Abstract
The zinc-finger antiviral protein (ZAP) specifically inhibits the replication of many viruses by preventing the accumulation of viral mRNAs in the cytoplasm. ZAP directly binds to the viral mRNAs and recruits the RNA exosome to degrade the target RNA. In the present study, we identified the p72 DEAD box RNA helicase, but not the highly similar RNA helicase p68, as a ZAP-interacting protein. The binding domain of ZAP was mapped to its N-terminal portion, whereas both the N- and C-terminal domains of p72 bound to ZAP. Overexpression of the C-terminal domain of p72 reduced ZAP's activity, whereas overexpression of the full-length p72 enhanced ZAP's activity. The RNA helicase activity was required for p72 to promote ZAP-mediated RNA degradation. Depletion of p72 by RNAi also reduced ZAP's activity but did not affect tristetraprolin-mediated RNA degradation. We conclude that p72 is required for the optimal activity of ZAP, and we propose that p72 helps to restructure the ZAP-bound target mRNA for efficient degradation.
Keywords: restriction, RNA degredation, retrovirus, alphavirus
The zinc-finger antiviral protein (ZAP) was initially recovered as a host factor that inhibited the infection of cells by Moloney murine leukemia virus (MLV) (1). In addition to MLV, overexpression of ZAP also inhibits the replication of Ebola virus and Marburg virus (2), and several members of the Alphavirus genus, including Sindbis virus (SINV) (3). IFN treatment or SINV infection of murine bone marrow-derived dendritic cells significantly up-regulates the expression level of ZAP (4), suggesting that ZAP also may function as an antiviral effector in vivo. However, ZAP is not a universal antiviral factor; some viruses, including herpes simplex virus type 1 and yellow fever virus, grow normally in ZAP-expressing cells (3).
ZAP inhibits virus replication by preventing the accumulation of the viral mRNA in the cytoplasm (1–3). In the N terminus of ZAP, there are four CCCH-type zinc-finger motifs (1). ZAP directly binds to specific viral mRNA sequences [ZAP-responsive element (ZRE)] through the CCCH-type zinc fingers (5). Furthermore, ZAP directly interacts with the RNA-processing exosome (6), a 3′–5′ exoribonucleases complex consisting of at least nine components (7). The current working model is that ZAP promotes the degradation of ZRE-containing mRNAs by directly binding to the target RNA and recruiting the exosome to degrade the RNA (6).
The p72 RNA helicase is a member of the DEAD box family of RNA helicases, which are characterized by a conserved motif including Asp-Glu-Ala-Asp (DEAD) and involved in various biological processes (8, 9). Compared with other DEAD box RNA helicases, p72 has a unique N-terminal domain containing repeats of the sequence RGG and a C-terminal domain rich in serine and glycine and terminating with a polyproline region (10). p72 is highly related to the better known p68 RNA helicase; they share ≈90% sequence identity in the core region spanning the conserved motifs characteristic of this family and 69.7% overall homology (10). The p72 and p68 RNA helicases have been shown to exhibit RNA-dependent ATPase and RNA helicase activities and to catalyze RNA structure rearrangement (11). Both proteins play important roles in transcriptional regulation (12) and RNA splicing (13–15). They also have been reported to be involved in the processing of a set of microRNAs and rRNAs (16). Gene disruption of p68 or p72 in mice results in early lethality (16).
The yeast homologue of p68/p72 (Dbp2p), which shares 55% and 46% sequence identity with human p68 and p72, respectively, has been shown to be required for nonsense-mediated mRNA decay and rRNA processing (17). In Dbp2p-depleted yeast cells, the defects in ribosome biogenesis, but not in mRNA decay, can be rescued by human p68 (17). Whether p68 or p72 is involved in RNA decay in mammalian cells has not been documented.
In the present study, we identified p72, but not p68, as a ZAP-interacting protein, and we provide evidence indicating that p72 is required for ZAP-mediated mRNA degradation.
Results
Identification of the p72 RNA Helicase as a Putative ZAP-Interacting Protein.
To search for cellular factors involved in ZAP-mediated mRNA degradation, we set out to identify proteins that coprecipitated with ZAP from cell lysates. Myc-tagged ZAP was expressed in 293TRex-ZAP cells, which expressed the protein upon tetracycline induction. Because ZAP primarily functions in the cytoplasm, the cells were lysed in a relatively mild lysis buffer (see Materials and Methods for details), leaving the nuclei intact. To prevent possible nonspecific RNA tethering, the cytoplasmic lysates were treated with RNase A. ZAP and its associated proteins were immunoprecipitated with anti-myc antibody and resolved on SDS/PAGE, and the proteins were visualized by Coomassie blue staining. Compared with the precipitates of 293TRex control cells, a specific band of ≈70 kDa was detected in the precipitates of the ZAP-expressing cells (data not shown). This band was excised and subject to matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) analysis to identify the protein. The results of database search identified the p72 DEAD box RNA helicase as a candidate.
Confirmation of the Interaction Between ZAP and p72.
