids Res. 25, 2205–2212]. In this report, we combine this procedure with direct sequencing of nested-deletion templates by using primers located in the transposon end to illustrate its value for position-specific single-nucleotide polymorphism (SNP) discovery from chosen regions of large insert clones. A simple ampicillin sensitivity screen was developed to facilitate identification and recovery of deletion clones free of transduced transposon plasmid. This directed approach requires minimal DNA sequencing, and no in vitro subclone library generation; positionally oriented SNPs are a consequence of the method. The procedure is used to discover new SNPs as well as physically map those identified from random subcloned libraries or sequence databases. The deletion templates, positioned SNPs, and markers are also used to orient large insert clones into a contig. The deletion clone can serve as a ready resource for future functional genomic studies because each carries a mammalian cell-specific antibiotic resistance gene from the transposon. Furthermore, the technique should be especially applicable to the analysis of genomes for which a full genome sequence or radiation hybrid cell lines are unavailable.
Identifying polymorphic sites in the genome is a basic aspect of molecular genetics and genomics. The process is needed for a variety of purposes, ranging from the development of polymorphic marker sets useful as a tool for genetic analysis of a chromosomal region or full genome scan, to the initial identification of variants or mutations in a newly discovered gene (1, 2). In most cases, the identity of base differences and their location relative to a gene or other polymorphic sites is either useful or required. Recent estimates of the number of single-nucleotide polymorphisms (SNPs) needed for whole genome association studies in humans vary from several thousand to several hundred thousand (1, 3); thus, efficient and cost-effective methods for identifying a large number of SNPs with the required characteristics of dense yet even spacing, and of known order over large uncharacterized regions of the genome, is of interest. A comparison of two methods to develop a densely ordered map of SNPs covering a 4-Mb region of the human genome was recently reported (4). In one approach, large-insert bacterial clones, bacterial artificial chromosomes (BACs) (5) and P1 artificial chromosomes (PACs) (6), spanning this region were fragmented and reconstructed in 2-kb plasmid libraries, which were then sequenced. This shotgun procedure is efficient in identifying SNPs; however, to approach a map of 30-kb average SNP spacing, bidirectional sequencing of approximately 500 randomly chosen subclones per 100 kb of genomic sequence was required. Multiple BAC and PAC clones mapping to the region were pooled during DNA preparation before the in vitro generation of high-quality subclone libraries by using a multistep process. After identification, the SNPs were ordered by PCR-based testing on radiation hybrid DNAs. In the second approach, an end-deletion strategy of yeast artificial chromosomes using Alu repeats offered a more directed approach to locus-specific SNP identification (7, 8). Although relative order and orientation are never compromised in this approach, the method depends on the spacing of Alu sequences within the original yeast artificial chromosome. The largest gap found between SNPs in either approach was 200 kb; this gap presumably arises because of chromosomal regions that are either refractile to the Alu recombination in yeast artificial chromosomes or relatively underrepresented in 2-kb plasmid libraries in the shotgun procedure.
Here we describe an alternative directed approach that uses direct sequencing of PAC- and BAC-nested deletions for SNP discovery, potentially at very high densities with a minimal amount of sequencing. The method retains information relevant to spacing and order of the SNP during the identification process, thereby eliminating the need to sequence many randomly chosen clones from a subclone library and the subsequent use of radiation hybrid mapping for SNP order determination. In applications where an ordered set of overlapping large-insert bacterial clones or a contig has been previously assembled, it eliminates generation of SNPs from that portion of a recombinant clone already covered within the contig.
We have used a loxP sequence containing transposon 10 (Tn10) minitransposon insertion strategy (retrofitting) to generate nested deletions from one end of the genomic insert in BACs and PACs (9). A small, transposon-containing plasmid is introduced into the clone of interest, followed by a single-step and single-tube process for generating deletions in vivo. After isolation and sizing of DNA from the deleted clone series, a 500-nucleotide sequence is generated directly from a chosen BAC or PAC deletant by using a common sequencing primer located within the transposon end. The result is a DNA sequence generated from chosen regions at defined intervals of the original recombinant clone. This sequence is then used for PCR primer pair design so that the genome of multiple individuals can be sequenced directly in search of polymorphic variants.
