Abstract
Unraveling of the unified networking characteristics of complex networking phenomena is of great interest yet a formidable task. There is currently no simple strategy with a rigorous framework. Using an analogy to the exact algebraic property for a transition matrix of a master equation in statistical physics, we propose a method based on a Laplacian matrix for the discovery and prediction of new classes in the unsupervised complex networking phenomena where the class of each sample is completely unknown. Using this proposed Laplacian approach, we can simultaneously discover different classes and determine the identity of each class. Through an illustrative test of the Laplacian approach applied to real datasets of gene expression profiles, leukemia data [Golub TR, et al. (1999) Science 286:531–537], and lymphoma data [Alizadeh AA, et al. (2000) Nature 403:503–511], we demonstrate that this approach is accurate and robust with a mathematical and physical realization. It offers a general framework for characterizing any kind of complex networking phenomenon in broad areas irrespective of whether they are supervised or unsupervised.
Keywords: complex networking phenomenon, leukemia, lymphoma
Uncovering the common essence of complex networking phenomena occurring in the broad range of nature, for example social networks (1, 2), biological networks, metabolic networks (3–7), the Internet, disordered networks (8, 9), the stock market (10), etc., is an important endeavor for grasping the unified description for various networking phenomena. Although developing such a unified mathematical or numerical framework is nontrivial and highly challenging, graph theory and its application to the network facilitated the investigation of the underlying networking characteristics of various complex networking phenomena (11). Among many interesting phenomena, of particular importance to our life is the application of complex network analyses for the treatment of cancer (3, 4). The number of studies involving gene expression profiles based on DNA microarray technology in biomedical laboratories is increasing (12–15). The identification of new tumor classes using gene expression profiles is very important to the successful and efficient treatment of cancer, especially when the number of different classes of tumors is unknown. To achieve this goal, the clustering method is the usual statistical tool in the analysis of a complex network (16–18). Cluster analysis partitions a set of objects into groups or clusters where each of the clustered objects is as similar as possible sharing common characters. Among many clustering methods, the K-means clustering (12) and self-organizing maps (19) are widely used. However, clustering conditions such as the number of different classes K and the number of grids are unknown a priori and should be predetermined or chosen artificially. Therefore, arbitrary specification of clustering conditions in the beginning of the analysis of a complex network may easily mislead researchers in the essential clustering nature of the underlying complexity and moreover hamper the correct interpretation of the associated complex phenomenon. This is a serious obstacle for the exact analysis and success of new class discovery and prediction.
To build up a logical and general strategy for clustering of complex networking phenomena based on the rigorous mathematical and physical model, we propose one method, not by an ad hoc assumption or an iterative calculation (13, 14) from experience, but by using the simple and exact algebraic properties of a Laplacian matrix (11, 20) for correlated complex networking phenomena represented by complex networks. Our method eliminates fundamental difficulties found in the existing clustering methods by both estimating the number of different classes in a data-driven manner and classifying the class of each sample simultaneously. The eigenvector for the nonzero smallest eigenvalue of a Laplacian matrix is called the Fiedler vector (21, 22). This vector has been used for several graph manipulations such as partitioning (23, 24), linear labeling (25), and envelope minimization (11, 20, 25, 26). In this study, after identifying an exact analogy of a Laplacian matrix with the transition matrix of a master equation in nonequilibrium statistical physics, we propose a Laplacian clustering method based on the unique character of the Fiedler vector for the discovery and prediction of new classes in complex networking phenomena. The idea behind the proposed method is quite simple, exact, and robust so that it can be broadly used, in the same spirit for any kinds of complex networking phenomena in general, irrespective of whether they are supervised or unsupervised. We illustrate the practical application of this Laplacian clustering method to gene expression profiles of tumors.
Results and Discussion
The Laplacian Matrix and Motivation for Clustering.