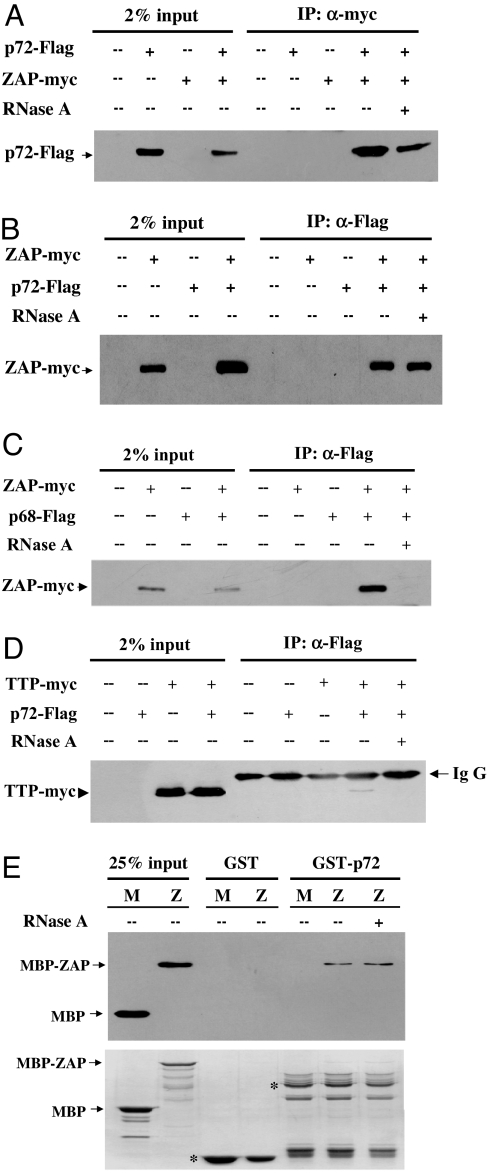
To confirm the interaction between ZAP and p72, coimmunoprecipitation assays were performed. Flag-tagged p72 and myc-tagged ZAP were coexpressed in HEK293T cells. Immunoprecipitation of ZAP using the anti-myc antibody coprecipitated Flag-tagged p72 in the absence or presence of RNase A (Fig. 1A). In a reverse experiment, immunoprecipitation of Flag-tagged p72 coprecipitated ZAP in the absence or presence of RNase A (Fig. 1B). In contrast, immunoprecipitation of the RNA helicase p68, which is highly related to p72, coprecipitated ZAP only in the absence of RNase A (Fig. 1C). To further evaluate the specificity of the interaction between p72 and ZAP, we tested whether p72 bound to tristetraprolin (TTP), which also contains CCCH-type zinc fingers, but lacks extensive sequence homology with ZAP (18). Immunoprecipitation of p72 coprecipitated a relatively low level of TTP only in the absence of RNase A (Fig. 1D). These results indicated that the RNase A treatment efficiently disrupted nonspecific RNA tethering and that the interaction between ZAP and p72 was specific and RNase-resistant. To further confirm the direct interaction between p72 and ZAP, p72 and ZAP were expressed in Escherichia coli as GST and maltose-binding protein (MBP) fusion proteins, respectively. The partially purified proteins were analyzed for their ability to bind to each other by pull-down assays. Indeed, MBP-ZAP bound to GST-p72 with or without RNase A treatment (Fig. 1E). Collectively, these results established that ZAP directly interacted with p72 in an RNA-independent manner.
Fig. 1.
ZAP interacts with the p72 RNA helicase. (A) The plasmids expressing myc-tagged ZAP and Flag-tagged p72 were transiently cotransfected into HEK293T cells. Forty-eight hours after transfection, the cells were lysed. The cell lysates were immunoprecipitated with the anti-myc antibody in the presence (+) or absence (−) of RNase A and Western blotted with the anti-Flag antibody. (B) The plasmids expressing Flag-tagged p72 and myc-tagged ZAP were transiently cotransfected into HEK293T cells. The cell lysates were immunoprecipitated with the anti-Flag antibody in the presence (+) or absence (−) of RNase A and Western blotted with the anti-myc antibody. (C) The plasmids expressing Flag-tagged p68 and myc-tagged ZAP were transiently cotransfected into HEK293T cells. The cell lysates were immunoprecipitated with the anti-Flag antibody in the presence (+) or absence (−) of RNase A and Western blotted with the anti-myc antibody. (D) The plasmids expressing Flag-tagged p72 and myc-tagged TTP were transiently cotransfected into HEK293T cells. The cell lysates were immunoprecipitated with the anti-Flag antibody in the presence (+) or absence (−) of RNase A and Western blotted with the anti-myc antibody. (E) Bacterially expressed GST or GST-p72 was immobilized onto glutathione-Sepharose 4B resin and incubated with bacterially expressed and partially purified myc-tagged MBP or ZAP, which was fused to myc-tag at the C terminus and to MBP at the N terminus (MBP-ZAP). The resins were washed and boiled in the sample loading buffer. The proteins were resolved by SDS/PAGE and detected by Western blotting with the anti-myc antibody (Upper) or by Coomassie blue staining (Lower). The positions of MBP and MBP-ZAP are indicated by arrows, and the positions of GST and GST-p72 are indicated by asterisks. M, MBP; Z, MBP-ZAP.
Mapping the Binding Domains of ZAP and p72.
