Abstract
We performed a three-phase genome-wide association study (GWAS) using cases and controls from a genetically isolated population, Ashkenazi Jews (AJ), to identify loci associated with breast cancer risk. In the first phase, we compared allele frequencies of 150,080 SNPs in 249 high-risk, BRCA1/2 mutation-negative AJ familial cases and 299 cancer-free AJ controls using χ2 and the Cochran–Armitage trend tests. In the second phase, we genotyped 343 SNPs from 123 regions most significantly associated from stage 1, including 4 SNPs from the FGFR2 region, in 950 consecutive AJ breast cancer cases and 979 age-matched AJ controls. We replicated major associations in a third independent set of 243 AJ cases and 187 controls. We obtained a significant allele P value of association with AJ breast cancer in the FGFR2 region (P = 1.5 × 10−5, odds ratio (OR) 1.26, 95% confidence interval (CI) 1.13–1.40 at rs1078806 for all phases combined). In addition, we found a risk locus in a region of chromosome 6q22.33 (P = 2.9 × 10−8, OR 1.41, 95% CI 1.25–1.59 at rs2180341). Using several SNPs at each implicated locus, we were able to verify associations and impute haplotypes. The major haplotype at the 6q22.33 locus conferred protection from disease, whereas the minor haplotype conferred risk. Candidate genes in the 6q22.33 region include ECHDC1, which encodes a protein involved in mitochondrial fatty acid oxidation, and also RNF146, which encodes a ubiquitin protein ligase, both known pathways in breast cancer pathogenesis.
Keywords: genomics, mapping, disease, predisposition, SNP
Cohort and twin studies have indicated that 5–15% of incident breast cancer cases result from autosomal-dominant cancer susceptibility (1–5). However, only ≈40% of the familial aggregation of breast cancers can be explained by mutations in BRCA1, BRCA2, or other identified cancer susceptibility genes (6). Attempts to use linkage strategies to localize other genes associated with an inherited predisposition to cancer have been hampered by genetic heterogeneity, decreased penetrance, and chance clustering (7–12). Candidate gene studies in multiplex kindreds affected by breast cancer have implicated rare variants of CHEK2, ATM, BRIP1, and PALB2 in the subset of families lacking BRCA mutations, but in most cases, the rarity and small effect sizes of these associations have precluded clinical application (13). Association studies of biologically plausible candidate genes have identified low-penetrance susceptibility alleles in pathways of carcinogen metabolism, inflammation and immune response, DNA metabolism and DNA repair as well as other known oncogenes and tumor suppressor genes (14–17). Most recently, two groups have carried out genome-wide association studies (GWAS) of selected kindreds and unselected individuals affected by breast cancer (18, 19). These studies have implicated a locus near FGFR2 as associated with an ≈1.2-fold increased risk of the disease. To add to the potential power of the GWAS approach, we have proposed and validated the use of a genetic isolate, in which larger regions of linkage disequilibrium surrounding known and putative “founder” mutations should increase the ability to map previously unidentified loci (20). As a first test of this approach, we have performed a GWAS study with 249 Ashkenazi Jewish (AJ) kindreds containing multiple cases of breast cancer but lacking BRCA1 or BRCA2 mutations and then replicated our findings in an independently ascertained cohort of nearly 1,000 AJ breast cancer cases and matched AJ controls. This approach successfully confirms the previously reported FGFR2 locus and also identifies a locus not seen in prior studies.
Results
GWAS in 249 Familial Breast Cancer Cases.
In phase 1, we analyzed 435,632 genotypes of SNPs in 249 probands from AJ kindreds with three or more cases of breast cancer but no identifiable mutations in BRCA1 or BRCA2 and in 299 cancer-free AJ controls. Genotyping was performed on the Affymetrix Early Access Version 3 (EAv3) 500K SNP platform as described in Material and Methods. As an initial data quality control, we filtered out SNPs that were out of Hardy–Weinberg equilibrium (HWE) in the controls; quantile–quantile plot analysis showed that SNPs with Fisher exact test P values <0.02 were not in HWE [supporting information (SI) Fig. 3 in SI Appendix], leaving 391,467 SNPs. Next, we compared allele frequencies in cases versus controls. χ2 and Cochran–Armitage tests produced closely comparable results, with the number of significant SNPs and level of significance far exceeding expectation, a finding similar to that reported by Easton et al. (18), who used a genotyping platform similar to ours. In view of reported discordances at large numbers of SNPs surveyed between genotype calls made with the BRLMM algorithm developed by Affymetrix and the fluorescence intensity values, we elected to graph genotypes versus relative fluorescence intensities for all SNPs (these data are available at our browser online at: http://theta2.ncifcrf.gov/cgi-bin/gbrowse/gold1/). These analyses showed that miscalled genotypes were responsible for the exceedingly low P values for most of these SNPs.
