Abstract
We report here that the alternatively spliced nuclear factors associated with double-stranded RNA, NFAR-1 (90 kDa) and -2 (110 kDa), are involved in retaining cellular transcripts in intranuclear foci and can regulate the export of mRNA to the cytoplasm. Furthermore, the NFAR proteins were found to remain associated with exported ribonucleoprotein complexes. Loss of NFAR function, which was embryonic-lethal, caused an increase in protein synthesis rates, an effect augmented by the presence of the mRNA export factors TAP, p15, or Rae1. Significantly, NFAR depletion in normal murine fibroblasts rendered these cells dramatically susceptible to vesicular stomatitis virus replication. Collectively, our data demonstrate that the NFARs exert influence on mRNA trafficking and the modulation of translation rates and may constitute an innate immune translational surveillance mechanism important in host defense countermeasures against virus infection.
Keywords: innate immunity, mRNA export, vesicular stomatitis virus
Double-stranded RNA (dsRNA)-binding domains (DRBDs) are evolutionarily conserved 65- to 68-aa motifs found in dsRNA-binding proteins (DRBPs) that interact in a sequence-nonspecific manner with dsRNA species and regulate a diverse array of cellular processes in the cell (1, 2). Members of this group include Dicer, the adenosine deaminase acting on RNA (ADARs), RNase III, and the dsRNA-dependent protein kinase, PKR. We have previously identified a single human gene (NFAR) on chromosome 19 that produces two spliced variants encoding products of 90 and 110 kDa, referred to as NFAR-1 and -2, respectively (3, 4). NFAR-1 and -2 each contain two DRBDs in their C-terminus regions and exhibit 98% homology to one another at the amino acid level (Fig. 1A) (3, 5–7). NFAR-2 contains an extra 192 amino acids at the C-terminal region that are encoded by an extra three exons present in its corresponding transcript (3). The NFAR proteins are conserved, exhibiting 91% and 74% identity at the amino acid level to counterparts found in murine and Xenopus, respectively (8, 9). Although the exact function of the mammalian NFARs remains to be clarified, it is apparent that these DRBPs are ubiquitously expressed, predominantly reside in the nucleus of the cell, and can associate with both pre-mRNA and spliced mRNA in vitro, as well as with purified spliceosomes, suggesting that these molecules may play a potential role in the processing of newly synthesized transcripts [supporting information (SI) Fig. 5] (4, 10). The NFARs also have been reported to be substrates for PKR, which, after binding to viral dsRNA species, can inhibit translation by phosphorylating the α-subunit of eIF2 (4). Furthermore, the NFARs have been shown to bind to exportin-5, a member of the karyopherin-β family, to facilitate the export of minihelix containing viral RNAs, such as adenovirus VA1 RNA (11). Finally, other reports have indicated that the NFARs may be involved in the transcription and/or stabilization of selected mRNAs such as IL-2 and MyoD (12, 13).
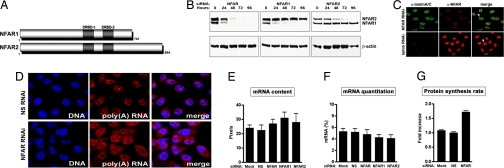
Fig. 1.
Depletion of NFAR proteins in the cell results in the redistribution of nuclear poly(A) foci and in increased protein synthesis. (A) Schematic of NFAR-1 and -2. (B) Depletion of NFARs using RNAi. HeLa cells were treated with siRNAs specific for NFAR-1, -2, or both. The levels of NFAR-1 and -2 were assessed by immunoblot analysis up to 96 h after the start of treatment. (C) NFAR levels in HeLa cells after treatment with NFAR- or lamin-specific siRNA (72 h) were visualized by confocal microscopy using rabbit anti-NFAR and Texas red-conjugated anti-rabbit antibody as described in Materials and Methods. (D) HeLa cells treated with NFAR siRNA for 72 h were subjected to in situ hybridization of total mRNA using biotinylated oligo(dT) 45-mers and Cy3-labeled streptavidine. Nuclei were stained with DAPI (blue). (E) Fluorescence intensities of poly(A) mRNA (pixels) at the equatorial section of siRNA-treated HeLa cells were quantified by using a Zeiss LSM510 confocal microscope. (F) Detection and quantification of total mRNA in siRNA-treated HeLa cells. Levels of poly(A) mRNA (as percentage of total RNA) were determined through hybridization to oligo(dT) primers and a series of enzymatic reactions as described in Materials and Methods. (G) Protein synthesis rates in siRNA-treated HeLa cells as determined by [35S]-Met labeling. Rates are graphed as fold increases over the average rate observed in nonspecific siRNA-treated cells, which was arbitrarily set to 1.