Materials and Methods
Creating Nested Deletions in BACs and PACs.
The virulent form of P1 phage (P1 vir), the bacterial Cre− host used to transfer PAC/BAC-nested deletions (Escherichia coli strain NS3516), and procedures to grow and titer P1 phage have been described (9–11). Super Broth (Advanced Biotechnologies, Columbia, MD) was used to grow BAC/PAC deletion clones to high cell densities for plasmid DNA isolation.
Transposon plasmids pTnBAC/loxP or pTnPGKpuro/loxP described earlier were introduced into a BAC or PAC clone, respectively, by calcium chloride transformation (9, 12). Procedures for inducing transposon insertions, transduction of retrofitted BAC or PAC plasmid with P1 phage, and isolating DNA carrying nested deletions have been described (9, 10) with the exception that 2 mM IPTG was used to induce transposition in our current study.
Screening Retrofitted Clones Sensitive to Ampicillin.
Clones with PAC or BAC deletions were transferred manually or robotically from LB plates containing kanamycin plus chloramphenicol into 96-well microtiter dishes containing 120 μl of LB with 25 μg/ml each of kanamycin and chloramphenicol per well. Overnight cultures were grown without shaking at 37°C, and glycerol was added to 12% for storage at −20°C. These master stocks were replica-plated in parallel with a 96-pin replicator (GENETIX, Christchurch, Dorset, U.K.) onto LB agar plates or into 100 μl of liquid medium per well containing either 25 μg/ml chloramphenicol or 60 μg/ml ampicillin. Colonies unable to grow in ampicillin were identified for isolating plasmid DNA. Procedures for isolating DNA for NotI digestions from the parent BAC/PAC clone and its nested deletions have been described (10). High throughput procedures using R.E.A.L Prep Plasmid Kits (Qiagen, Chatsworth, CA) for simultaneous handling of multiples of 96 clones are described below.
Field Inversion Gel Electrophoresis (FIGE) Analysis of DNA from Nested Deletions.
NotI-digested DNA from BAC/PAC deletions was analyzed by using a BIORAD–FIGE mapper system with the preprogrammed Program 6 pulse-ramp switch time (Bio-Rad). Samples (10 μl; 15% of total amount) were electrophoresed in a 1% SeaKem LE agarose gel in 0.5× TBE buffer with circulation of buffer.
Methods for Preparing BAC/PAC DNA from Nested Deletions.
Qiagen column purification of BAC/PAC DNA for end sequencing.
When only a few BAC/PAC template DNAs were prepared, the manufacturer’s procedure for purifying small supercoiled plasmids was used with the following changes.
1. The cell lysate containing PAC/BAC DNA was precipitated at room temperature with an equal volume of isopropanol before applying to the Qiagen tip. The precipitate was resuspended in column equilibration buffer QBT.
2. BAC/PAC DNA was eluted from the Qiagen tip with elution buffer (QF) made 1.4 M in sodium chloride and prewarmed to 70°C.
3. The resuspended isopropanol precipitate, containing PAC/BAC DNA after Qiagen tip purification, was extracted once with a 50:50 mixture of phenol and chloroform and once with chloroform. This last step enabled a linear increase in signal strength during Applied Biosystems sequencing with an increase in template DNA.
High throughput purification of BAC/PAC DNA for end sequencing by using the Qiagen R.E.A.L Prep 96 Plasmid Kit.
Colonies containing BAC/PAC deletions were grown with shaking for 16 h in 96-well blocks in 1.2 ml of media per well containing Super Broth and LB (1:2) with 12.5 μg/ml chloramphenicol. Cells were processed exactly as specified by the manufacturer, except an additional wash of the DNA pellet with 70% ethanol was incorporated at the end. The final pellet of PAC/BAC DNA in each well was resuspended in 35 μl of TE (10 mM Tris·HCl, pH 7.6/0.1 mM EDTA); 12 μl of this was used in each sequencing reaction and 5 μl was used for digestion with NotI and FIGE analysis.
DNA Sequencing.