To ensure genotype accuracy, we decided to exclude any SNP with more than two no calls, because we observed that the discordance between BRLMM genotype calls and fluorescence-intensity values increased dramatically at the threshold of three or more miscalls. Setting the miscall threshold at two or less restricted the analysis to those SNPs with >99.7% call rate. This procedure reduced the effective size of the survey to 150,080 SNPs, but now the BRLMM genotypes correlated well with the relative fluorescence-intensity values. It also furnished a more realistic estimate of the number of SNPs exceeding χ2 distribution expectations (Table 1 and SI Fig. 4 in SI Appendix). We attribute the observed excess of smaller than expected χ2 P values (points above the gray line in SI Fig. 4 in SI Appendix) to significant associations with disease, rather than population stratification or genotyping miscalls. As can be seen in Table 1, the bin with the greatest credible P value excess was that between 10−5 and 10−4, where we see a ratio of 1.75 in observed to expected P values that translates to 21 observed SNPs versus 12 expected in that category.
Table 1.
Number of significant associations after initial AJ familial GWAS
| Level of significance | Observed* | Expected | Ratio |
|---|---|---|---|
| 0.01–0.05 | 6,232 | 5,977 | 1.04 |
| 0.001–0.01 | 1,438 | 1,362 | 1.06 |
| 0.0001–0.001 | 133 | 137 | 0.97 |
| 0.00001–0.0001 | 21 | 12 | 1.75 |
| <0.00001 | 1 | 1 | 1 |
| All P < 0.05 | 7,825 | 7,489 | 1.04 |
*Observed includes only values with H-W χ 2 in 299 controls with P > 0.02; the number of finite P values of this category were 150,080 of the 167,676 SNPs with call rates >99.7% surveyed.
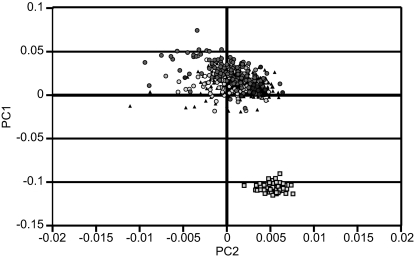
To determine whether the case-control groups were sufficiently similar to study them by using SNP association analysis, we applied the numerical methods developed by Price et al. (21) and implemented in the Eigenstrat package. This analysis (Fig. 1) confirmed cluster overlap between the AJ cases and controls used in phase 1 of this study and revealed distinct differences from a European–American reference population (see Material and Methods for a description of reference population). Filtering of outliers through the application of the PCA reduced the familial study to 299 research subject controls and 249 familial cases.
Fig. 1.
Principal components cluster analysis of phase 1 cases (triangles), controls (circles) of Ashkenazi origin, and a reference set of northern Europeans (squares).
Replication Analysis Using a Custom SNP Array.
In phase 2, we selected from the phase 1 analysis of 249 BRCA wild-type breast cancer kindreds the top-ranking 123 chromosomal regions (each region spanning 200 kb). To achieve satisfactory density of SNPs in candidate regions for haplotype analysis, we added from 2 to 4 additional SNPs per region and an additional 18 SNPs that also showed strong association (P < 0.001) and mapped within the distance of 200 kb from the top 123 loci. In total, there were 343 SNPs selected for genotyping in a larger replication cohort that consisted of a fully independent set of 950 consecutive AJ breast cancer cases and 979 age-matched cancer-free AJ controls. This analysis was performed on the Illumina “GoldenGate” platform as described in Materials and Methods. For the 343 SNPs, we compared allele frequencies in the phase 2 breast cancer cases and controls using both the χ2 test and the Cochran–Armitage trend test, which produced closely comparable results.
Replication in a Third Cohort and Aggregate Analysis.
We used a third group of 243 sporadic AJ breast cancer cases and 187 cancer-free AJ controls, which were also genotyped on the Affymetrix 500K SNP platform as an independent replication cohort of the phase 1 findings (Table 2, Phase 3). This cohort was ascertained from the New York metropolitan area as described in Materials and Methods and, hence, are derived from the same source population as the study's other cohorts. We also calculated χ2 P values from an aggregate analysis consisting of a total of 1,442 cases and 1,465 controls from all three phases of the study. Table 2 lists seven SNPs, including those with the most significant P values by using the allele χ2 test in the aggregate statistics. Table 2 also lists the odds ratios (ORs) and 95% confidence intervals (CIs) for the top-ranked SNPs calculated by using the χ2 test based on a dominant or a recessive genetic model. Analysis using Cochran–Armitage trend tests produced comparable results (data not shown).