A graph G = G(V, E) consists of a set of vertices V and a set of edges E. Two vertices vi and vj of a graph G are said to be adjacent if there exists an edge connecting vi and vj with the nonzero weight eij. The adjacency matrix A = (aij) of a graph G with n vertices is defined as a n × n symmetric matrix with components aij =eij or aij = 0 if there is no connecting edge, where the diagonal elements ajj are equal to zero for all j = 1, 2, …, n. The Laplacian matrix of a graph G is defined as L = D − A, where D, called the degree matrix, is a diagonal matrix with the jth diagonal element djj = Σi=1naij (11, 18, 20).
We assume that there are n samples x1, …, xn, where xi is a vector of g-dimension. For example, in the gene expression matrix, xi denotes an expression of g genes for the ith sample. To construct the Laplacian matrix for clustering, we first define the adjacency matrix from some similarity measures between x1, …, xn. For example, for two vectors x and y, the Euclidean distance [{Σi=1g(xi − yi)2}1/2] and Manhattan distance (Σi=1g|xi − yi|) are often used among others (16–18). In general the component of the adjacency matrix aij should reveal the closeness or degree of connectivity between xi and xj. The goal is to partition n samples into an arbitrary number of groups such that samples belonging to the same group have higher correlation or stronger connectivity sharing the common characteristics than those in the other group.
Let z = (z1, …, zn)T be an unknown argument, where the T denotes the transpose of a vector or a matrix and zi contains information on the group where the ith sample belongs, i.e., if zi = zj, then xi and xj belong to the same group. Therefore, the classification of n samples corresponds to obtaining the solution z. This goal can be achieved by minimizing the weighted sum of squares
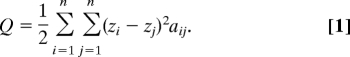 |
After we set S = Σi=1nzi and R = Σi=1nzi2, we impose two constraints R = 1 to avoid the trivial solution zi = 0 for all i and S = 0 to keep the invariance of the minimum in Q. We take advantage of the relation Q = zTLz. We use Lagrangian multiplier methods to minimize Q subject to R = 1 with a Lagrangian multiplier λ and then obtain the eigenvalue equation Lz = λz. This equation yields a nontrivial solution z if and only if λ is an eigenvalue of L, and z is the corresponding eigenvector. Now we have zTLz = λ by multiplying zT on both sides of Lz = λz. Therefore, the nonzero smallest eigenvalue and its associated eigenvector (Fiedler vector) yields the optimal solution (11, 20–24).
Analogy of the Laplacian Matrix with a Transition Matrix in a Master Equation.
Because the Laplacian matrix is constructed such that the sum of elements in each raw vector is always zero, it provides an exact analogy with a transition matrix of a master equation in nonequilibrium statistical physics (27, 28). Let us consider a particle moving on a network with n vertices. The motion of a particle on this network could be described as a hopping between adjacent vertices. Assuming that the hopping probability of this particle from a site j to i is given by mij, where M = (mij) is a transition matrix, and that p(t) = (p1, p2, …, pn) is a probability vector of finding a particle on the vertices (1, 2, …, n) at a time t, then the time evolution of the probability vector p(t) satisfies a master equation dp(t)/dt = −Mp(t) with the detailed balance condition, where mjj = −Σi≠jnmij such that Σi=1nmij = 0 for each row j. This master equation describes how a system that starts from a nonequilibrium state evolves into an equilibrium state at the asymptotic time limit. The time evolution (relaxation rate) toward an equilibrium stationary state is governed by the eigenvalues of a transition matrix M and the relaxation mode is determined by the eigenvectors of M. The exact algebraic property of the solution of a master equation (27, 28), in particular, is that (i) there is always one zero eigenvalue. Its eigenvector describes the equilibrium (stationary) probability of finding a particle on the vertices of a network, (ii) the sum of the eigenvector elements for each of the nonzero eigenvalues is always zero, which governs the relaxation mode of a probability toward that of an equilibrium one. Most importantly, the nonzero smallest eigenvalue and its eigenvector dictate the dominant mode of time evolution (relaxation) of p(t) to an equilibrium one with the longest relaxation time. Bearing in mind the exact properties of eigenvalues and eigenvectors of a master equation, it is important to recognize that the Laplacian matrix L in our setting for a cluster analysis and a transition matrix M in a master equation are constructed in the same way. Therefore, the Fiedler vector of the Laplacian matrix L shares the same exact property of the eigenvector for the nonzero smallest eigenvalue of a transition matrix M (21, 22, 27, 28).