We report here that ablation of NFAR function leads to a loss in nuclear mRNA retention mediated through the TAP–p15 export pathway. Our data also indicate that the NFARs are retained on polyribosomes and act to govern translation rates. Furthermore, the loss of NFAR function resulted in a dramatic increase in vesicular stomatitis virus (VSV) and influenza virus replication, indicating that the NFAR proteins play an important role in innate immune defense to virus infection.
Results
Loss of NFARs Leads to the Redistribution of mRNA.
To examine NFAR's function, attempts were made to analyze the consequences of disrupting the NFAR gene in murine models through gene targeting. However, despite using two independent ES clones with confirmed NFAR gene disruption, no viable heterozygous mice were detected from >70 transfers of each ES clone into C57BL/6 blastocysts. Indeed, confirmed chimeric embryos were only detected through embryonic day 14.5 (SI Fig. 6). As an alternative approach to extend our analysis into the function of NFAR-1 and -2, we used siRNA to deplete the expression of these proteins in mammalian cells. Treatment of HeLa cells with siRNA directed to transcripts representing both NFARs, or specifically to transcripts of NFAR-1 or NFAR-2 alone, resulted in the loss of transcript and the depletion of 90–95% of the targeted protein(s) 72 h after treatment, as determined by immunoblotting and confocal microscopy using a polyclonal antibody recognizing both species (Fig. 1 B and C).
Given that the NFARs are known to interact with dsRNA and ssRNA species, it seemed possible that these proteins could be associated with ribosomal RNA (rRNA) or mRNA (4). We therefore analyzed the integrity of mRNA distribution in cells lacking both NFAR-1 and -2. NFAR-depleted cells were subjected to in situ hybridization with biotinylated oligo(dT) probes. As expected, mock treatment of HeLa cells or treatment with control siRNAs demonstrated the presence of discrete intranuclear domains rich in poly(A) mRNA, which are potentially major sources of transcriptional activity, as well as diffuse mRNA staining in the cytoplasm (Fig. 1D and SI Fig. 7 A and B) (14, 15). However, the redistribution of nuclear poly(A) foci was observed in NFAR-depleted cells. Repeated in situ confocal measurements (quantitation of pixel intensity) indicated no significant loss of poly(A) RNA in cells lacking NFAR (Fig. 1E). To confirm that nuclear mRNA was being redistributed, rather than degraded, quantitative mRNA measurements were carried out in cells lacking or containing the NFAR proteins. Analysis of total cellular mRNA confirmed that the loss of the NFARs does not result in a decrease in cellular mRNA levels (Fig. 1F). Finally, a number of cellular mRNAs, including PGK1, GAPDH, actin, and TFRC, as well as the ribosomal 18S RNA in cells lacking or containing the NFAR proteins, were quantitatively analyzed by RT-PCR; no significant differences in the levels of selective mRNAs analyzed were found (SI Fig. 7C). Analysis of the levels of actin and GAPDH transcripts after actinomycin D treatment of cells containing or lacking the NFARs further indicated that loss of NFAR function did not result in higher mRNA turnover rates (SI Fig. 7C). However, [35S]-Met incorporation into protein indicated that total cellular protein synthesis rates were actually moderately increased in NFAR-lacking cells compared with controls (Fig. 1G). Collectively, our data would suggest that the NFARs regulate the dispersion of newly synthesized transcripts, a consequence that may influence translation rates.
The NFARs Regulate Translation Rates.
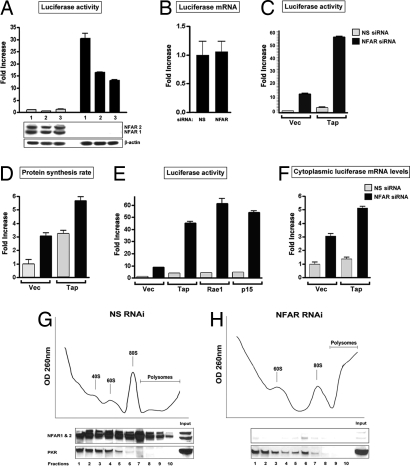
To further investigate the role of the NFARs in the regulation of gene expression, a luciferase reporter gene under the control of the SV40 promoter was transfected into control and NFAR-lacking cells. Significantly, we noted that luciferase activity increased ≈10- to ≈30-fold in NFAR-lacking cells, compared with cells treated with nonspecific siRNAs (Fig. 2A). These experiments were carried out in triplicate and were confirmed by using three independent siRNAs specific for the NFARs, as well as three independent control siRNAs. Immunoblot analysis was carried out for every experiment to ensure NFAR knockdown, and assays were monitored to ensure equal transfection efficiencies (Fig. 2A) (data not shown). Reconstitution of the NFAR proteins into siRNA-treated cells rescued the increase in luciferase expression (SI Fig. 8). Similar experiments using a SEAP-based reporter construct or with the luciferase gene under a different promoter (CMV) also indicated >10-fold increases in reporter expression in NFAR-lacking cells, compared with controls, indicating that the effect was not restricted to genes under the control of specific promoter elements (data not shown). Indeed, luciferase mRNA levels in the control and NFAR-depleted cells remained essentially equivalent with no loss, as determined by quantitative real-time PCR analysis, essentially confirming our earlier results and indicating that the NFARs' effect on gene regulation is likely to be at the posttranscriptional level (Fig. 2B).