Table 2.
Regions of the genome that showed the strongest associations with AJ breast cancer after phase 2 of the study
| SNP* | Chr† | Gene/region‡ | χ2P value of allele frequency (top rank no.)§ |
χ2P value aggregate¶ | OR allele for risk‖ (95% CI) | OR dom.** (95% CI) | OR rec.†† (95% CI) | MAFcs | MAFcn | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phase 1 | Phase 2 | Phase3 | |||||||||
| rs6569479 | 6q22.33 | ECHDC1; RNF146 | 6.0 × 10−3 (1,358) | 9.8 × 10−5 (1) | 2.2 × 10−2 | 1.2 × 10−7 | 1.39 (1.23–1.57) | 1.50 (1.29–1.74) | 1.48 (1.07–2.04) | 0.271 | 0.211 |
| rs7776136 | 6q22.33 | ECHDC1; RNF146 | 2.7 × 10−3 (742) | 9.9 × 10−5 (2) | 2 × 10−2 | 6.6 × 10−8 | 1.39 (1.23–1.57) | 1.51 (1.31–1.76) | 1.42 (1.05–1.90) | 0.278 | 0.217 |
| rs2180341 | 6q22.33 | ECHDC1; RNF146 | 8.9 × 10−4 (205) | 1.1 × 10−4 (3) | 1.8 × 10−2 | 2.9 × 10−8 | 1.41 (1.25–1.59) | 1.53 (1.32–1.77) | 1.51 (1.10–2.08) | 0.273 | 0.211 |
| rs6569480 | 6q22.33 | ECHDC1; RNF146 | 2.2 × 10−3 (616) | 1.2 × 10−4 (4) | 2 × 10−2 | 6.1 × 10−8 | 1.40 (1.24–1.58) | 1.51 (1.30–1.75) | 1.51 (1.10–2.08) | 0.272 | 0.211 |
| rs1078806 | 10q26.13 | FGFR2 | 4.5 × 10−2 (10,062) | 8.6 × 10−4 (5) | 4 × 10−2 | 1.5 × 10−5 | 1.26 (1.13–1.40) | 1.32 (1.13–1.54) | 1.40 (1.16–1.69) | 0.455 | 0.399 |
| rs3012642 | 23q13.1 | PHKA1; HDAC8 | 2.1 × 10−2 (4,751) | 2.1 × 10−3 (6) | 7.7 × 10−2 | NS | NS | NS | NS | 0.034 | 0.039 |
| rs7203563 | 16p13.3 | A2BP1 | 2.5 × 10−2 (5,573) | 3.3 × 10−3 (7) | 0.57 | 1.8 × 10−3 | 1.32 (1.11–1.57) | 1.36 (1.13–1.64) | NS | 0.110 | 0.086 |
NS, not significant; dom., dominant; rec., recessive; MAFcs, minor allele frequency in cases; MAFcn, minor allele frequency in controls.
*Included are all SNPs that had P < 0.01 based on the analysis of the phase 2 case-control data (see §).
†Chromosome position by cytogenetic band.
‡Genes identified in the genome browser that are within 100 kb on either side of the SNP indicated.
§Phase 1 consisted of 249 AJ probands from multiplex families in whom a mutation in BRCA1 and BRCA2 was excluded versus 299 cancer-free AJ controls; phase 2 consisted of 950 consecutive AJ breast cancer cases versus 979 cancer-free AJ controls; phase 3 consisted of an additional 243 AJ breast cancer cases from MSKCC and an independent 187 AJ cancer-free controls (rank no. represents the top order of the SNP of the total SNPs analyzed in phases 1 and 2).
¶Aggregate data consisted of the combined phases 1, 2, and 3.
‖OR calculated by using the aggregate data and based on the χ 2 test of the alleles inverted to express the risk allele when necessary; 95% CI.
**OR calculated by using the aggregate data and based on the χ 2 test of the dominant model.
††OR calculated by using the aggregate data and based on the χ 2 test of the recessive model.
During the course of the study, we became aware of the findings by other groups of associations in the FGFR2 region in association with breast cancer. Although we saw a weak signal for FGFR2 region in our familial study at rs1078806 (P = 0.045), to replicate the findings of Easton (18) and Hunter (19) in AJ breast cancer, we chose to genotype rs1078806 and three other SNPs from this region (rs1047111, rs12776781, and rs10886927) in the phase 2 breast cancer cases and controls. Of these four SNPs, the most significant P value in the allele frequency test was obtained at rs1078806 (P = 8.6 × 10−4; OR = 1.24; 95% CI 1.09–1.41). We also determined that rs1078806 was in high linkage disequilibrium (D′ = 0.9778, r2 = 0.9354) to other SNPs used in the study by Hunter et al.: rs2981582, rs1219648, rs2420946, and rs2981579 (data not shown). As shown in Table 2, the allele frequency test for this SNP obtained a P value of 1.5 × 10−5 in the aggregate analysis. Along with rs1078806, rs10886927 obtained a significant χ2 P value in the comparison of the phase 2 cases and controls (P = 0.036; OR = 1.15; 95% CI 1.01–1.31), but the P value in the aggregate data was not significant.