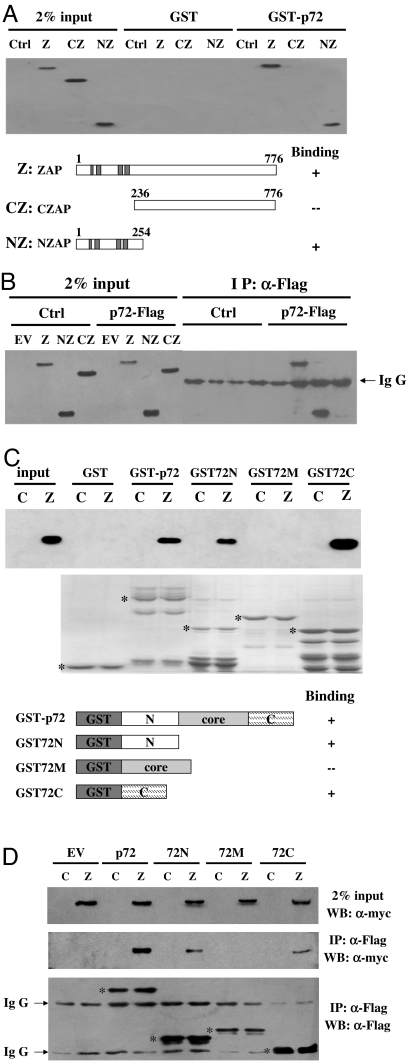
The N-terminal domain of ZAP fused with the product of the zeocin resistance gene at the C terminus (NZAP-Zeo) displayed the same antiviral activity as the full-length ZAP (1). We reasoned that if the interaction between ZAP and p72 were important for ZAP's antiviral activity, p72 also should bind to NZAP. ZAP-truncation mutants were analyzed for their abilities to bind to p72 in the presence of RNaseA using pull-down and coimmunoprecipitation assays. Indeed, NZAP bound to p72 as well as the full-length ZAP in both assays (Fig. 2 A and B). In contrast, the C-terminal domain of ZAP (CZAP) failed to do so (Fig. 2 A and B).
Fig. 2.
Mapping the binding domains of ZAP and the p72 RNA helicase. (A) Bacterially expressed GST or GST-p72 was immobilized onto glutathione-Sepharose 4B resin and incubated with the lysates of the cells expressing the indicated myc-tagged ZAP proteins in the presence of RNase A at 4°C for 2 h. The resins were washed and boiled in the sample loading buffer. (Upper) The proteins were resolved by SDS/PAGE and detected by Western blotting using the anti-myc antibody. (Lower) Schematic representations of the ZAP proteins are shown, and their binding activities are summarized. Input, total cell lysate; Ctrl, control cells. (B) The plasmid expressing the indicated myc-tagged ZAP protein and the plasmid expressing Flag-tagged p72 were transiently cotransfected into HEK293T cells. The cell lysates were immunoprecipitated with the anti-Flag antibody in the presence of RNase A and Western-blotted with the anti-myc antibody. Input, total cell lysate; Ctrl, control plasmid pCMVHF; EV, empty vector pcDNA4. (C) Bacterially expressed recombinant proteins of the indicated p72-truncation mutants were immobilized onto glutathioneSepharose 4B resin and incubated with the lysates of control cells or ZAP-expressing cells in the presence of RNase A at 4°C for 2 h. The resins were washed and boiled in the sample loading buffer. (Upper) The proteins were resolved by SDS/PAGE and detected by Western blotting using the anti-myc antibody. (Lower) Schematic representations of the p72 proteins are shown and their binding activities are summarized. Input, total cell lysate; C, control cells; Z, ZAP-expressing cells. The positions of the GST proteins are indicated by asterisks. (D) The plasmid expressing the indicated Flag-tagged p72 proteins were transiently transfected into ZAP-expressing cells or control cells. The cell lysates were immunoprecipitated with the anti-Flag antibody in the presence of RNase A and Western-blotted with the indicated antibodies. Input, total cell lysate; C, control cells; Z, ZAP-expressing cells. EV, empty vector. The positions of the p72 proteins are indicated by asterisks.
To determine the domains of p72 responsible for binding to ZAP, a series of p72-deletion mutants was constructed. These proteins were bacterially expressed with GST fused at the N terminus and used to pull down ZAP expressed in the 293TRex-ZAP cells. Both the N- and C-terminal domains (GST72N and GST72C, respectively) bound to ZAP (Fig. 2C). In contrast, the central domain (GST72M) did not display any detectable binding (Fig. 2C). These results were confirmed in coimmunoprecipation assays using Flag-tagged p72-deletion mutants (Fig. 2D).
Effects of the Overexpression of the p72 Mutants on ZAP's Activity.