Fig. 2.
TAP augments the stimulatory effect of NFAR knockdown on mRNA export and translation. (A) HeLa cells treated with three independent siRNAs to the NFARs or three independent siRNA controls were transfected with a luciferase reporter plasmid and a GFP-expressing plasmid to control for transfection efficiency. Each experiment was carried out in triplicate. NFAR depletion was confirmed by immunoblot analysis. Luciferase activities are expressed as fold increases over the average activity observed in nonspecific siRNA-treated cells, which was arbitrarily set to 1. (B) Luciferase mRNA levels were determined by quantitative real-time PCR analysis in NFAR and control siRNA-treated cells. RNA levels are expressed as fold increases over the level measured in nonspecific siRNA-treated cells, which was arbitrarily set to 1. (C) NFAR or NS siRNA-treated HeLa cells were cotransfected with a luciferase-expressing reporter vector and either a negative control vector (Vec) or a TAP-expressing vector. Luciferase activities are expressed as fold increases over the average activity observed in NS siRNA-treated cells that were transfected with the empty control vector, which was arbitrarily set to 1. (D) Protein synthesis rates were determined by [35S]-Met labeling in cells treated as described in C. Rates are graphed as fold increases over the average rate observed in nonspecific siRNA-treated cells transfected with Vec, which was arbitrarily set to 1. (E) Same as in C. Cells were additionally cotransfected with a Rae1- or p15-expressing plasmid. (F) Cytoplasmic luciferase mRNA levels were measured in cells treated as described in C by fluorescence real-time PCR analysis after cellular fractionation. RNA levels are expressed as fold increases over the average mRNA level observed in NS siRNA-treated cells transfected with vector alone, which was arbitrarily set to 1. (G and H) Polysomal profiles of HeLa cells treated with NS or NFAR-specific siRNA. Profiles were produced as described in Material and Methods by sucrose gradient centrifugation. Ten gradient fractions were collected and examined by poly(IC) agarose pull down and subsequent immunoblot analysis for the presence of NFAR and PKR. RNA was isolated from each fraction and analyzed to identify the 60S peak of the polysome profile. Each profile represents the average profile of three separate experiments.
The NFARs have been previously reported to be associated with purified spliceosome complexes (10). Thus, we next analyzed whether splicing was aberrant in NFAR-depleted cells by using a transfected β-globin construct (wild-type human β-globin-splicing cassette, pUCβ128SV). However, no significant differences in splicing were observed in either control or NFAR-lacking cells (SI Fig. 9A). To confirm this observation, we treated NFAR-depleted cells with type I IFN and monitored STAT 1 protein expression, an IFN-inducible gene comprising at least 24 exons and introns (16). That STAT 1 appeared to be translated in NFAR-lacking cells emphasizes that, although the NFAR proteins could conceivably comprise part of spliceosome–RNP complexes and have been reported to bind to selected intron and exon features in vitro, they do not appear to be critically required for the splicing process (SI Fig. 9B).
The NFARs Influence TAP/RAE1 Activity.
Correctly processed mRNAs are committed for export in the form of large ribonucleoprotein complexes (mRNPs), which our data indicate may incorporate the NFARs. The control of mRNA export is governed predominantly (≈75%) by the nuclear export factor NXF1, also called TAP in mammals and MEX67 in yeast (15, 17). TAP function is mediated by a cofactor, p15 (NXT1), and the mRNA export factor, Rae1/mrnp41/Gle2 (15, 18–21). Rae1 is thought to facilitate the delivery of TAP–RNP complexes to selected nucleoporins (Nups), such as Nup98, and are then directed through the NPC to the cytoplasm, with TAP remaining associated with polysome complexes (22, 23). Notably, the overexpression of TAP and/or p15 has been shown to moderately enhance gene expression by facilitating mRNA export, translation, or both (22, 24). To further evaluate the molecular mechanisms of NFAR's role in the control of mRNA distribution and translation, we examined the effect of TAP overexpression in NFAR-depleted cells. As shown in Fig. 2A, treatment of cells with NFAR-specific siRNA alone resulted in a 10- to 12-fold increase in expression of a transfected luciferase reporter gene (Fig. 2C). Overexpression of TAP alone (in control siRNA-treated cells) was found to increase the expression of luciferase ≈2- to 3-fold. In NFAR siRNA-treated cells, however, TAP's ability to increase luciferase expression was augmented ≈20-fold (Fig. 2C). Global protein synthesis rates in NFAR-depleted and TAP-overexpressing cells also were found to be elevated (2-fold) when compared with either NFAR-depleted or TAP-overexpressing cells alone, indicating that the effects were unlikely to be luciferase-dependent (Fig. 2D). To extend this analysis, we similarly depleted the NFARs by using siRNA and heterologously coexpressed Rae1 and p15 with a luciferase reporter construct. This analysis demonstrated that, in the absence of the NFARs, Rae1 or p15 also could greatly enhance the expression of luciferase (Fig. 2E). Whereas the total luciferase mRNA levels remained approximately equivalent in NFAR-depleted cells in the presence of TAP, compared with controls, cytoplasmic luciferase mRNA fractions were increased, again indicative of a change in mRNA localization (Fig. 2F) (data not shown).