Big dye chain terminator chemistry described for sequencing small plasmids (4) was used for sequencing BAC/PAC-nested deletions. Each sequencing reaction contained 2–4 μg of BAC/PAC template DNA, 16 pmol of primer, and 16 μl of Big Dye terminator mix in a 30-μl reaction volume.
Physical Mapping with Deletion Clones.
A high-throughput PCR analysis of radiation hybrid (RH) markers and known SNPs was conducted by using whole-cell bacterial cultures containing the BAC/PAC deletions. Overnight cultures of starting full-length or deletion clones were grown in 96-well blocks. Cells were washed twice with distilled water by first pelleting and then resuspending. Cell suspensions were aliquoted into 96-well PCR plates (Applied Biosystems) by using a robot and the cultures dried in a 70°C oven for 2 h. PCR reactions were run by using reagents from Perkin–Elmer directly in these 96-well plates.
Results
Generating Nested Deletions in BACs and PACs.
LoxP-transposon retrofitting and subsequent deletion formation of BAC and PAC recombinant clones were performed by using the two transposon-containing plasmids pTnBAC/loxP and pTnPGKpuro/loxP, respectively (9). This process occurs in the same bacterial host containing the genomic plasmid isolated directly from a BAC/PAC library (9). Cre- mediated recombination between a transposon-inserted loxP site and that existing in vector sequences of a BAC or PAC clone can either delete or invert the intervening DNA; the outcome depends on the relative orientation of the two sites (13, 14). If the starting BAC/PAC plasmid is larger than the P1-headful size (110 kb), then deletions can selectively be isolated by using transduction with P1 phage (9, 15, 16). Cre protein, required for recombination of loxP sites, is supplied in trans by the P1 phage during transduction.
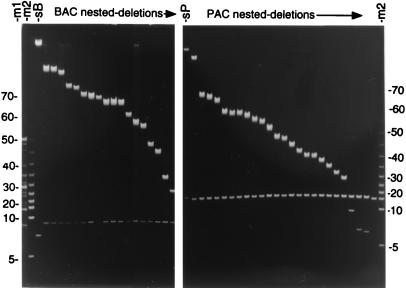
Fig. 1 Left displays a FIGE analysis of nested deletions typically obtained with a BAC plasmid larger than 110 kb by using pTnBAC/loxP. The DNA was digested with NotI before electrophoresis. This panel of 18 deletions was chosen from 24 randomly isolated clones initially sized by FIGE, from the plating of a small fraction of a thousand member, nested-deletion library. The starting BAC (lane sB) is 140 kb in size as compared with the standards in lanes m1 and m2.
Figure 1.
(Left) FIGE analysis of a NotI-digested 140-kb BAC clone (lane sB) and 18 of its deletions arrayed according to decreasing size of insert DNA. A 5-kb ladder and 8–48 kb molecular weight standards were applied to lanes m2 and m1, respectively. (Right) A similar analysis with a 140-kb PAC clone (lane sP) and 25 of its deletions. A 5-kb ladder is included in lane m2. The numbers on either side indicate the size in kb pairs of bands from the 5-kb ladder molecular weight standard.
An identical analysis with a 140-kb PAC plasmid (Fig. 1 Right) and with pTnPGKpuro/loxP indicates efficient deletion formation in PACs as well. The panel of 25 deletions shown here was chosen from 35 randomly isolated colonies. The NotI-digested starting PAC is shown in lane sP and a 5-kb ladder is shown in lane m2. As illustrated in Fig. 1, three-fourths of randomly isolated clones from BAC/PAC nested-deletion libraries were of unique size according to the initial FIGE analysis. Deletions of apparently similar size were often found to be from independent transposon insertion sites (unpublished data), confirming that the insertion of a retrofitting transposon into genomic DNA occurs with little sequence specificity (10); thus, a clone library of several thousand BAC/PAC deletions with a high percentage (75%) of unique sizes illustrates the power of this in vivo method to provide the several hundred deletion templates needed to initiate a region-specific search for SNPs, which on average are found approximately every 1.1 kb in the human genome (4).
Diagnostic Feature of BAC/PAC Deletions Formed by Recombination of loxP Sites.