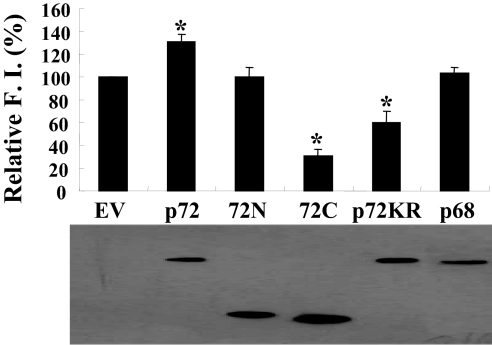
In an attempt to explore the role of p72 in ZAP's function, various versions of p72 were analyzed for their abilities to affect ZAP's activity. The plasmid expressing these proteins was cotranfected with the pMLV-Luc reporter into ZAP-expressing cells. ZAP's activity was measured by its ability to inhibit luciferase expression and was expressed as relative fold inhibition of the luciferase reporter (see Materials and Methods for details). Overexpression of the full-length p72 slightly enhanced ZAP's activity (Fig. 3A Upper). In contrast, the overexpression of a p72 mutant (p72-K142R), which has been reported to lack the ATPase and RNA helicase activities (14), reduced ZAP's activity (Fig. 3 Upper). Although both the N- and C-terminal domains of p72 (72N and 72C, respectively) bound to ZAP comparably well (Fig. 2 C and D), their effects on ZAP's activity were very different. Although the overexpression of 72C significantly reduced ZAP's activity, 72N had little effect (Fig. 3A Upper). Western blotting results revealed that the expression levels of these p72 proteins were roughly comparable (Fig. 3A Lower). Consistent with the observation that p68 did not interact with ZAP in an RNA-independent manner, the overexpression of p68 did not have any detectable effect on ZAP's activity (Fig. 3A). Collectively, these results suggested that p72 is involved in ZAP's function, and the RNA helicase activity of p72 is required for its involvement. The p72-K142R mutant and 72C might block ZAP and interfere with the function of endogenous p72.
Fig. 3.
Effects of the overexpression of the p72 mutants on ZAP's activity. The plasmid expressing the indicated Flag-tagged p72 proteins or Flag-tagged p68 was cotransfected with the pMLV-luc reporter into 293TRex-ZAP cells. Immediately after transfection, tetracycline was added to induce ZAP expression. Forty-eight hours after transfection, the cells were lysed. An aliquot of the lysate was analyzed for the luciferase activities, and another aliquot was analyzed for the expression of the p72 proteins or p68 by Western blotting. Fold inhibition was calculated as the luciferase activity in the mock-treated cells divided by the luciferase activity in the tetracycline-treated cells. (Upper) Relative fold inhibition was calculated as the fold inhibition in the presence of empty vector divided by the fold inhibition in the presence of the p68 or p72 proteins. The relative fold inhibition data are means + SE of three independent experiments, and the Western blotting result (Lower) is representative of three independent experiments. The asterisks denote to P < 0.05. F.I., fold inhibition; EV, empty vector; 72N, N-terminal domain of p72; 72C, C-terminal domain of p72; p72KR, p72-K142R mutant.
Depletion of p72 Reduced ZAP's Activity.
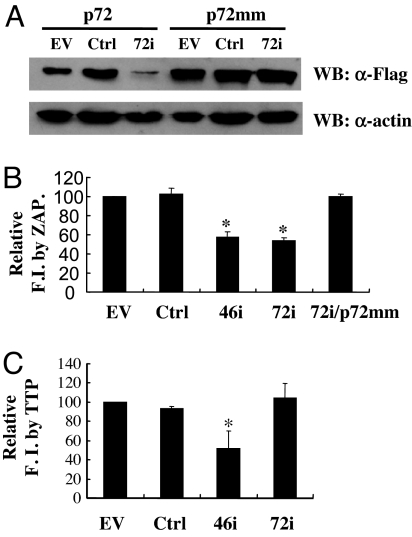
To further analyze whether p72 is required for ZAP's activity, p72 was depleted by the RNAi method. The plasmid expressing the shRNA directed against p72 (72i) was first tested for its ability to down-regulate the expression of p72 by cotransfection into 293TRex cells with the construct expressing the Flag-tagged p72. Compared with empty vector or control shRNA, 72i reduced the expression level of Flag-tagged p72 significantly (Fig. 4A). To confirm the specificity of the effect of 72i, a rescue p72-expressing plasmid (p72 mm) was constructed, wherein silent mutations were introduced in the coding sequence of p72 such that it could not be depleted by 72i. As expected, 72i had little effect on the expression level of p72 mm (Fig. 4A). To test the effect of the depletion of p72 on the activity of ZAP, 293TRex-ZAP cells were cotransfected with pMLV-luc reporter and 72i and assayed for inhibition of the reporter. The shRNA directed against a key component of the RNA exosome, hRrp46 (46i), which has been shown to reduce ZAP's activity (6), was used as a positive control. Indeed, 72i reduced ZAP's activity to a similar level as 46i, and the inhibitory effect of 72i could be rescued by the overexpression of p72 mm (Fig. 4B). These results further indicated that p72 is required for ZAP's activity.
Fig. 4.