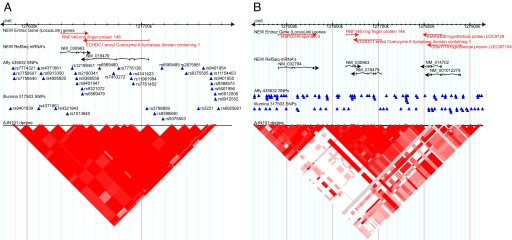
As shown in Table 2, the association with the RNF146; ECHDC1 region at 6q22 was the strongest and most consistent in this study. The association with rs2180341, rs6569479, rs6569480, and rs7776136 was initially found in the familial study and confirmed in the subsequent supporting studies, including the aggregate analysis of all 1,442 cases and 1,465 controls in the study where the P value for the allele frequency test was 2.9 × 10−8 for rs2180341. The major haplotype (H1) composed of the four SNPs was found to be protective at ≈5.53 × 10−5 level of significance (OR 0.564, 95% CI 0.422–0.752) (Table 3). This haplotype was confirmed in phase 2 at a significance threshold of 4 × 10−5 with nearly identical ORs and 95% CIs as the familial study (data not shown). Because of the high linkage disequilibrium (LD) in the region (see Fig. 2), it could not be determined whether the signal was arising from the RNF146 or the ECHDC1 gene.
Table 3.
Haplotype statistics using rs2180341, rs6569479, rs6569480, and rs7776136 in the RNF146; ECHDC1 locus
| Haplotype | Genotype | Phase 1 |
Phase 2 |
Aggregate study |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | n | OR (CI) | P | n | OR (CI) | P | n | OR (CI) | ||
| H1 | ACGT | 5.53 × 10−5 | 829 | 0.56 (0.42–0.75) | 1.30 × 10−4 | 2,889 | 0.75 (0.65–0.87) | 2.05 × 10−9 | 4,367 | 0.69 (0.61–0.78) |
| H2 | GTAA | 2.79 × 10−2 | 241 | 1.39 (1.03–1.87) | 1.11 × 10−4 | 939 | 1.34 (1.15–1.55) | 1.33 × 10−7 | 1,382 | 1.39 (1.23–1.57) |
In the phase 1 and aggregate studies, ≈4% of the subjects were imputed to possess five rare haplotypes not shown here. NS, not significant.
Fig. 2.
Significant linkage disequilibrium in the region at 6q22.33 associating with breast cancer in AJs. (A) A 200-kb window. (B) A 500-kb window. A and B show triangle plots using the 101 AJ controls. Red filled triangles represent a D′ of 1. The intensity of the box color is proportional to the strength of the linkage-disequilibrium property for the marker pair; white regions represent regions of low or no linkage disequilibrium; gray represents missing values; pink represents regions of D′ < 1.
There were also several other loci identified in phase 1 with associations and P values in the range of 10−3 to 10−6, which were confirmed in phase 2 with P values on the order of 10−2 and aggregate P values from 10−4 to 10−5 (data not shown). Other than FGFR2, these additional loci are not among the associations reported in prior WGAS (18, 19). These loci included 4q32.1, 3p21.31, 10q22.3, 17q25.3, and 5q12.2. Among these, the most significant P value found in the familial study was 5.7 × 10−6 for SNP rs6449674 at 5q12.2, but this SNP had only a marginally significant P value (0.06) by χ2 test of the allele frequencies in phase 2 (Table 2). Call rates for this SNP were high in the final stage (99.48%), genotype frequencies were in HWE in both cases and controls, and allele frequencies were stable in controls, suggesting that the relatively low relative risk associated with this SNP (and those surrounding it) may require larger studies to provide more accurate risk estimates. Analysis of additional SNPs surrounding this locus and the other loci yielded both protective and increased risk haplotypes (T.K. and B.G., unpublished work), but no haplotype thus far analyzed approaches the level of significance of the signal arising from the locus near the RNF146 and ECHDC1 genes.