A Strategy for Cluster Discovery and Prediction.
Here, we briefly illustrate the flow of the clustering strategy using a Laplacian matrix constructed from gene expression profiles. We first construct an n × n adjacency matrix A = (aij) from the g × n gene expression matrix X = (xji), where j = 1, …, g; i = 1, …, n. xji is the gene expression level of the jth gene for the ith sample. After we eliminate noises in the raw data aij by the thresholding procedure, we define a Laplacian matrix L. Then, n elements (samples) of a Fiedler vector of L are readily grouped into K groups where K is the number of distinct clusters. Few essential genes playing a significant role in clustering are selected based on the F-test following Dudoit et al. (29) and Lee and Lee (30). Based on these essential genes and n samples, we reconstruct a new Laplacian matrix and estimate the number K where m out of n samples are classified into their corresponding clusters by the unique character of the Fiedler vector for a new Laplacian matrix. Now we predict the classes of n − m unclassified samples in such a way that each unclassified sample has the highest correlation with those in the already classified class. (For the details of the clustering strategy, see Materials and Methods.)
Illustrative Examples of Cluster Discovery and Prediction.
A. Leukemia data.
Golub et al. (3) suggested gene expression monitoring for the classification of two types of acute leukemia; namely, acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). This dataset includes 6,817 genes for the 38 training set (patients) (27 ALL, 11 AML) and the 34 test set. The raw data are available at www.genome.wi.mit.edu/MPR. Golub et al. (3) proposed “a weighted gene voting scheme,” which turned out to be a variant of quadratic discriminant analysis. They applied it to training data based on 50 informative genes. The number of correct decisions was 36 genes of 38 training samples and 29 genes of 34 test samples. ALL can be further divided into finer subclasses such as B cell and T cell ALLs. Then, this problem can be also regarded as a three-class (ALLB/ALLT/AML) problem. In contrast, Lee and Lee (30) argued that gene expression patterns in ALLB are much closer to those in AML than ALLT by inspecting 20 top ranked genes. Therefore, the leukemia data can be classified into three different types.
 |
The raw microarray data contain a lot of abnormal samples or outliers, so it is conventional to perform data preprocessing before we make a further downstream analysis. Here, we perform the following data preprocessing as done in Dudoit et al. (29): i.e., (i) thresholding, with a floor of 100 and a ceiling of 16,000; (ii) filtering, with an exclusion of genes with max/min ≤5 or max–min ≤500, where max and min refer, respectively, to the maximum and minimum intensities for a particular gene across the samples; and (iii) a base 10 logarithmic transformation. This preprocessing of data resulted in 3,571 genes of 6,817 genes. Herein, we deal with a 3,571 × 72 gene expression matrix.
Clustering in an unsupervised set-up.
Here, we assume that the leukemia data are unsupervised—i.e., the number of different classes of tumor and the class of each sample are assumed to be completely unknown. We apply our proposed method to 72 samples. We demonstrate how well our Laplacian clustering method discovers and predicts the tumor class of each sample.
First, we construct a 3,571 × 72 gene expression matrix X with normalized elements and get a 72 × 72 adjacency matrix A = XTT − I. The criteria of WCC and ANC, defined in Materials and Methods, give the optimal value of δ such that δ = 0.800 for K = 2 and δ = 0.818 for K = 3. We note that the δ = 0.800 for the K = 2 case corresponds to the Type B classification and the δ = 0.818 for the K = 3 case corresponds to the Type C classification, respectively. The elements of each class are the same as those of Golub et al. (3) except for a couple of misclassification. Here, the number of selected essential genes is 50. We also tried 100 and 200 essential genes, and they gave similar results (δ = 0.790–0.802 for K = 2 and δ = 0.812–0.820 for K = 3, respectively). For the K = 2 case (Type B) only two samples (1 and 2) are misclassified of 72 samples, and for K = 3 case (Type C) only two samples (1 and 34) are misclassified. Clustering results based on our proposed method are as good as other clustering methods such as K-means or SOM where the number of different classes had to be predetermined for a reasonable cluster analysis. One thing to note is that the first sample is misclassified in both cases. This sample is also misclassified in the approach of Golub et al. (3). This patient seems to be either an outlier or incorrectly reported so that further examination or follow-up on that patient is required. Fig. 1 shows heat maps for the adjacency matrix for K = 2 (Type B) and K = 3 (Type C), respectively, based on 100 essential genes. We clearly observe two groups in Fig. 1a and three groups in Fig. 1b, whose detailed classification in terms of a connecting network is also shown in Fig. 2.