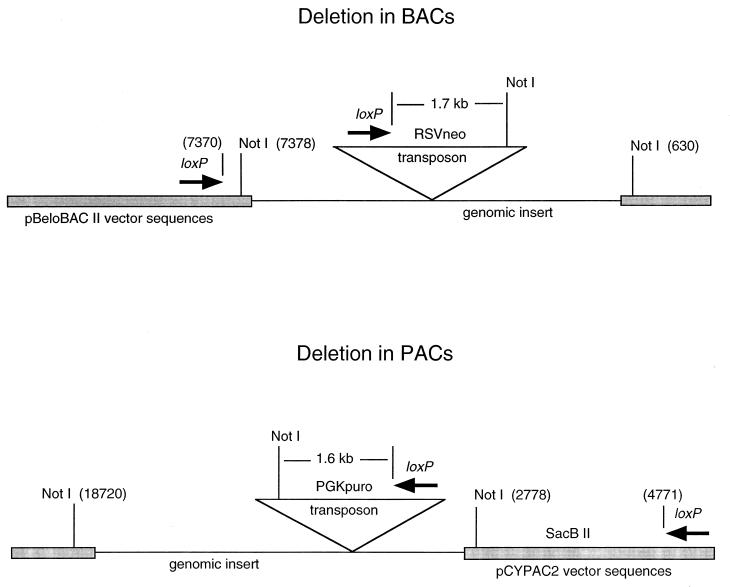
Products of authentic loxP-Cre recombination events can be readily distinguished from other, less frequent rearrangements of potential recombinogenic sequences within genomic inserts by analyzing the size of a vector DNA band in a NotI digest. This diagnostic tool is available for cloning vectors pBeloBAC II (17) and pCYPAC2 (18), referred to as simply BAC and PAC vectors, respectively, in discussions below. Both vectors contain two NotI sites that were designed to release insert DNA on digestion of genomic plasmid with the enzyme.
The NotI restriction map of the initial BAC/PAC vector and the resulting retrofitted deletion clone is shown schematically in Fig. 2. The vector DNA fragment in a NotI digest of a BAC clone is 6,748 bp, whereas that for a PAC clone is 15,942 bp. Such vector DNA fragments from the starting BACs and PACs can be seen in lanes sB and sP of Fig. 1, respectively. Transposons used to insert loxP sequences in BACs and PACs (pTnBAC/loxP and pTnPGKpuro/loxP) each contain a single NotI site located 1.7 kb and 1.6 kb, respectively, in front of the loxP site. Deletions generated through recombination of loxP sites replace the endogenous NotI site in front of the vector loxP sequence with that from the transposon in both BACs and PACs; thus, the NotI vector fragment in BAC-nested deletions would increase in size from 6.7 to 8.5 kb, whereas in PACs, it would decrease in size from 15.9 to 15.5 kb. These size shifts are discernable in Fig. 1, illustrating that initial FIGE analyses of NotI-digested nested-deletion clones can identify those deletions arising exclusively through loxP recombinations. It is worth noting that routine analysis of large numbers of deletion clones from both BACs and PACs reveals that unusual rearrangements do occur at a measurable frequency approaching 1% (e.g., lane 11 in Fig. 3B). BACs/PACs with internal NotI sites produce additional DNA bands on FIGE analysis (data not shown); however, this does not obscure identification of the vector band because of the high resolution of fragments in that size range.
Figure 2.
Schematic representation of loxP transposon-mediated deletion formation in BACs (Upper) and PACs (Lower). The shaded regions represent vector DNA sequences and the numbering of sites is as indicated for BAC (17) and PAC (18).
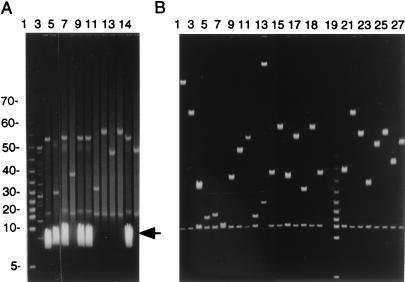
Figure 3.