A number of proteins such as TAP remain associated with RNP complexes after splicing and export and are known to regulate translation. Because translation seemed to be greatly affected by the presence or absence of the NFAR proteins, we examined polysome profiles in NFAR-lacking cells. Polysomes from NFAR-depleted or control cells were obtained by gradient centrifugation of cell lysates (equal RNA). Depletion of the NFARs was confirmed by immunoblot analysis after precipitation of fractions with agarose beads conjugated to poly(IC), which effectively bind DRBD-containing proteins (Fig. 2 G and H and SI Fig. 10) (3). Principally, in NS siRNA-treated cells, we observed an accumulation of 80S ribosomes and diminished polysomes when compared with NFAR siRNA-treated cells, which could reflect an increase in translation initiation in NFAR-depleted cells. Second, although both NFAR-1 and NFAR-2 are predominantly localized to the nucleus, a proportion of both of these proteins appear associated with ribosomes at various stages of the translation process, as determined by immunoblot analysis (Fig. 2 G and H). As a control, polysome blots were reprobed with anti-PKR antibody because this cytoplasmic kinase contains two classical DRBDs, similar to the NFARs, binds efficiently to poly(IC) agarose beads, and has been reported as being associated with 60S subunits (25). This analysis confirmed that, in control and NFAR-depleted cells, PKR was more evidently associated with ribosomal subunits engaged in the initiation of translation and not with polysomes engaged in the elongation process, as was evident with the NFARs (Fig. 2 G and H). Collectively, our data indicate that the NFARs are involved in controlling mRNA retention and export and plausibly retain association with ribosomes to influence the translation process, at the level of initiation or elongation, similar to TAP (22, 23).
Regulation of Translation by the NFARs.
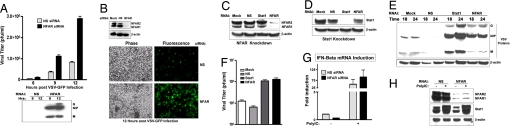
To further investigate the role of the NFARs in translational regulation, we evaluated the consequences of NFAR loss on replication of the cytoplasmic vesicular stomatitis virus (VSV). VSV provides an ideal model because it is not dependent on any nuclear export pathways for its replication. Thus, VSV mRNAs are exclusively synthesized and processed in the cytoplasm (26–28). Accordingly, HeLa cells lacking NFAR or controls treated with nonspecific siRNAs were infected with recombinant VSV expressing a GFP fluorescent gene (VSV-GFP) (29). This analysis indicated that VSV exhibited enhanced replication in NFAR-lacking HeLa cells (up to 4-fold), compared with controls (Fig. 3 A and B). These data are consistent with increased protein synthesis in NFAR-depleted cells.
Fig. 3.
NFAR depletion renders HeLa cells and primary MEFs susceptible to VSV. (A) NFAR or NS siRNA-treated HeLa cells were infected with VSV-GFP virus at an moi of 1. VSV-GFP titers (pfu/ml) were determined at 6, 9, and 12 h after infection. VSV protein synthesis was measured by immunoblot analysis using a mouse polyclonal anti-VSV serum. (B) The depletion of the NFAR proteins before VSV infection was confirmed by immunoblot analysis. GFP expression was visualized 12 h after VSV-GFP infection by fluorescence microscopy. (C and D) MEFs (C57BL/6 background) were treated with NS, NFAR, or STAT1 siRNA, and knockdown of NFAR and STAT1 was confirmed by immunblot analysis. (E and F) siRNA-treated MEFs were infected with VSV-GFP (moi = 1). Viral protein synthesis was assayed by immunoblot analysis at 18 and 24 h after infection. Viral titers (pfu/ml) were determined 24 h after infection. (G) siRNA-treated HeLa cells were transfected with poly(IC) for 6 h, and IFN-β mRNA levels were quantitated by RT-PCR. RNA levels are expressed as fold increases over the average IFN-β mRNA level observed in NS siRNA-treated cells not transfected with poly(IC). (H) siRNA-treated cells were analyzed by immunoblot for STAT1 induction after poly(IC) treatment.