Depletion of p72 by RNAi reduced ZAP's activity. (A) The plasmid expressing Flag-tagged p72 or the p72 rescue plasmid (p72 mm) was cotransfected into 293TRex cells with the plasmid expressing control shRNA (Ctrl) or the shRNA directed against p72 (72i). The expression levels of the p72 proteins were judged by Western blotting using anti-Flag and anti-β-actin antibodies. (B) The pMLV-luc reporter was cotransfected with the plasmid expressing the indicated shRNA or 72i plus p72 mm into 293TRex-ZAP cells. Immediately after transfection, tetracycline was added to induce ZAP expression. Forty-eight hours after transfection, the luciferase activities were measured. Fold inhibition was calculated as in Fig. 3. Relative fold inhibition was calculated as the fold inhibition in the absence of the shRNA (empty vector) divided by the fold inhibition in the presence of the indicated RNAi. The data are means + SE of four independent experiments. The asterisks denote P < 0.05. (C) The ARE-containing reporter, pGL3-ARE-Luc, was cotransfected with the plasmid expressing the indicated shRNA into 293TRex-TTP cells. Immediately after transfection, tetracycline was added to induce TTP expression. Forty-eight hours after transfection, the luciferase activities were measured. Fold inhibition was calculated as the luciferase activity in the mock-treated cells divided by the luciferase activity in the tetracycline-treated cells. Relative fold inhibition was calculated as the fold inhibition in the absence of the shRNA (empty vector) divided by the fold inhibition in the presence of the indicated shRNA. The data are means + SE of three independent experiments. The asterisks denote to P < 0.05. EV, empty vector; 46i, shRNA-directed against the exosome component hRrp46; FI, fold inhibition.
In an attempt to test whether p72 also is involved in other RNA-degradation processes, 72i was analyzed for its effect on the degradation of AU-rich element (ARE)-containing mRNA by TTP, which binds to ARE and recruits the RNA exosome to degrade the ARE-containing RNA (19). Although 46i effectively reduced TTP's activity, 72i had little effect (Fig. 4C), suggesting that the p72 RNA helicase is not involved in TTP-mediated degradation of ARE-containing mRNA.
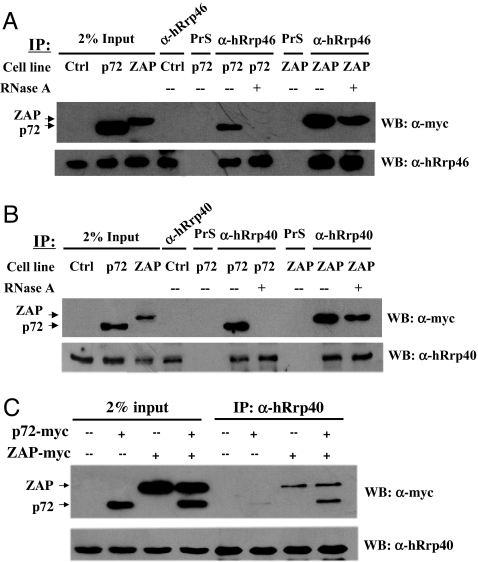
p72 Interacts with the Exosome in an RNA-Independent Manner Only in the Presence of ZAP.
To better understand the mechanism underlying the involvement of p72 in ZAP's function, coimmunoprecipitation assays were performed to analyze whether p72 interacted with the exosome. The myc-tagged p72 was expressed in 293TRex cells, and the myc-tagged ZAP, which has been demonstrated to interact with the exosome in an RNA-independent manner (6), was used as a positive control. Although immunoprecipitation of the endogenous exosome using either the polyclonal antibody against the exosome component hRrp46p (Fig. 5A) or hRrp40p (Fig. 5B) coprecipitated ZAP in the absence or presence of RNase A, immunoprecipitation of the exosome coprecipitated p72 only in the absence of RNase A (Fig. 5 A and B). These results indicated that p72 does not directly interact with the exosome in an RNA-independent manner. We then analyzed whether ZAP could mediate the interaction between p72 and the exosome. The myc-tagged p72 and ZAP were expressed alone or coexpressed in HEK293T cells. Consistent with the above results, in the presence of RNase, immunoprecipitation of the endogenous exosome coprecipitated only ZAP but not p72 (Fig. 5C). However, when p72 was coexpressed with ZAP, immunoprecipitation of the endogenous exosome coprecipitated p72 in an RNA-independent manner (Fig. 5C). These results suggested that ZAP could bind to both p72 and the exosome to form a tertiary complex.
Fig. 5.
The p72 RNA helicase does not directly interact with the exosome. (A and B) The 293TRex cells expressing myc-tagged p72 or myc-tagged ZAP were treated with tetracycline to induce ZAP or p72 expression and lysed in the lysis buffer in the presence (+) or absence (−) of 50 μg/ml RNase A. The proteins were immunoprecipitated with anti-hRrp46p (A) or anti-hRrp40p (B) antibody and Western blotted with the indicated antibodies. Input, total cell lysate; Ctrl, 293TRex cells; p72, 293TRex cells expressing p72; ZAP, 293TRex cells expressing ZAP. (C) The HEK293T cells expressing myc-tagged p72, myc-tagged ZAP, or both were lysed in the lysis buffer in the presence of 50 μg/ml RNase A. The proteins were immunoprecipitated with anti-hRrp40p antibody and Western blotted with the anti-myc antibody (Upper) or with the anti-hRrp40p antibody (Lower).