(A) FIGE analysis of NotI-digested DNAs from two independent nested-deletion libraries from PACs are shown in lanes 2–6 and 7–14. The heavy band marked with the arrowhead represents transposon plasmid DNA. (B) An identical analysis of clones isolated from the same two deletion libraries (lanes 1–18 and 20–27, respectively) after the clones passed through the ampicillin sensitivity screen. Lane 19 displays a 5-kb ladder. Lane 11 shows an example of a PAC deletion without the 15.5-kb vector band; therefore, it does not arise through authentic loxP recombination.
Screening Deletion Clones That Are Free of Transposon-Donating Plasmid.
Recovering a loxP-transposon-inserted BAC or PAC with P1 transduction has the potential drawback of copurifying the transposon-donating plasmid (9). The frequency of recovering the transposon plasmid with a BAC/PAC deletion varies widely (<1 to as high as 80%). A simple antibiotic screen was devised to identify clones that contain the transposon-donating plasmid, such that these could be discarded early on and avoid the more tedious method published earlier (9). The new screen relies on the expression of ampicillin resistance by the full-length transposon plasmid but not by that part of the transposon which is inserted into a BAC or PAC. The beta-lactamase gene is outside the transposon boundaries and adjacent to the origin of replication of the transposon plasmid in both pTnBAC/loxP and pTnPGKpuro/loxP (9); thus, BAC/PAC deletion clones that were resistant to kanamycin and chloramphenicol were picked robotically and transferred to two 96-well plates in parallel. One plate contained chloramphenicol (or kanamycin), whereas the other contained ampicillin. Colonies sensitive to ampicillin, indicating loss of the transposon donor plasmid, were identified for further analysis. The effect of such a screen is shown by the FIGE analysis after NotI digestion of two different PAC nested-deletion libraries prepared without (Fig. 3A lanes 3–5, 7, 8, and 13) or with (Fig. 3B lanes 1–18 and 20–27) such a selection in place.
Direct Sequencing of BAC/PAC-Nested Deletions with Primers in the Transposon End.
A set of sequencing primers useful for sequencing the ends of either PAC or BAC deletion clones was designed from the transposon end remaining in the deletions (Table 1). SP6II and T7L are primers from the end of BAC and PAC vectors, respectively, opposite to the loxP site. BAC/PAC deletion templates were prepared for direct sequencing in one of two ways: (i) alkaline lysis followed by Qiagen tip purification of the BAC/PAC plasmid DNA, or (ii) by using the Qiagen 96-well R.E.A.L Kit. Templates prepared by the first method give high signal strengths in sequencing (1,200 on the Applied Biosystems scale on average) but has the drawback of being low-throughput (10–20 deletion templates can be handled at a time). The 96-well format of the latter procedure guarantees high-throughput albeit a significantly lower signal intensity (around 500). Read-lengths range between 400 and 600 bases in either case. A phred analysis of sequences obtained by using the Qiagen tip purification procedure for a nested deletion series is shown in column 2 of Table 1.
Table 1.
Characteristics of primers for sequencing BAC/PAC-nested deletions
| Sequencing primer | Distance from Tn end, bases | Sequence read length, bases called by PHRED (trim mode) | BAC/PAC deletion template size, kb |
|---|---|---|---|
| Seq 1 5′-GACAAGATGTGTATCCACCTTAAC-3′ | 38 | 503 | 84 |
| 497 | 65 | ||
| 321 | 106 | ||
| Seq 4 5′-CTCCCAGAGCCTGATAAAAACGG-3′ | 87 | 552 | 42 |
| 528 | 66 | ||
| 353 | 106 | ||
| Seq 6 5′-CCGTGGAGGTAATAATTGACGATATG-3′ | 124 | 534 | 62 |
| 458 | 70 | ||
| 241 | 106 | ||
| Seq 9 5′-GGTTCAGGGCAGGGTCGTTAAATAGC-3′ | 240 | 521 | 84 |
| 408 | 58 | ||
| 272 | 106 | ||
| SP6II 5′-TTACGCCAAGCTATTTAGGTGACACTATAGAATAC-3′ | N/A | 544 | 54 |
| 518 | 46 | ||
| 312 | 106 | ||
| T7L 5′-ATTGTAATACGACTCACTATAGGG-3′ | N/A | 540 | 90 |
| 438 | 72 | ||
| 415 | 61 |
Processing Sequence Reads for SNP Discovery.