We originally isolated the NFARs through yeast two-hybrid screening by using the antiviral IFN-inducible protein kinase PKR as bait, and we have demonstrated that the NFARs also are substrates for PKR (4). Speculatively, this finding would infer that the NFARs could be regulated by PKR in the event of virus infection and plausibly that the NFARs also play a role in host defense. Accordingly, to extend our analysis into the potential innate immune role carried out by the NFAR proteins, we ablated NFAR from normal murine embryonic fibroblasts (MEFs), which, unlike transformed HeLa cells, exhibit significant resistance to VSV infection (Fig. 3C) (29). As a control, we similarly depleted STAT1 by RNAi because ablation of this protein inhibits IFN signaling and renders cells susceptible to virus infection (Fig. 3D) (30, 31). Significantly, using this method, we demonstrated that loss of the NFARs, as well as STAT1, in MEFs lead to a dramatic increase in VSV protein synthesis and virus replication (Fig. 3 E and F and SI Fig. 11A). Mock-treated or control RNAi-treated cells infected with VSV exhibited significant resistance to virus replication (greater than a log difference). One explanation for these NFAR-mediated antiviral functions may include that the NFARs could act as intracellular sensors to recognize viral mRNA species and mediate the activation of IFN-β, a consequence that could inhibit viral replication. Thus, loss of the NFARs would lead to an increase in viral replication through a defect in IFN-β production. However, our data indicate that ablation of the NFARs did not prevent the production of IFN-β mRNA in response to poly(IC) treatment, neither in HeLa cells nor in MEFs, indicating that they do not exert their antiviral function through IFN-β-signaling mechanisms (Fig. 3G and SI Fig. 11B). Furthermore, the reduction of the NFARs did not inhibit the JAK–STAT pathway responsible for the induction of IFN-induced genes because STAT1, a key IFN-inducible gene, was robustly expressed in NFAR-depleted cells treated with dsRNA (Fig. 3H).
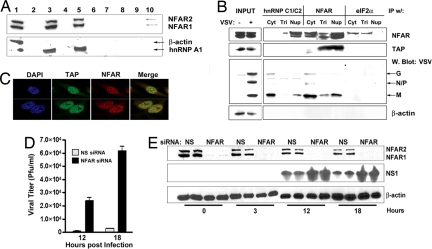
Previous data have indicated that VSV mRNAs may hijack host mRNP complexes to facilitate their translation (32, 33). Plausibly, the NFARs could function within exported RNP complexes as sentinels that can sense foreign RNAs through their RNA-binding motifs. To evaluate whether VSV mRNAs can associate with the NFARs, biotinylated VSV G or M mRNAs were generated, and extracts of HeLa cells were incubated with the labeled nucleotides. After precipitation with agarose-avidin beads, VSV G- or M mRNA-interacting proteins were electrophoresed and immunoblotted by using anti-NFAR serum. These data demonstrated that both VSV G and M mRNA could robustly associate with the NFAR proteins, compared with control RNA (Fig. 4A, lanes 3, 5, and 10). As a further control, we reprobed the blots by using antiserum to hnRNP A1, a known component of RNP complexes (34). This analysis indicated that hnRNP A1 coprecipitated with the NFAR proteins, indicating that VSV mRNAs bind directly to hnRNP A1 or indirectly through NFAR association. To further evaluate the association of the NFARs with RNP complexes, we infected HeLa cells with VSV and, after cell lysis under varying conditions of stringency, immunoprecipitated protein complexes with antibody to hnRNP proteins, the NFAR proteins, or, as a negative control, eIF2α. Precipitates were then subjected to immunoblot analysis by using anti-TAP or anti-NFAR antibody or serum raised to VSV. These analyses indicated that hnRNP C1/C2 directly or indirectly associated with the NFAR proteins (Fig. 4B) (data not shown). Moreover, TAP also was found to be part of heterogeneous nuclear RNP (hnRNP)–NFAR complexes; indeed, evidence of TAP and NFAR colocalization was observed by confocal microscopy (Fig. 4C). Interestingly, only the M protein of VSV was able to significantly coprecipitate with hnRNP molecular complexes or the NFAR proteins. These associations were not observed in complexes precipitated with eIF2α antibody.
Fig. 4.