Discussion
This study constitutes the second whole-genome analysis to date of a large number of kindreds with multiple cases of familial breast cancer and wild-type BRCA mutation status. Compared with the study of Easton (18), where 390 familial breast cancer cases were screened with 227,876 SNPs, here, we used 249 familial breast cancer cases at approximately the same SNP density. In contrast to the Easton study, this study was performed on a relatively isolated population, AJs, in which we have demonstrated a significant increase in power to detect founder mutations in other genes (20). A principal components analysis (PCA) was also applied as an exclusion criteria for the familial study, in contrast to genomic control or a statistical-significance adjustment criterion method.
We have compared our results with the National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) study in which DNA from 1,183 postmenopausal sporadic breast cancer cases were typed against 1,185 individually matched controls from the Nurses Health Study (19) and have also sought to reconcile the observations of Easton (18) detailing numerous statistically significant associations emerging from an experimental design similar to ours. Our study noted a risk locus in the region of chromosome 6q22.33, not seen in the Hunter et al. (19) study, whereas we also confirmed the breast cancer association with the FGFR2 locus noted in the two prior studies. Easton et al. (18) implicated 10 SNPs in seven other genomic regions as associated with breast cancer risk. Three of the SNPS in that study were in or near the TNRC9 locus on chromosome 16q. In our familial study, there were two reliable SNPs (rs3803662 and rs3112625) with allele P values in the range of P ≈ 0.01, however, these were on the 5′ end of the gene, and in the Hunter et al. (19) study, there was only one SNP within 200 kb of TNRC9 that was significant, with P ≈ 0.02 (rs8049226) by using an allele test. Similarly, at the MAP3K1 locus on chromosome 5q, where Easton reported P values in the range of 10−6 to 10−20 (18), we saw no significant SNPs by allele test, and Hunter found only one SNP (rs726501) with a P value in the range of P ≈ 0.01 by allele test (19). Near the LSP1 region, our data showed two SNPs (rs3817198, rs498337) with P values in the range of P ≈ 0.01 by allele test, where Easton reported P values in the range 10−5 to 10−9 (18); the Hunter et al. (19) data provided evidence for one SNP (rs7120258) in the region with a P value ≈0.01. In the nearby H19 region on chromosome 11p, where Easton et al. (18), reported P values in the range 0.01–10−5, we saw no signal, whereas Hunter found two SNPs (rs7120258, rs7578974), with association P values in the range of 0.01, with one additional SNP, rs217228, with a P value in the range of 0.02 (19). Thus, with the exception of the common findings regarding the FGFR2 locus, there is relatively little overlap in findings between each of the two published studies and the current study.
We also searched within 200 kb of each of four other regions (2p, 5p, 5q, and 8q) implicated by Easton with P values in the range of 0.02–10−12 (18) and, with one exception, found results similar to those reported above for the genic regions. When we searched near SNP rs981782, we found only modest signals in our study (P ≈ 0.02 at rs6451793), but Hunter's data revealed a P value of 6 × 10−5 at rs4866929 in HCN1 (hyperpolarization activated cyclic) (19). Easton apparently failed to map their significant SNP to this locus name (18).
It is possible that the differences observed between these studies as described above are a result of population stratification, sample-size differences, or genetic heterogeneity in the setting of differing genotyping platforms and different algorithms for “filtering” the data. That is, there may be a different spectrum of breast cancer-susceptibility gene mutations in the AJs, in the American Nurses, or in the cohorts used by the U.K.-led consortium. These differences may alternatively have arisen as a result of chance effects and different sample sizes, because the final phase of the U.K.-led consortium was an order of magnitude greater than our phase 2. However, the P values seen in the CGEMS study for FGFR2, for example, were similar to P values in the U.K.-led consortium, and yet the overlap of significantly associated SNPs between the two studies was minimal. Similarly, our study phase 2 is comparable in size with the CGEMS study, and, although the latter study used a GWAS for nonfamilial cases, the overlap in findings with our study was modest. Thus, an explanation other than population stratification is that the differences observed in the three studies to date may be a result of differential choice of SNPs used in the Perlegen, Affymetrix, and Illumina platforms used in these studies. In the study using the Perlegen platform, and in our study using the Affymetrix platform, “filtering” removed from 25% to 75% of the SNPs on the original arrays. Although this resulted in significantly reduced likelihood of “false positive” associations, it could also have allowed “false negatives,” i.e., missed associations. Similarly, although there was often tight linkage between the SNPs used in this study of AJ compared with non-AJ, e.g., in the FGFR2 region, and we have shown only modest global differences (by FST) in AJ (22), the failure to confirm other associations in this study may be due to SNP choice and differences in AJ LD structure.