Discussion
ZAP inhibits the replication of MLV and SINV by degrading the viral mRNA in the cytoplasm (1, 3, 5, 6). mRNA decay is a highly organized process, in which multiple protein complexes are involved (20). In eukaryotic cells, mRNA decay begins with shortening the poly(A) tail by deadenylases. The deadenylated RNA is either degraded by the 3′–5′ RNA-processing exosome or decapped by decapping enzymes and then degraded by 5′–3′ exonucleases. In mammalian cells, the 3′–5′ pathway is the major pathway (21, 22). A variety of cis-acting motifs can promote mRNA degradation (23–27). ZAP directly binds to one such element, the ZRE, and recruits the exosome to degrade the ZRE-containing mRNA (5, 6).
The core of the human exosome is composed of nine subunits, including six RNase PH domain-containing proteins and three RNA-binding proteins. The reconstituted human exosome using these nine proteins displayed processive hydrolytic activity on AU-rich or generic RNAs in vitro (28). When structured RNAs containing a stem loop were used as substrates, the reconstituted exosome could degrade only the extended sequence, but not the stem-loop structure (28). These results strongly suggest that RNA helicase(s) are needed to disrupt the secondary structure for efficient RNA degradation by the exosome. It is not yet clear how many and by what mechanisms RNA helicases are used in exosome-mediated RNA degradation. It is conceivable that RNA helicases could be constitutively associated with the exosome as cofactors, temporarily recruited to the exosome to facilitate RNA degradation, or both. Some RNA helicases, such as RHAU and KIAA0052, have been reported to associate with the exosome (19, 29). RHAU was reported to be involved in the degradation of ARE-containing mRNAs mediated by HuR and NAFR1 (29). Although the yeast homologue of KIAA0052, Mtr4p, in the TRAMP complex was reported to enhance the activity of nuclear exosome (30), the mechanism by which mammalian KIAA0052 participates in exosome-mediated RNA degradation has not been documented.
In this article, we identified the p72 RNA helicase as a ZAP-interacting protein and provide evidence indicating that p72 is required for the optimal function of ZAP. p72 alone did not directly interact with the exosome (Fig. 5 A and B), suggesting that p72 is not a constitutive cofactor of the exosome. However, p72 can be recruited by ZAP to form a complex with the exosome (Fig. 5C). Considering that the minimum length of ZRE so far characterized is ≈500 nt (5), it is tempting to propose a working model for ZAP-mediated RNA degradation in which ZAP binds to the target RNA, employs p72 to unwind the RNA, and recruits the exosome to degrade the RNA. Depletion of p72 by the RNAi method resulted in ≈50% reduction in ZAP's activity (Fig. 4), and overexpression of the C-terminal domain of p72 reduced ZAP's activity by ≈70% (Fig. 3). Such incomplete inhibition to ZAP's activity could be accounted for by the possibility that the inhibition to the endogenous p72 by the RNAi method or the overexpression of 72C was not complete. Alternatively, the incomplete inhibition to ZAP's activity suggested that p72-independent ZAP-mediated RNA-degradation mechanisms might exist. Our preliminary results suggest that other RNA helicases also may be involved in ZAP-mediated RNA degradation, and one common feature of these RNA helicases seems to be that they directly interact with ZAP in an RNA-independent manner (Z. Wang and G.G., unpublished data). Despite its high similarity to p72, the RNA helicase p68 did not directly bind to ZAP and seemed not to be involved in ZAP-mediated RNA degradation (Figs. 1C and 3). These results suggest that ZAP recruits RNA helicases through direct interactions as cofactors for its optimal function. The functional relationship of these RNA helicases in ZAP-mediated RNA degradation is being investigated.
p72 also may be involved in other steps in the process of ZAP-mediated mRNA degradation, such as decapping and deadenylation. It would be interesting to analyze whether p72 interacts with other components of the RNA-degradation machinery.
In this study, we also analyzed the function of p72 in TTP-mediated degradation of the ARE-containing mRNA. Weak interaction between p72 and TTP was detected only in the absence of RNase, and the depletion of p72 by RNAi had no effect on TTP's activity, suggesting that p72 may not be a universal factor involved in RNA degradation. Further work will determine whether p72 is involved in other RNA-degradation pathways.
Materials and Methods
Plasmids.
The plasmids pcDNA4TO/myc-ZAP, pcDNA4TO/myc-NZAP, pcDNA4TO/myc-CZAP, and pcDNA4TO/myc-TTP, which expressed myc-tagged full-length ZAP, NZAP254 (amino acids 1–254), CZAP236 (amino acids 236–776), and TTP, respectively, have been described previously (5, 6).