VSV RNAs associate with RNP complexes harboring the NFAR proteins. NFAR-lacking cells are susceptible to influenza A infection. (A) Biotinylated VSV G or M mRNAs were incubated with HeLa cell extracts and subsequently precipitated with streptavidin agarose. RNA-interacting proteins were electrophoresed and immunoblotted by using anti-NFAR serum and antiserum to hnRNP A1. Lane 1, input; lane 2, protein marker; lane 3, biotinylated VSV G mRNA + lysates; lane 4, nonbiotinylated G mRNA + lysates; lane 5, biotinylated VSV M mRNA + lysates; lane 6, nonbiotinylated M mRNA + lysates; lane 7, biotinylated G mRNA + lysis buffer; lane 8, biotinylated M mRNA + lysis buffer; lane 9, no RNA + lysates; lane 10, biotinylated nonspecific mRNA + lysate. (B) Three fractions, an RNP fraction (Nup), a cytoplasmic fraction (Cyt), and a Triton wash fraction (Tri), were separated from VSV-infected 293T cell lysates and then subjected to coimmunoprecipitation (co-IP) with hnRNP C1/C2, NFAR, or eIF2α anti-serum conjugated to polyG agarose beads. After the co-IPs, elutants from all fractions were examined by immunoblot for the presence of NFAR, TAP, and the VSV proteins. (C) The nuclear colocalization of NFAR and TAP was determined by immunofluorescence. (D) NFAR- or NS siRNA-treated HeLa cells were infected with influenza A virus (INV) at an moi of 1. INV titers (pfu/ml) were determined at 12 and 18 h after infection. (E) Immunoblot analysis confirming depletion of the NFAR proteins and expression of viral NS1 protein in HeLa cells up to 18 h after influenza infection.
To evaluate whether the NFARs exerted influence on the replication of viruses other than VSV, we infected HeLa cells lacking the NFARs, or control cells, with influenza virus and monitored virus replication at various stages after infection. Our analysis indicated that by 12 h after infection, influenza virus had replicated higher (36-fold) in cells lacking the NFARs, compared with cells treated with nonspecific siRNA (Fig. 4 D and E). This effect was more pronounced 18 h after infection. Collectively, aside from establishing a role for the NFARs in mRNA export and translational regulation, these data additionally suggest that the NFARs, similar to its DRBD members, PKR and DICER, are important for efficient host defense.
Discussion
Our data presented here indicate that loss of NFAR activity does not significantly affect transcription or splicing, but leads to the redistribution of nuclear mRNA to the cytoplasm as well as to increased protein synthesis rates. NFAR activity was determined not to be restricted to T cell subsets or to specific mRNAs, although we found that NFAR loss was extremely detrimental to the host (3, 7, 10, 12, 13). Overexpression of TAP, Rae1, or p15 enhanced translation rates in the absence of the NFARs. These data would indicate that the NFARs negatively regulate the export of cellular mRNA governed by the TAP–p15 pathway. The NFARs largely localize to the nucleus, but we have shown here that both factors are shuttling proteins and are able to associate with RNPs and polysomes. Plausibly, the NFARs may be tethered to ribosomes or nuclear mRNAs within RNPs for editing or other purposes. Collectively, our studies indicate that, similar to TAP, a proportion of the NFARs may be retained by RNPs and polysomes to negatively influence translation rates as well as RNA export. This finding was most clearly seen by demonstrating that the loss of the NFARs leads to a dramatic increase in VSV protein synthesis, a virus that replicates exclusively in the cytoplasm. These data further suggest the possibility that the NFARs may play an important innate immune role in host defense against virus infection and may even provide a potential mechanism to explain viral oncolysis (29). A number of antiviral checkpoints at the translational level, such as those involving the PKR–eIF2α pathway, have evolved to impede viral translation suffice to allow the host time to activate antiviral genes (35). The NFARs may act similarly and function, in part, to directly detect and inhibit viral translation. Alternatively, the NFARs may be substrates and respond to antiviral molecules such as PKR to repress viral translation. Nearly all RNA viruses replicate in the cytoplasm, and so one possible mechanism of triggering antiviral pathways could involve host defense machinery recognizing foreign transcripts not associated with specific cellular proteins that are tethered to cellular mRNA and that accompany the transcript from the nucleus to the cytoplasm.
Materials and Methods
Plasmids.
pEGFP-C1, the EGFP-expressing negative control vector (Vec), was purchased from Clontech, and pGL3, the Firefly luciferase-expressing plasmid, was purchased from Promega. Plasmids pEGFP-C1-TAP and pEGFP-C1-p15 were received from Elisa Izaurralde (EMBL, Heidelberg, Germany), and plasmid pcDNA-Rae1 was sent to us by Jan van Deursen (Department of Pediatrics and Adolescent Medicine, Mayo Clinic, Rochester, MN).
Knockdown of NFARs by RNAi.
The 21-nt siRNA duplexes were purchased from Dharmacon. The constructs targeting both NFARs were: no. 1 (aagccacugaugcuauugggc), no. 2 (gcucaaagcuguguccgacugga), and no. 3 (gucgacgaugcugcgauugugau). We used no. 1 for all experiments unless otherwise stated. Constructs specific to either NFAR-1 or NFAR-2 were the following: NFAR-1 (aagacugcuacggcuaucaug) and NFAR-2 (aaggcaaacaaggaggcuacu and aacuacagugguaguggaggc; mismatched, aacuucagugguagucgaggc). Control siRNAs were specific to mFADD (acgaucugauggagcucaa), GFP (P-2048), and mouse STAT1 (D-058881–05) or were nonspecific (NS) (D-001206–01-80). HeLa cells were transfected with 240 pmoles of siRNA according to the manufacturer's instructions. MEFs were transfected by using an Amaxa nucleofector apparatus (program A-023) and Amaxz MEF nucleofector kit 1 according to the manufacturer's recommendations. Cells were incubated for 72 h. NFAR depletion was always confirmed by immunoblot analysis.