The association with the RNF146; ECHDC1 region at 6q22.33 was the strongest and most consistent in this study. However, it could not be determined whether the association noted in this study was arising from the ECHDC1 gene, RNF146, or another locus in linkage disequilibrium. ECHDC1 has a related domain to the mitochondrial enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase, a trifunctional protein that plays a major role in mitochondrial fatty acid oxidation (23). Although ECHDC1 has been little studied in breast cancer, it is well established that fatty acid synthase-dependent endogenous fatty acid synthetic activity is abnormally elevated in a biologically aggressive subset of breast carcinomas and that inhibition of fatty acid oxidation can induce apoptosis in breast cancer cell lines, an effect that is increased 300-fold in TP53-silenced cell lines (24, 25). RNF146, also called dactylidin, is differentially expressed in neurodegenerative diseases; it encodes a polypeptide containing an amino-terminal C3HC4 RING finger domain, and is ubiquitously expressed, with cytoplasmic localization. Based on known cytoplasmic RING finger proteins, dactylidin likely functions as a ubiquitin protein ligase (E3). Protein degradation through the ubiquitin proteasome system regulates such processes as cell cycle, apoptosis, transcription, protein trafficking, signaling, DNA replication and repair, and angiogenesis; defects in this pathway have been well documented in breast cancer (26). Well known examples are the deregulation of the ubiquitin ligases BRCA1, BRCA2, BARD1, and MDM2 in subsets of human breast cancers (27). Although dactylidin has been little studied in cancer, it's membership in a class of genes including BRCA1, BRCA2, and BARD1 suggest that it could play a role in tumorigenesis of breast or other malignancies.
As with the prior study by Easton (18), we used a case enrichment of families where the incidence of breast cancer was high and confirmed the observed associations in a larger, independent cohort of sporadic cases and age-matched controls. In contrast to that report, this study was performed on a relatively isolated population, AJs. Residual effects of population heterogeneity were addressed through use of a principal components analysis. It remains to be seen whether further study in non-Ashkenazi cohorts, using these and other SNPs, will confirm the association with the RNF146/ECHDC1 region. As is currently the case for the loci mapped for breast cancer, and the 8q24 locus recently linked to risk for prostate and other cancers, follow-up studies including sequencing of candidate genes and measurement of RNA expression in breast tumors, as well as functional studies, will be required to achieve further insight into the biology underlying these associations.
It is important also to emphasize the clinical challenges posed by the relatively modest magnitude of relative risk associated with, for example, the H2 haplotype at the RNF146; ECHDC1 locus. The 1.4 relative risk documented here is small compared with the 20- to 40-fold increase in risk for early-onset breast cancer associated with BRCA mutations. The high frequency of these risk factors (23% of the population studied carried the H2 haplotype), combined with the observed relative risk produces a calculated proportion of breast cancer attributable to this risk factor in this population of ≈7%. However, such calculations do not take into account possible interactions between multiple loci. As noted in the context of prior associations of FGFR2 and CASP8, it will be important to look for multiplicative effects involving these and other low-penetrance alleles and cancer risk. Such interactions, which will impact both the attributable fraction and the relative risk of breast cancer, have thus far not been observed among known candidate loci (17, 18). In the absence of such interactive effects, the finding of individual low-penetrance genetic risk factors for breast cancer, such as the chromosome 6q22 locus reported here, will be of limited clinical utility compared with known high-penetrance mutations of genes such as BRCA1 or BRCA2 (13). Nonetheless, continued genetic epidemiologic inquiry and functional studies of candidate genes will shed further light on the polygenic nature of this common human malignancy.
Materials and Methods
Study Population.
As part of the first phase of this study, one hundred eighty-eight AJ women who presented at Memorial Sloan–Kettering Cancer Center (MSKCC) with breast cancer and a family history of three or more breast cancers in a single lineage were enrolled. Two additional AJ probands were ascertained at the Dana–Farber Cancer Institute in Boston, 14 AJ probands were ascertained in Toronto, and 45 AJ probands were ascertained at Beth Sheva Medical Center in Israel. Of these 249 kindreds, in 153 kindreds there were no unaffected (“nonpenetrant”) females in the affected lineage; in 72 kindreds there was one such female, and in 24 kindreds there were two such individuals. All affected probands tested negative for the three Ashkenazi BRCA founder mutations; 47 had additional full-sequence analysis. Kindreds were not ascertained if there was a case of ovarian cancer in the lineage affected by breast cancer. Participants completed a self-administered questionnaire about their medical history, date of birth, date of last mammogram, race, religious affiliation, country of birth, and religious affiliation of grandparents. To be eligible for enrollment in this study, either as a case or as a control, individuals must have indicated that all four grandparents were Jewish and of Eastern European ancestry. Mean age of the patients affected by breast cancer was 55 years, median 55 (range 25–95).