pCMV-HA-Flag-p72 expresses Flag-tagged p72. The coding sequence of p72 was amplified from a human fetal liver cDNA library by using forward primer p72-FP bearing an EcoRI site and reverse primer p72-RP bearing a SalI site and cloned into the expression vector pCMV-HA-Flag (6) using these two sites: p72-FP, 5′-TATGAATTCATGCGCGGAGGAGGCTTTG-3′; and p72-RP, 5′-TATGTCGACTCATTTACGTGAAGGAGG-3′. pCMV-HA-Flag-p68 expresses Flag-tagged p68. The coding sequence of p68 was amplified from a human fetal liver cDNA library by using forward primer p68-FP bearing a SalI site and reverse primer p68-RP bearing a BglII site and cloned into the expression vector pCMV-HA-Flag by using these two sites: p68-FP, ATATGTCGACCATGTCGGGTTATTCGAGTG; and p68-RP, GTAAGATCTTTATTGGGAATATCCTGTTG. The p72-deletion mutants were generated by cloning PCR-derived p72 fragments into pGEX-5x-3 (Amersham Pharmacia). A BamHI site was built in the forward primers, and an XhoI site was built in the reverse primers. The following primers are listed with the restriction sites italicized: p72 forward primer, 5′-TATAGGATCCCCATGCGCGGAGGAGGCTTTGGGG-3′; p72 reverse primer, 5′-TATCTCGAGTCATTTACGTGAAGGAGG-3′; p72N reverse primer, 5′-ATACTCGAGTCAGGGACCTTTAGGAGTACC-3′; p72M forward primer, 5′-TATAGGATCCCCCAGATTCGAGACTTGGAAAG-3′; p72M reverse primer, 5′-ATACTCGAGTCAGCCTCCTCTGTGGTCCAC-3′; and p72C reverse primer, 5′-TATAGGATCCCCCTTGTGGACCACAGAGGAG-3′.
The fragments encoding the p72-deletion mutants were excised from the plasmids listed above by using BamHI and XhoI and cloned into pCMV-HA-Flag digested with BglII and XhoI to construct plasmids for expressing the Flag-tagged deletion mutants in mammalian cells.
The p72 mutant p72-K142R was generated by cloning EcoRI-BamHI and BamHI-SalI PCR-derived fragments into pCMV-HA-Flag by using EcoRI and SalI sites. The EcoRI-BamHI fragment was generated by using forward primer p72m-FP and reverse primer p72KR-RP bearing silent mutations to create a BamHI site. The BamHI-SalI fragment was generated by using reverse primer p72m-RP and forward primer p72KR-FP, in which the mutation codon and a BamHI site were built in. The following primers are listed with the restriction sites italicized and the mutation codon in bold type: p72m-FP, 5′-AGCGCTAGCTGACAGAATGCTTG-3′; p72KR-RP, 5′-AGAGGATCCAGTCTGAGCAATGCCCACC-3′; p72KR-FP, 5′-AGAGGATCCGGGCGAACGTTGGCGTATCTCCTGCC-3′; and p72m-RP, 5′-TCAGCTAGCGCCAATACAAGGTAAGTAC-3′.
To generate the p72-expressing construct that cannot be down-regulated by 72i, silent mutations were introduced into pCMV-HA-Flag-p72. The PCR fragments generated by using primers p72FP/p72mmRP and p72mmFP/p72RP were mixed and amplified by using PCR primers p72FP and p72RP. The resulting fragment was cloned into pCMV-HA-Flag: p72 mm FP, 5′-GGACGTGCTTATGGACCAGCACTTTACAGAACCAACTCCAATTC-3′; and p72 mm RP, 5′-CTGGTCCATAAGCACGTCCATTACATATTGTGGGAAGTTAGC-3′.
The plasmid-expressing myc-tagged ZAP with MBP fused at the N terminus of ZAP in E. coli has been described previously (6).
pBabe-Superp72 RNAi expresses shRNA directed against p72. Oligonucleotides p72-RNAi FP and p72-RNAi RP were annealed and cloned into pBabe-super (6) by using the BglII and HindIII sites to generate pBabe-Super-p72 RNAi. pBabe-Supercontrol RNAi were constructed by using the same strategy. The sequences of the primers were: p72-RNAi-FP, 5′-GATCCCCTGGATGTGTTGATGGATCATTCAAGAGATGATCCATCAACACATCCATTTTTGGAAA-3′; p72-RNAi-RP, 5′-AGCTTTTCCAAAAATGGATGTGTTGATGGATCATCTCTTGAATGATCCATCAACACATCCAGGG-3′; control RNAi-FP, 5′-GATCCCCGAGCACTCTGAACTACCTGTTCAAGAGACAGGTAGTTCAGAGTGCTCTTTTTGGAAA-3′; and control RNAi-RP: 5′-AGCTTTTCCAAAAAGAGCACTCTGAACTACCTGTCTCTTGAACAGGTAGTTCAGAGTGCTCGGG-3′.
Cell Culture.
All of the cells were maintained in DMEM supplemented with 10% FBS. Transfection was performed by using Fugene 6 (Roche Diagnostics) following the manufacturer's instructions. 293TRex and 293TRex-ZAP cell lines have been described previously (5). To establish the 293TRex-TTP cell line, 293TRex cells were stably transfected with pcDNA4/TO/myc-TTP and selected for Zeocin resistance. Individual clones were picked, expanded, and tested for tetracycline-inducible expression of TTP by Western blotting. The method to assay ZAP's activity has been described previously (5). To analyze the effects of the RNAi or p72 mutants on ZAP's activity, 293TRex-ZAP cells were transfected with the pMLV-Luc reporter pRL-TK (for normalizing transfection efficiency) and the effector-expressing plasmid. Immediately after transfection, the cells were mock-treated or treated with tetracycline to induce ZAP expression. Forty-eight hours after transfection, luciferase activities were measured and normalized by dividing the firefly luciferase activity with the Renilla luciferase activity. Fold inhibition by ZAP was calculated as the normalized luciferase activity in the mock-treated cells divided by the normalized luciferase activity in the tetracycline-treated cells. To assay the effects of RNAi on the activity of TTP, 293TRex-TTP cells were cotransfected with a type II ARE-containing reporter, pGL3-ARE-Luc (5), pRL-TK, and the shRNA-expressing plasmid. Immediately after transfection, the cells were treated with tetracycline to induce TTP expression. Forty-eight hours after transfection, the luciferase activities were measured. Fold inhibition by TTP was calculated as the normalized luciferase activity in the mock-treated cells divided by the normalized luciferase activity in the tetracycline-treated cells. The statistical significance of the data is analyzed with an SPSS program.