For those regions where there was interest in identifying a new SNP, sequence data were obtained from the genomic clone as described above. These sequence data were passed through standard programs to remove possible contaminating BAC/PAC vector and transposon sequences. Software programs for primer design were applied to the sequence reads to identify primer pairs suitable for SNP discovery as in an earlier study (4). Single primers from sequence reads on different deletions are also useful here when the plasmids differ in size by <1 kb. It allows a larger fraction of the raw sequence data to be used for primer design and produces longer PCR products spanning the gap between the deletions for SNP discovery.
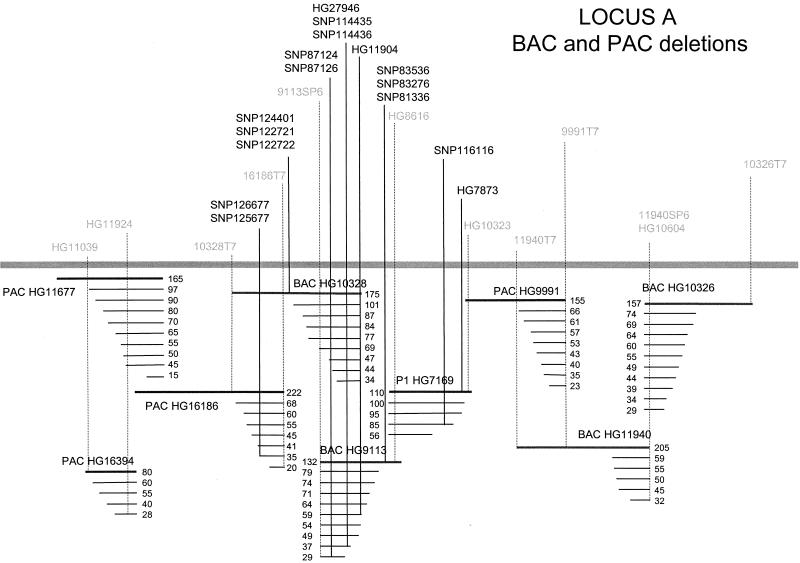
Genomic DNAs from 15 patients were used as templates with these primers to obtain 250- to 350-bp PCR products. Both strands of each PCR product DNA were sequenced as described (4). Sequence reads from both the Qiagen tip-purified and the 96-well R.E.A.L. Kit-isolated BAC/PAC DNAs produced SNPs, including new ones in regions previously explored by using the random sequencing approach. Fig. 4 illustrates the use of a nested deletion series for a combined approach of both physically mapping previously identified SNPs and polymorphic markers as well as new SNP discovery. This combined approach has also been applied to a region around the apoE gene since the previous publication (ref 4; data not shown).
Figure 4.
Schematic representation of SNPs (bold) and markers (gray) positioned on a region of cloned human genomic DNA as determined by nested deletion analysis of the corresponding PAC, BAC, and P1 clones. SNPs were generated from either the deletion-end sequencing approach described in this manuscript, the random-subclone library approach (4), or from inter-Alu libraries. The positions and relative orientation of the large insert clones were obtained by localizing the SNPs and other markers on the nested deletion series of the originating clone, together with a positive PCR amplification from clones other than the source clone. The overlap between PAC HG16394 and PAC HG16186 was determined by dual-color FISH. The solid bar represents a genomic insert, and the series of thinner lines immediately below it represent the deletions generated from that particular clone, with the sizes of the inserts as determined by FIGE analysis given to the right of each clone. This is not a comprehensive SNP map, for only regions resulting in the identification of a SNP are indicated by a vertical line, and the entire region covered by these clones was not tested for the presence of SNPs.
Determining the Orientation of Insert DNA in PACs and BACs from a Contig.
Insert DNA in PACs and BACs can be readily oriented by scoring for a marker in the starting clone and the nested-deletion series generated from it. Progressive deletions starting from the loxP end of the insert allows one to simultaneously determine the location of the marker and identify the relative orientation of the insert with respect to other clones in the contig containing the same marker.