Fig. 1.
Heat maps for the adjacency matrix for K = 2 (Type B) and k = 3 (Type C), respectively, based on 100 essential genes in the leukemia data.
Fig. 2.
Connection networks for samples in the leukemia data. a and b correspond to Type B and Type C, respectively.
Classification in a supervised set-up.
Here, we assume that the leukemia data are supervised—i.e., the class of each sample in the training data is known. Using the information in the training dataset, we predict the class of each sample in the test dataset. There are no widely accepted guidelines for choosing the relative sizes of the training sets and test sets. We choose a 2:1 cross-validation (CV) scheme (one-third of the datasets are assigned to the test sets) as done by Dudoit et al. (29). Specifically, we choose 24 samples numbered by 7–9, 27–34, and 60–72 of 72 samples. After applying the proposed method to the 48 training samples, we obtain classification results for the 24 test samples in three different types. For better validation we perform LOOCV (leave-one-out cross-validation), where only one sample is used for the test dataset and others are used for the training dataset. Both results are summarized in Table 1. The classification results are very good. The number of selected genes does not seriously affect classification results.
Table 1.
Misclassification results for two kinds of cross-validation (2:1 CV and LOOCV) in leukemia data
| Type of classification | No. of selected genes | 2:1 CV (24 samples) | LOOCV (72 samples) |
|---|---|---|---|
| Type A | 50 | 1/24 | 1/72 |
| 100 | 1/24 | 1/72 | |
| 200 | 3/24 | 2/72 | |
| Type B | 50 | 1/24 | 1/72 |
| 100 | 1/24 | 1/72 | |
| 200 | 1/24 | 1/72 | |
| Type C | 50 | 3/24 | 1/72 |
| 100 | 2/24 | 2/72 | |
| 200 | 2/24 | 1/72 |
For the type A using 50 selected genes, 2:1 CV (LOOCV) gives 1 (1) misclassified samples out of 24 (72) samples.
B. Lymphoma data.
The lymphoma dataset (4) consists of gene expression profiles in the three most prevalent adult lymphoid malignancies: diffuse large B cell lymphoma (DLBCL), follicular lymphoma (FL), and B cell chronic lymphocytic leukemia (B-CLL). The original gene expression matrix consists of p = 4,682 genes and n = 81 samples. Here, we take n = 62 samples: 42 samples of DLBCL, 9 samples of FL, and 11 samples of B-CLL. For convenience, we label each sample in each class as DLBCL (1–42), FL (43–51), and B-CLL (52- 62).
Clustering in an unsupervised set-up.
Assume that the lymphoma data are unsupervised—i.e., we do not know the number of different classes and the class of each sample a priori. By using the same steps applied to the leukemia data, we obtained two types of classification: (i) two classes DLBCL and FL/B-CLL (samples 1–42 and 43–62) with one misclassified sample (sample 42) and (ii) three classes DLBCL, FL, and B-CLL (samples 1–42, 43–51, and 52–62) with three misclassified samples (samples 1, 41, and 42). Hence, the first type of classification does not distinguish samples in FL and B-CLL classes. To see whether samples in the FL and B-CLL are really nondistinguishable, we applied our Laplacian matrix method to 20 samples in FL and B-CLL (43–62). We obtained two classes (43–51 and 52–62) without any misclassified samples. Therefore, we may conclude that the lymphoma data consist primarily of two classes (DLBCL and FL/B-CLL) and that FL and B-CLL are secondary classes—i.e., samples in FL and B-CLL are closer to each other compared with samples in DLBCL. Heat maps for the adjacency matrix and connection networks are shown in supporting information (SI) Figs. 3–6.