Identification of ZAP-Interacting Proteins.
293TRex-ZAP cells were treated with tetracycline at a final concentration of 1 μg/ml to induce ZAP expression. Thirty-six hours after induction, the cells were lysed with lysis buffer B [30 mM Hepes (pH 7.6), 100 mM NaCl, 0.5% Nonidet P-40, and protease inhibitors mixture]. The lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4°C in a microcentrifuge (Sorvall Fresco), treated with 50 μg/ml RNase A for 15 min at 37°C, and incubated with protein G plus agarose (Santa Cruz Biotechnology) and 9E10 anti-myc antibody for 2 h at 4°C to precipitate ZAP and associated proteins. After washing three times with lysis buffer B, the precipitates were resuspended in SDS-loading buffer and resolved on SDS/10% PAGE. The proteins were visualized by Coomassie blue staining. The specific bands from the ZAP-expressing cells were excised from the gel and digested with trypsin. The resulting peptides were analyzed by MALDI-TOF MS. The acquired MS data were analyzed with a human nucleotide/protein database by using a MASCOT database search tool for peptide identification. The protein was identified when multiple peptides corresponding to the ORF of a protein were identified.
Antibodies.
The mouse monoclonal antibody 9E10 (Santa Cruz Biotechnology) was used to immunoprecipitate or detect proteins with the myc epitope, and the mouse monoclonal antibody M2 (Sigma–Aldrich) was used to immunoprecipitate or detect proteins with the Flag epitope. Bacterially expressed hRrp46p or hRrp40p was used to immunize rabbits to generate polyclonal antibody against hRrp46p or hRrp40p, respectively. The antibodies were affinity-purified by using the cognate proteins.
Coimmunoprecipitation.
Cells were lysed in lysis buffer B [30 mM Hepes (pH 7.6), 100 mM NaCl, 0.5% Nonidet P-40, and protease inhibitors mixture] on ice for 10 min, and the lysates were clarified by centrifugation at 4°C for 10 min at 13,000 rpm. The supernatant was mixed with protein G plus agarose (Santa Cruz Biotechnology) and the antibody and incubated at 4°C for 2 h. The resins were then washed three times with lysis buffer B, and the bound proteins were detected by Western blotting.
Acknowledgments.
We thank Yiping Zhu, Xinlu Wang, and Jing Ma for technical support and Dr. Fuquan Yang for help with the MALDI-TOF analyses. This work was supported in part by National Science Foundation Grants 30470092 and 30530020 (to G.G.) and Ministry of Science and Technology 973 Program 2006CB504302 of China.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 2.Müller S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2006;10:1601–1606. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryman KD, et al. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J Virol. 2005;79:1487–1499. doi: 10.1128/JVI.79.3.1487-1499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X, et al. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buttner K, Wenig K, Hopfner KP. The exosome: A macromolecular cage for controlled RNA degradation. Mol Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- 8.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Rocak S, Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 10.Lamm GM, Nicol SM, Fuller-Pace FV, Lamond AI. p72: A human nuclear DEAD box protein highly related to p68. Nucleic Acids Res. 1996;24:3739–3747. doi: 10.1093/nar/24.19.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossler OG, Straka A, Stahl H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001;29:2088–2096. doi: 10.1093/nar/29.10.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guil S, et al. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23:2927–2941. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honig A, et al. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol Cell Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C, et al. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 17.Bond AT, Mangus DA, He F, Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 19.Chen CY, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 20.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 21.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 22.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 23.D'Orso I, Frasch AC. Functionally different AU- and G-rich cis-elements confer developmentally regulated mRNA stability in Trypanosoma cruzi by interaction with specific RNA-binding proteins. J Biol Chem. 2001;276:15783–15793. doi: 10.1074/jbc.M010959200. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, et al. CA repeats in the 3′-untranslated region of bcl-2 mRNA mediate constitutive decay of bcl-2 mRNA. J Biol Chem. 2004;279:42758–42764. doi: 10.1074/jbc.M407357200. [DOI] [PubMed] [Google Scholar]
- 25.Paste M, Huez G, Kruys V. Deadenylation of interferon-beta mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur J Biochem. 2003;270:1590–1597. doi: 10.1046/j.1432-1033.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- 26.Stoecklin G, Lu M, Rattenbacher B, Moroni C. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol Cell Biol. 2003;23:3506–3515. doi: 10.1128/MCB.23.10.3506-3515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Tran H, et al. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 30.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]