As controls to phase 1, the study enrolled 300 healthy AJ women who either accompanied male urology patients identified through the Urology Clinic or who were participating in cancer screening at MSKCC and who were cancer free and did not have a family history of breast cancer. The controls also included 29 healthy AJ women enrolled at Sheba Medical Center, Tel-Hashomer, Israel. For the controls, any woman who indicated a prior diagnosis of breast cancer, atypical hyperplasia, or lobular carcinoma in situ was not eligible for this study. Informed consent and blood specimens were obtained from these women under an institutional review board-approved protocol at MSKCC. One MSKCC control research subject withdrew permission to use her DNA before laboratory analysis. That sample and related records were redacted from the study. The remaining control subjects enrolled in the study were the first 299 control individuals from the ongoing study.
For a phase 2 validation of implicated regions, we typed an additional 950 breast cancer cases seen at MSKCC and unselected for family history cases and 979 age matched controls from the New York Cancer Project (NYCP). The NYCP is a cohort study involving consent for biospecimen collection and follow-up of 8,000 healthy volunteers in the same geographical region as the cases used in this study (28). MSKCC cases and NYCP controls were matched for age at diagnosis of breast cancer (cases) and age at genotyping (controls) to be within 2 years, and all were of AJ ancestry, and cases did not demonstrate any of the BRCA founder mutations.
As a third phase, and second replication set, we have included data from an additional and nonoverlapping cohort of 243 AJ women who presented at the MSKCC clinic with “sporadic” breast cancer. Absent was a first-degree family history of disease in these women. As an addition to the control group, we included 187 additional disease-free AJ females obtained from the ongoing NYCP who were not included in our second phase control cohort. These additional cases were genotyped on the EA Affymetrix 500K SNP array, and the controls were genotyped on the Affymetrix Commercial Version 500K Genotyping Chips for a separate study; because both of these platforms included the key loci replicated in phase 2 of the current study, this cohort was included for separate analysis of these loci, and these data were added to the aggregate analysis.
Genotyping.
Preparation of genomic DNA from blood was performed as previously described (29). Genotyping was carried out by using Affymetrix GeneChip Early Access Version 3 (EAv3) Human Mapping Arrays. Use of Affymetrix EAv3 chips for genotyping was performed as described in the Gtype 4.0 manual www.affymetrix.com/Auth/support/download/manuals/gtype_user_guide.pdf, except that 150 ng of all genomic DNA samples were evaluated for quality by gel electrophoresis. After qualification of the DNA samples, each sample was then divided into two aliquots. Sequence complexity was reduced by restriction enzyme digestion with either NspI or StyI, and a biotin-labeling primer amplification assay was performed on each DNA aliquot. Hybridization of the amplified probes was then performed on specific NspI or StyI arrays, as appropriate. Genotyping of 187 AJ control samples for phase 3 obtained from NY Academic Medicine Development Company (AMDeC) was essentially identical, except that these were applied to the Affymetrix Commercial Version 500K Chip, which had overlapping dbSNP ids at 435,632 sites.
For phase 2, we genotyped 384 custom-selected SNPs assembled in a 96-well microtiter plate format using the Illumina GoldenGate assay according to the manufacturer's protocol (30). Briefly, allele-specific primers were hybridized directly to genomic DNA that was immobilized on a solid support. In case of a perfect match, the primer was extended, and the extension product was ligated to a probe hybridized downstream of the SNP position. The ligated product was amplified by PCR by using universal primers that are complementary to a universal sequence in the 3′ end of the ligation probes and 5′ end of the allele-specific primers, respectively. The ligation probe contains a SNP-specific Tag-sequence, and the universal allele-specific primers carry an allele-specific fluorescent label in their 5′ end. After PCR, the amplified products were captured on beads carrying complementary target sequences for the SNP-specific Tag of the ligation probe. The beads are kept in fiber-optic array bundles in a format compatible with 96-well microtiter plates. In our assay format, 36,864 genotypes were generated on a single microtiter plate.
Analytic Pipeline Description.
We developed an efficient pipeline for GWAS analysis, minimizing the use of high-performance hardware and proprietary software. The main feature of the pipeline was the concentration of all available information into a single portable prettybase (PB) text file that includes structured SNP calls, confidences, and dbSNP references. Files with genotype calls will be available from our browser at http://theta2.ncifcrf.gov/cgi-bin/gbrowse/gold1/ at time of publication.
Additional Datasets Examined.
Subsequent to detailed examination of the 101 AJ and 60 CEU (European samples from Centre d'Etude du Polymorphisme Humain) genotype dataset described in Olshen (22), we sought to verify our linkage disequilibrium results. For this purpose, we culled any SNP with more than two no calls based on a theoretical optimization of our heterozygosity detection. This resulted in a dataset of 167,676 SNPs on which we performed each analysis. In addition, we carried out a cross-examination with additional data. For this purpose, we examined the ≈317,000 genotypes from 151 Jewish women included in the Seldin study (31). We ran HaploView on this dataset using the default parameters as previously described. The results of each of these validation datasets are displayed on our public browser.