Classification in a supervised set-up.
When we apply our proposed method to the lymphoma data with a supervised set-up, it gives two classes: class 1 (1–42) and class 2 (43–62), with no misclassification even though the given number of classes was three (1–42, 43–51, and 52–62). As in the unsupervised set-up, we applied the same method to the samples in class 2 (43–62). We obtained two classes: 43–51 and 52–62, with one misclassified sample. Conclusively, in the supervised set-up, we obtained the same results and have the same interpretations as those in the unsupervised set-up.
C. Comparison with other methods.
We have noted other clustering methods such as coupled two-way clustering (CTWC) analysis (13, 14) and the stochastic dynamic model (10), which share the similar objective of clustering as ours. CTWC is an iterative clustering process by looking for pairs of a relatively small subset of samples and genes because the “signal” may be masked by the “noise” generated by the uncorrelated data. CTWC can be performed with any clustering method, but the superparamagnetic clustering (SPC) algorithm (31, 32) is especially suitable for the analysis of gene microarray data. The input for SPC is a distance matrix that corresponds to the adjacency matrix A in the Laplacian clustering method. In SPC, the resolution of clustering is governed by a tunable parameter T, called temperature, which has a similar role to a thresholding parameter δ in our method. T is chosen as that above where the cluster is stable. Our thresholding parameter δ is determined by WCC and ANC criteria in a data-driven manner. The CTWC algorithm provides a broad list of stable gene and sample clusters so that an appropriate discovery of a meaningful process or interpretation of identified clusters is chosen. However, the Laplacian algorithm gives an estimate of number of different clusters and the corresponding members of each cluster. Despite some differences between the two methods, they gave similar results from the analysis of leukemia data. For example, both methods successfully detected ALLB and ALLT clusters that can hardly be found by other conventional clustering methods.
Another relevant method is the stochastic model of coupled random walks for stock–stock correlations (10). This model consists of a system of n walks at g different times that corresponds to n samples and g genes, respectively, in microarray data. This method is similar to the Laplacian clustering method in using the information contained in the eigenvector corresponding to the primal eigenvalues of the correlation matrix. However, it is different from the Laplacian clustering method in using all of the information contained in g times, whereas the Laplacian clustering method uses information contained in the essential genes only. In addition, like the CTWC method, the stochastic model does not estimate the number of clusters.
Summary and Conclusion.
We proposed a Laplacian clustering method for the discovery and prediction of new classes in unsupervised complex networking phenomena where the class of each sample is completely unknown. Our method is based on using an analogy of a Laplacian matrix of correlated complex networking phenomena with a transition matrix of a master equation in nonequilibrium statistical physics and applying their exact algebraic properties. Our approach differs fundamentally from previous methods in that it can classify both the unsupervised data and the supervised data without prior knowledge of the clustering condition, and discover different classes and determine the identity of each class simultaneously. When we applied the proposed method to the leukemia data (3), it produced successful results that could hardly be achieved by the existing clustering methods in the unsupervised set-up. Throughout the lymphoma data (4), we also obtained accurate results. Furthermore, we noticed the hierarchical property of our method—i.e., the lymphoma data consist primarily of two classes (DLBCL and FL/B-CLL), and FL and B-CLL are secondary classes.
In conclusion, the Laplacian clustering method is an excellent strategy for class discovery and prediction of unknown classes in unsupervised complex networking phenomena. This method is robust in the sense that the prediction results are not sensitive to the choice of the number of essential genes in the feature selection. It is simple with the mathematical basis and physical realization. It offers a general framework for analyzing any kind of complex networking phenomenon in the broad range of life science, sociology, the Internet, disordered complex networks, etc., irrespective of whether they are supervised or unsupervised.
Materials and Methods
Cluster Analysis by Using the Laplacian Matrix Method.