Fig. 4 illustrates the result of a nested-deletion series generated from large-insert clones of multiple types (BAC, PAC, and P1) in a 1.4-Mb contig from locus A. Markers such as SNP83536, SNP83276, SNP81336, and HG8616 were shown to exist in the full-length starting clone BAC HG9113 but not in its characterized deletions. In contrast, both full-length and deletion clones from BAC HG9113 and BAC HG10328 contained markers HG11904, HG27946, SNP87124, and SNP87126; therefore, the loxP sequence in BAC HG9113 is opposite in orientation to that of BAC HG10328 with the partial overlap as shown in Fig. 4. Similarly, the absence of markers 16186T7, SNP126677, and SNP125677 in deletions from BAC HG10328 but not PAC HG16186 allows us to orient the loxP sites and the overlap in these clones. Such pairwise comparisons of several markers between the full length and deletions were used to orient the remaining insert DNAs in the contig shown in Fig. 4.
Discussion
Creating a nested-deletion series in BACs, PACs, or P1 clones allows one to generate ordered-sequence reads at desired intervals throughout the clones. Sequence reads 2–5 kb apart are easily obtained with this approach (Fig. 1), with the ability to have smaller intervals determined by the number of clones one is willing to screen. Although sequence specificity for insertion of transposon 10 were reported in earlier studies with bacterial DNA (19), we have found no evidence that those conclusions hold for the mammalian DNA used in our experiments. There is also no data to suggest that the transposase gene present on the transposon plasmids has mutated to give rise to the relaxed specificity.
Clones from BAC/PAC libraries do not express the Cre protein. The Cre protein requirement for loxP recombination is provided in trans efficiently by P1 vir phage. The recovery of deletion plasmids as a packaged phage, and the ability to perform several of the steps in a single tube are additional advantages of using this P1 transduction procedure. Deletion clones are picked robotically into 96-well plates. We demonstrate that a 96-well format DNA preparation method for these clones provides sufficient high- quality DNA for Big Dye terminator sequencing used for PCR primer design, and FIGE analysis. Note that end sequencing of BAC/PAC deletions in our case is limited to clones <110 kb in size. Table 1 illustrates that there is a trend toward increased read lengths as the clone size is reduced; however, the need for sequencing a much smaller number of clones, as compared with the random subclone sequencing approach, allows this technique to be adopted by the average laboratory which is not equipped for high throughput DNA sequencing.
Although truncations that generate larger than 110-kb DNA cannot be recovered by this method, this does not appear to be a serious drawback because the genomic inserts of overlapping BACs/PACs in a contig are expected to be in either orientation with respect to the loxP site; therefore, that end of a large insert that fails to be represented in deletions from a particular clone is likely to be covered in an overlapping clone. Thus, all regions of a contig of >110-kb clones should be represented in nested-deletion libraries. This potential size restriction may also be overcome by electroporating rather than P1 packaging the loxP-inserted BAC/PAC DNA into Cre-expressing cells (20).
A direct comparison of efficiency and relative cost between the nested deletion and shotgun approaches for SNP identification on the same genetic locus has not been conducted and is likely to be laboratory dependent. Furthermore, as illustrated in Fig. 4, the two approaches are not mutually exclusive. The arrayed-end deletions in BACs and PACs are useful in determining or verifying the order of SNPs/sequence tagged sites obtained with other less-directed approaches including shotgun sequencing or sequencing from transposon insertions into small plasmids (21, 22). From a practical standpoint, generating an in vivo library of nested deletions is faster and easier than the multistep process of in vitro ligation of gel-purified, genomic insert DNA that has been sized to provide 2-kb plasmids from a given BAC/PAC. One does not require genomic clone DNA of high purity or highly skilled technical expertise to obtain the nested-deletion library. Both methods allow for processing multiple clones in parallel. This upfront gel-sizing of deletion clones essentially replaces the need for statistical mapping of the polymorphism at a later stage by using the gel-based radiation hybrid mapping technology. Considering all of the above issues, we believe that the nested-deletion approach is a valuable tool to augment the building of an ideal SNP map with high density, even spacing, and positionally oriented SNPs. This is otherwise difficult with the shotgun procedure alone.