Transient Transfection of siRNA-Treated Cells.
siRNA-treated HeLa cells were transiently transfected with pGL3 in combination with pEGFP-C1, pEGFP-C1-TAP, pEGFP-C1-p15, or pcDNA-Rae1 at 72 h after the start of the siRNA transfection. EGFP and/or luciferase expression was analyzed 24 h after transfection. Experiments were carried out in triplicates. Luciferase activities were corrected for total protein content, and results were expressed as fold increases over the average activity observed in control siRNA-treated cells. Transfection efficiencies were evaluated by EGFP fluorescence.
Viral Infection/Poly(IC) Transfection of siRNA-Treated Cells and Determination of Viral Titers.
VSV (Indiana strain) and influenza A virus (WSN) infections and plaque assays were performed as described previously (36). Cells were infected with virus at a multiplicity of infection (moi) of 1. At particular time points, supernatant, total RNA, and total protein were collected, and GFP fluorescence (VSV-GFP only) was analyzed. Experiments were carried out in duplicates. Poly(IC) transfections were carried out as described (37).
Western Blotting.
The following primary antibodies were used: NFAR polyclonal rabbit antiserum (1:10,000), NS1 polyclonal antiserum (1:1,000), mouse polyclonal antiserum to VSV (1:5,000), goat polyclonal eIF2α (1:1,000; Santa Cruz Biotechnology), rabbit phospho-eIF2α (1:1,000; Santa Cruz Biotechnology), rabbit polyclonal Stat1 (1:1,000; Abcam), rabbit polyclonal PKR (1:1,000; Santa Cruz Biotechnology), or β-actin monoclonal antibody (1:10,000; Sigma–Aldrich). For detection, secondary HRP-conjugated anti-mouse or anti-rabbit IgG (Jackson Immunochemicals) was used.
Measurement of Protein Synthesis by [35S]-Met Labeling.
Protein synthesis rates were determined as described (38). The [35S] counts were measured in a scintillation counter and were corrected for total protein concentration as determined by the Bradford assay using a commercial kit (Pierce). Protein synthesis rates were expressed as fold increases over the average rate observed in control siRNA-treated cells.
RNA Measurements.
Total or cytoplasmic RNA was isolated from HeLa cells by using either TRIzol reagent (Invitrogen) or the RNeasy kit (Qiagen) following the manufacturer's recommendations. Total mRNA was quantified by using the poly(A) mRNA-detection system (Promega). Quantitative real-time PCR assays for luciferase were carried out by using the following primers: 5′-GCCCGCGAACGACATTT-3′ and 5′-CCACGGTAGGCTGCGAAAT-3′. Assays for GAPDH, actin, 18S, PGK1, and TFRC were carried out similarly by using total RNA and primers as described (39). Fluorescence real-time PCR analysis was performed by using a LightCycler 2.0 instrument (Roche) and TaqMan gene expression assays specific for Firefly luciferase and human IFN-β (Applied Biosystems). Relative amounts of mRNA were normalized to the 18S ribosomal RNA levels in each sample. The preceding cDNA synthesis was performed by using the first-strand cDNA synthesis kit for RT-PCR (Roche).
Confocal Microscopy and in Situ Hybridization.
Confocal microscopy and in situ hybridization of siRNA-treated HeLa cells with a biotinylated oligo(dT) 45-mer was performed as described previously (28). For immunofluorescence, NFAR rabbit polyclonal antiserum (1:100), lamin A/C mouse monoclonal antibody (1:10) (sc-7292; Santa Cruz Biotechnology), and monoclonal mouse TAP IgG1 (1:10; BD Biosciences) were used. The secondary antibodies used were fluorescein-conjugated anti-mouse antibody (1:100) and Texas red-conjugated anti-rabbit antibody (1:100).
Polysomal Profiles.
Cell extracts were prepared and polysome profiles analyzed essentially as described previously (40) for HeLa cells pretreated with NS or NFAR siRNA. Lysates (equal RNA) were loaded onto 47–20% linear sucrose density gradients (centrifuged at 29,000 rpm for 3.5 h at 4°C in a Sorvall StepSaver 65V13 Rotor, Cole–Parmer). RNA was extracted from each fraction by using TRIzol reagent (Invitrogen) and analyzed by using an Agilent 2100 Bioanalyzer to identify 18s and 28s ribosomal RNA.
Measurement of NFAR Expression in Polysome Fractions.