Recall that xji, j = 1, …, g; i = 1, …, n denotes the gene expression level of the jth gene for the ith sample, and let X = (xji) be the g × n gene expression matrix. We describe the details of a clustering method for the unsupervised data in the following steps.
Step 1. Construction of a Laplacian matrix.
We normalize the gene expression level xji as xji ← (xji − x̄.i)/si, where x̄.i = Σj=1gxji/g and si2 = Σj=1g(xji − x̄.i)2. Let A = XTX − I be the adjacency matrix that is the same as the correlation matrix except all of the diagonal terms aii = 0, i = 1, …, n. To remove noise in the raw data, we modify the adjacency matrix as R = (rij), rij = aijI (|aij| ≥ δ), for some δ > 0, called a thresholding parameter. Finally, we define a Laplacian matrix L = D − R, where D = (dij) is the degree matrix with djj = Σi=1nrij and dij = 0 (i ≠ j).
Step 2. Estimation of the number of classes.
The general from of Fiedler vector for L is
Then n samples are classified into K groups for a given δ; that is, B1 = (b11, …, b1n1), …, BK = (bK1, …, bKnK), where nk ≥ 2, k = 1, …, K, and Bk is the index set of the kth class. Note that m = Σl=1Knl is the number of classified samples and n − m is the number of unclassified samples. We apply two criteria to determine a proper thresholding parameter δ. First, it is appropriate to select δ that maximizes the average of within the class correlations (WCC) defined as all possible correlations between two samples in the same class. Second, we select δ that maximizes the average number of samples (ANC) per class, ANC = m/K. Therefore, considering both WCC and ANC will give a moderate-sized K with relatively large members in each class.
Step 3. Selection of the essential genes.
Even though the gene expression profiles consist of thousands of genes, most of them are uninformative for classification. Therefore, it is highly recommended to select a few essential genes before clustering. Assume that m samples are classified as B1, B2, …, BK based on δ chosen in Step 2. To test whether the mean intensities for each class at the jth gene are the same, we apply the F-test defined as the ratio of the between sum of squares (BSS) and the within sum of squares (WSS), which was used by Dudoit et al. (29) and Lee and Lee (30). Here, we selected 50, 100, and 200 essential genes. The clustering results were not seriously affected by the number of essential genes.
Step 4. Prediction for the unclassified samples.
We repeat Step 1 and Step 2 based on essential genes selected from Step 3, then m of n samples are classified into K classes. Now, we predict the classes of n − m unclassified samples in the following way. Let r̄j(k) = Σi∈Bkrij/nk be the average correlation between an unclassified sample j and classified samples in a class k, then the class of sample j is declared as the class satisfying maxkr̄j(k).
Application to Classification of Supervised Data.
Because the supervised data already has information on the class of each sample, the clustering strategy for the supervised data are the same as the unsupervised case except in determining δ. We minimize the misclassification rate, defined as the proportion of incorrectly predicted samples out of the training dataset to determine δ.
Supplementary Material
Acknowledgments.
We acknowledge the valuable help of E. Moon and W. Yu in the first stage of this work. We thank K. Han (Molecular Cancer Center/KRIBB), J. Bhak (Korea Bioinformatics Center/KRIBB), and S. Kim (Chemistry/Pusan Nat'l Univ.) for their careful reading of and critical feedback on the manuscript. This work was supported by the Korea Science and Engineering Foundation under National Research Laboratory Program M104-333-06J-33310 (M.C. and I.C.) and Grant R14-2003-002-01000-0 (to C.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708598105/DC1.