Finally, this capability is likely to facilitate marker analyses in specific regions of the genome in many species, where tools of radiation-hybrid libraries or dense SNP maps may lag behind that of the human genome and association with a large genome center is not possible. Functional mapping of genes and other regulatory elements in BACs and PACs is likely to be an attractive application of nested deletions subject to the availability of suitable mammalian cell lines/animals for complementation studies. The loxP transposons used to generate these deletions also deliver a mammalian cell-selectable antibiotic-resistance gene to the BAC/PAC deletions that is suitable for selecting transformants when this DNA is introduced into these cells.
Acknowledgments
We thank Tai-He Xia for PCR primer design and Jonathan Charnecki, Philip Rivers, Susan Brady, Beth Harris, Sein Yin See, Ray Thomas, and Thalia Taylor for DNA sequencing. We gratefully acknowledge the valuable suggestions and encouragement of Sandra Stinnett, Donald Cain, Terri Fleming, Arash Afshari, Zheng Yu Xue, Anita Nelsen, Nathalie Godinot, Linda Briley, Jingwen Chen, Eric Lai, Jinghui Zhang, Karin Au, and Stephanie Shouse. We thank Quan Nguyen for helpful discussions and a critical reading of the manuscript; P.K.C. gratefully acknowledges the support and encouragement of Drs. Sakti Mookherjee, Steve Scheinman, and Ken Harewood at various stages of the work. All plasmids and strains used in this study are available on request.
Abbreviations
- BAC
bacterial artificial chromosome
- PAC
P1 artificial chromosome
- SNP
single-nucleotide polymorphism
- FIGE
field inversion gel electrophoresis
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schafer A J, Hawkins J R. Nat Biotechnol. 1998;16:33–39. doi: 10.1038/nbt0198-33. [DOI] [PubMed] [Google Scholar]
- 2.Kruglyak L. Nat Genet. 1997;1:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- 3.Collins F S, Guyer M S, Chakravarti A. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 4.Lai E, Riley J, Purvis I, Roses A. Genomics. 1998;54:31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- 5.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 7.Hermonson G G, Hoekstra M F, McElligott D L, Evans G A. Nucleic Acids Res. 1991;19:4943–4948. doi: 10.1093/nar/19.18.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell C, Gulati R, Nandi A K, Floy K, Heiter P, Kucherlapati R S. Proc Natl Acad Sci USA. 1991;88:5744–5748. doi: 10.1073/pnas.88.13.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee P K, Coren J S. Nucleic Acids Res. 1997;25:2205–2212. doi: 10.1093/nar/25.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee P K, Sternberg N L. Genet Anal Biomol Eng. 1996;13:33–42. doi: 10.1016/1050-3862(95)00147-6. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg N L, Shepherd N S. In: Current Protocols in Human Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. New York: Wiley; 1996. pp. 5.3.1–5.3.26. [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 1.1.82–1.1.84. [Google Scholar]
- 13.Abremski K, Hoess R, Sternberg N. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 14.Hoess R H, Abremski K. Proc Natl Acad Sci USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg N, Smoller D, Braden T. Genet Anal Tech Appl. 1994;11:171–180. doi: 10.1016/1050-3862(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 16.Coren J S, Pierce J C, Sternberg N. J Mol Biol. 1995;249:176–184. doi: 10.1006/jmbi.1995.0287. [DOI] [PubMed] [Google Scholar]
- 17.Kim U-J, Birren B W, Slepak T, Mancino V, Boysen C, Kang H-L, Simon M I, Shizuya H. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 18.Ioannou P A, de Jong P J. In: Current Protocols in Human Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. New York: Wiley; 1996. pp. 5.15.1–5.15.24. [Google Scholar]
- 19.Kleckner N. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- 20.Sheng Y, Mancino V, Birren B. Nucleic Acids Res. 1995;23:1990–1996. doi: 10.1093/nar/23.11.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellwagen A E, Craig N L. EMBO J. 1997;16:6823–6834. doi: 10.1093/emboj/16.22.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devine S E, Chissoe S L, Eby Y, Wilson R K, Boeke J D. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]