Additional description of genotyping methods, software utilized, and adjustments for population heterogeneity as well as acknowledgments for research support are in SI Text in SI Appendix.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800441105/DC1.
References
- 1.Claus EB, Schildkraut JM, Thompson WD, Risch NJ. The genetic attributable risk of breast and ovarian cancer. Cancer. 1996;77:2318–2324. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2318::AID-CNCR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Colditz GA, et al. Family history, age, and risk of breast cancer. Prospective data from the Nurses' Health Study. J Am Med Assoc. 1993;270:338–343. [PubMed] [Google Scholar]
- 3.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Locatelli I, Rosina A, Lichtenstein P, Yashin AI. A correlated frailty model with long term survivors for estimating the heritability of breast cancer. Stat Med. 2007;26:3722–3734. doi: 10.1002/sim.2761. [DOI] [PubMed] [Google Scholar]
- 5.Slattery ML, Kerber RA. A comprehensive evaluation of family history and breast cancer risk. The Utah Population Database. J Am Med Assoc. 1993;270:1563–1568. [PubMed] [Google Scholar]
- 6.Ford D, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kainu T, et al. Somatic deletions in hereditary breast cancers implicate 13q21 as a putative novel breast cancer susceptibility locus. Proc Natl Acad Sci USA. 2000;97:9603–9608. doi: 10.1073/pnas.97.17.9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerangueven F, et al. Loss of heterozygosity and linkage analysis in breast carcinoma: Indication for a putative third susceptibility gene on the short arm of chromosome 8. Oncogene. 1995;10:1023–1026. [PubMed] [Google Scholar]
- 9.Rahman N, et al. Absence of evidence for a familial breast cancer susceptibility gene at chromosome 8p12–p22. Oncogene. 2000;19:4170–4173. doi: 10.1038/sj.onc.1203735. [DOI] [PubMed] [Google Scholar]
- 10.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 11.Seitz S, et al. Strong indication for a breast cancer susceptibility gene on chromosome 8p12–p22: Linkage analysis in German breast cancer families. Oncogene. 1997;14:741–743. doi: 10.1038/sj.onc.1200881. [DOI] [PubMed] [Google Scholar]
- 12.Thompson D, et al. Evaluation of linkage of breast cancer to the putative BRCA3 locus on chromosome 13q21 in 128 multiple case families from the Breast Cancer Linkage Consortium. Proc Natl Acad Sci USA. 2002;99:827–831. doi: 10.1073/pnas.012584499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Offit K, Garber JE. Time to check CHEK2 in families with breast cancer? J Clin Oncol. 2008;26:519–520. doi: 10.1200/JCO.2007.13.8503. [DOI] [PubMed] [Google Scholar]
- 14.Dunning AM, et al. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8:843–854. [PubMed] [Google Scholar]
- 15.Friedberg T. Cytochrome P450 polymorphisms as risk factors for steroid hormone-related cancers. Am J Pharmacogenom. 2001;1:83–91. doi: 10.2165/00129785-200101020-00001. [DOI] [PubMed] [Google Scholar]
- 16.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 17.Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis NA, et al. Localization of breast cancer susceptibility loci by genome-wide SNP linkage disequilibrium mapping. Genet Epidemiol. 2006;30:48–61. doi: 10.1002/gepi.20101. [DOI] [PubMed] [Google Scholar]
- 21.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Olshen A, et al. Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping. BMC Genet. 2008;9:14. doi: 10.1186/1471-2156-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto T, Shindo Y, Souri M, Baldwin GS. A new inhibitor of mitochondrial fatty acid oxidation. J Biochem. 1996;119:1196–1201. doi: 10.1093/oxfordjournals.jbchem.a021368. [DOI] [PubMed] [Google Scholar]
- 24.Menendez JA, Lupu R. RNA interference-mediated silencing of the p53 tumor-suppressor protein drastically increases apoptosis after inhibition of endogenous fatty acid metabolism in breast cancer cells. Int J Mol Med. 2005;15:33–40. [PubMed] [Google Scholar]
- 25.Zhou W, et al. Fatty acid synthase inhibition triggers apoptosis during S phase in human cancer cells. Cancer Res. 2003;63:7330–7337. [PubMed] [Google Scholar]
- 26.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer Project: Rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterlongo P, et al. MSH6 germline mutations are rare in colorectal cancer families. Int J Cancer. 2003;107:571–579. doi: 10.1002/ijc.11415. [DOI] [PubMed] [Google Scholar]
- 30.Shen R, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Seldin MF, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.