References
- 1.Amaral LAN, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci USA. 2000;97:1149–1152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liljeros F, Edling CR, Amaral LAN, Stanley HE, Aberg Y. The web of human sexual contacts. Nature. 2001;411:907–908. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- 3.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh AA, et al. Different types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 5.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 6.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 7.Rho K, Jeong H, Kahng B. Identification of lethal cluster of genes in the yeast transcription network. Physica A. 2006;364:557–564. [Google Scholar]
- 8.Braunstein LA, Buldyrev SV, Cohen R, Havlin S, Stanley HE. Optimal paths in disordered complex networks. Phys Rev Lett. 2003;91:168701. doi: 10.1103/PhysRevLett.91.168701. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Lopez E, Havlin S, Stanley HE. Universal behavior of optimal paths in weighted networks with general disorder. Phys Rev Lett. 2006;96 doi: 10.1103/PhysRevLett.96.068702. 068702. [DOI] [PubMed] [Google Scholar]
- 10.Ma WJ, Hu CK, Amritkar RE. Stochastic dynamical model for stock-stock correlations. Phys Rev E. 2004;70 doi: 10.1103/PhysRevE.70.026101. 026101. [DOI] [PubMed] [Google Scholar]
- 11.Mohar B. The Laplacian spectrum of graphs. In: Alavi Y, Chartrand G, Oellermann OR, Schwenk AJ, editors. Graph Theory, Combinatorics and Applications. New York: Wiley; 1991. pp. 871–898. [Google Scholar]
- 12.Eisen MB, Spellman PT, Brown PO, Botstein D. Clustering analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domany E. Cluster analysis of gene expression data. Physica A. 1999;263:158–169. [Google Scholar]
- 14.Getz G, Levine E, Domany E. Coupled two-way clustering of gene microarray data. Proc Natl Acad Sci USA. 2000;97:12079–12084. doi: 10.1073/pnas.210134797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speed T. Statistical Analysis of Gene Expression Microarray Data. New York: Chapman & Hall; 2003. [Google Scholar]
- 16.Everitt B. Cluster Analysis. New York: Halstead; 1990. [Google Scholar]
- 17.Gordon A. Classification. 2nd Ed. London: Chapman & Hall; 1999. [Google Scholar]
- 18.Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- 19.Kohonen T. Self-Organizing Maps. Berlin: Springer; 1997. [Google Scholar]
- 20.Mohar B. Laplace eigenvalues of graphs—A survey. Discrete Math. 1992;109:171–183. [Google Scholar]
- 21.Fiedler M. Algebraic connectivity of graphs. Czech Math J. 1973;23:298–305. [Google Scholar]
- 22.Fiedler M. A property of eigenvectors of nonnegative symmetric matrices and its application to graph theory. Czech Math J. 1975;25:619–633. [Google Scholar]
- 23.Pothen A, Simon HD, Liou KP. Partitioning sparse matrices with eigenvectors of graph. SIAM J Matrix Anal Appl. 1990;11:430–452. [Google Scholar]
- 24.Vishveshwara K, Brinda KV, Kannan J. Protein structure: Insight from graph theory. J Theor Comput Chem. 2002;1:187–211. [Google Scholar]
- 25.Juvan M, Mohar B. Optimal linear labelling and eigenvalues of graphs. Discrete Appl Math. 1992;36:153–168. [Google Scholar]
- 26.Barnard ST, Pothen A, Simon HD. A spectral algorithm for envelope reduction of sparse matrices. Proc Supercomput. 1993;93:493–502. [Google Scholar]
- 27.Cieplak M, Henkel M, Karbowski J, Banavar JR. Master equation approach to protein folding kinetics and kinetic traps. Phys Rev Lett. 1998;80:3654–3657. [Google Scholar]
- 28.Chang I, Cieplak M, Banavar JR, Maritan A. What one can learn from experiments about the elusive transition state. Protein Sci. 2004;13:2446–2457. doi: 10.1110/ps.04713804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudoit S, Fridlyand J, Speed T. Comparison of methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–87. [Google Scholar]
- 30.Lee Y, Lee C-K. Classification of multiple cancer types by multicategory support vector machines using gene expression data. Bioinformatics. 2003;19:1132–1139. doi: 10.1093/bioinformatics/btg102. [DOI] [PubMed] [Google Scholar]
- 31.Blatt M, Weisman S, Domany E. Superparamagnetic clustering of data. Phys Rev Lett. 1996;76:3251–3255. doi: 10.1103/PhysRevLett.76.3251. [DOI] [PubMed] [Google Scholar]
- 32.Blatt M, Weisman S, Domany E. Data clustering using model granular magnet. Neural Comput. 1997;9:1805–1842. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