Poly(IC) agarose beads (Amersham) were used to pull down NFAR in each polysome fraction as described (4).
RNA Pull Down.
Labeled (biotin-UTP, 1:5 ratio to UTP; Roche) VSV G and M RNAs, as well as the negative control RNA (singed, Drosophila) were generated by in vitro transcription. The integrity and size of the RNAs were confirmed by denaturing agarose gel electrophoresis. The pull-down experiment was essentially carried out as described (41).
RNP Isolation.
RNPs were isolated as described (42). All fractions were then immunoprecipitated with NFAR, eIF2α, or hnRNP C1/C2 (Santa Cruz Biotechnology) antibodies conjugated to polyG agarose beads. Bound proteins were identified by Western blot analysis.
Supplementary Material
Acknowledgments.
We thank Gina Spruill, Laura Saunders, Nancy Gutgsell, and Isildinha M. Reis for technical help; Akila Mayeda (Institute for Comprehensive Medical Science, Fujita Health University, Aichi, Japan) for β-globin plasmids; Elisa Izaurralde (EMBL, Heidelberg, Germany) for the TAP plasmid; and Jan van Deursen (Department of Pediatrics and Adolescent Medicine, Mayo Clinic, Rochester, MN) for the Rae1 plasmid. This work was supported by Grant no. R01CA086431, from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711222105/DC1.
References
- 1.St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders LR, Barber GN. The evolutionarily conserved dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 3.Saunders LR, Jurecic V, Barber GN. The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics. 2001;71:256–259. doi: 10.1006/geno.2000.6423. [DOI] [PubMed] [Google Scholar]
- 4.Saunders LR, et al. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and −2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 2001;276:32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 5.Buaas FW, Lee K, Edelhoff S, Disteche C, Braun RE. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm Genome. 1999;10:451–456. doi: 10.1007/s003359901022. [DOI] [PubMed] [Google Scholar]
- 6.Patel RC, et al. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 7.Kao PN, et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 8.Orford RL, Robinson C, Haydon JM, Patient RK, Guille MJ. The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double- stranded-RNA-binding subunit. Mol Cell Biol. 1998;18:5557–5566. doi: 10.1128/mcb.18.9.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzostowski J, et al. RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. EMBO J. 2000;19:3683–3693. doi: 10.1093/emboj/19.14.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 11.Gwizdek C, et al. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279:884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- 12.Shim J, Lim H, R Yates J, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 13.Shi L, et al. NF90 regulates cell cycle exit and terminal miogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 14.Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiegand HL, et al. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yar R, Qureshi S, Zhong Z, Wen Z, Darnell JE., Jr The genomic structure of the STAT genes: Mulitple exons in coincident sites in Stat1 and Stat2. Nucleic Acids Res. 1995;23:459–463. doi: 10.1093/nar/23.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends in Cell Bio. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 18.Forler D, et al. RanBP2/Nup358 provides a major binding site for NXF1–p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol. 2004;24:1155–1167. doi: 10.1128/MCB.24.3.1155-1167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 21.Brown JA, et al. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- 24.Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 25.Kumar KU, Srivastava SP, Kaufman RJ. Double-stranded RNA-activated protein kinase (PKR) is negatively regulated by 60S ribosomal subunit protein L18. Mol Cell Biol. 1999;19:1116–1125. doi: 10.1128/mcb.19.2.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Her LS, Lund E, Dahlberg JE. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 27.Von Kobbe C, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 28.Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez M, Porosnicu M, Markovic D, Barber GN. Genetically engineered vesicular stomatitis virus in gene therapy: Application for treatment of malignant disease. J Virol. 2002;76:895–904. doi: 10.1128/JVI.76.2.895-904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meraz MA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 31.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 32.Adam SA, Choi YD, Dreyfuss G. Interaction of mRNA with proteins in vesicular stomatitis virus-infected cells. J Virol. 1986;57:616–622. doi: 10.1128/jvi.57.2.614-622.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen CA, Ennis HL, Cohen PS. Translational control of vesicular stomatitis virus protein synthesis: Isolation of an mRNA-sequestering particle. J Virol. 1982;44:932–938. doi: 10.1128/jvi.44.3.932-938.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mili S, Shu HJ, Zhao Y, Pino-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balachandran S, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 36.Balachandran S, et al. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J Virol. 2000;74:1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balachandran S, Venkataraman T, Fisher PB, Barber GN. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178:2429–2439. doi: 10.4049/jimmunol.178.4.2429. [DOI] [PubMed] [Google Scholar]
- 38.Balachandran S, Barber GN. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell. 2004;5:51–65. doi: 10.1016/s1535-6108(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 39.Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia. 2003;17:789–795. doi: 10.1038/sj.leu.2402880. [DOI] [PubMed] [Google Scholar]
- 40.Galbán S, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 42.Lal A, et al. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–4102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.