Abstract
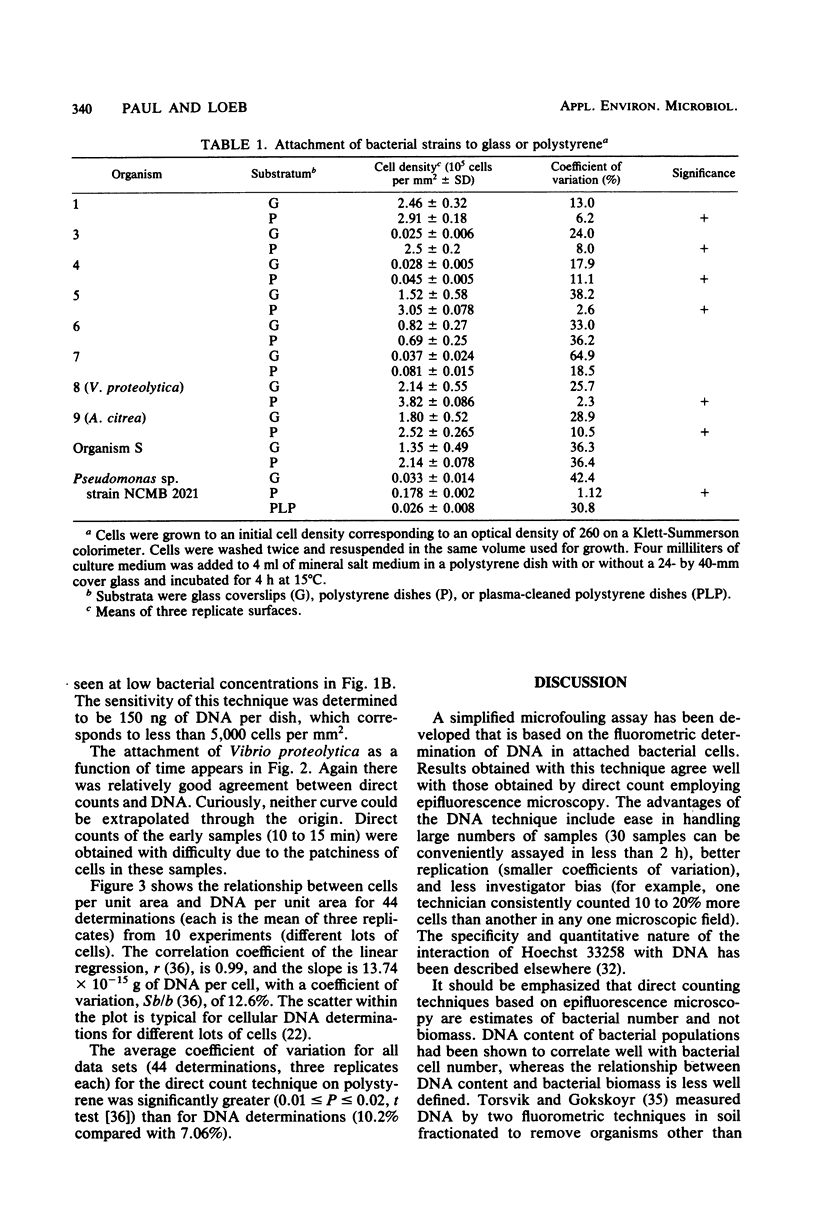
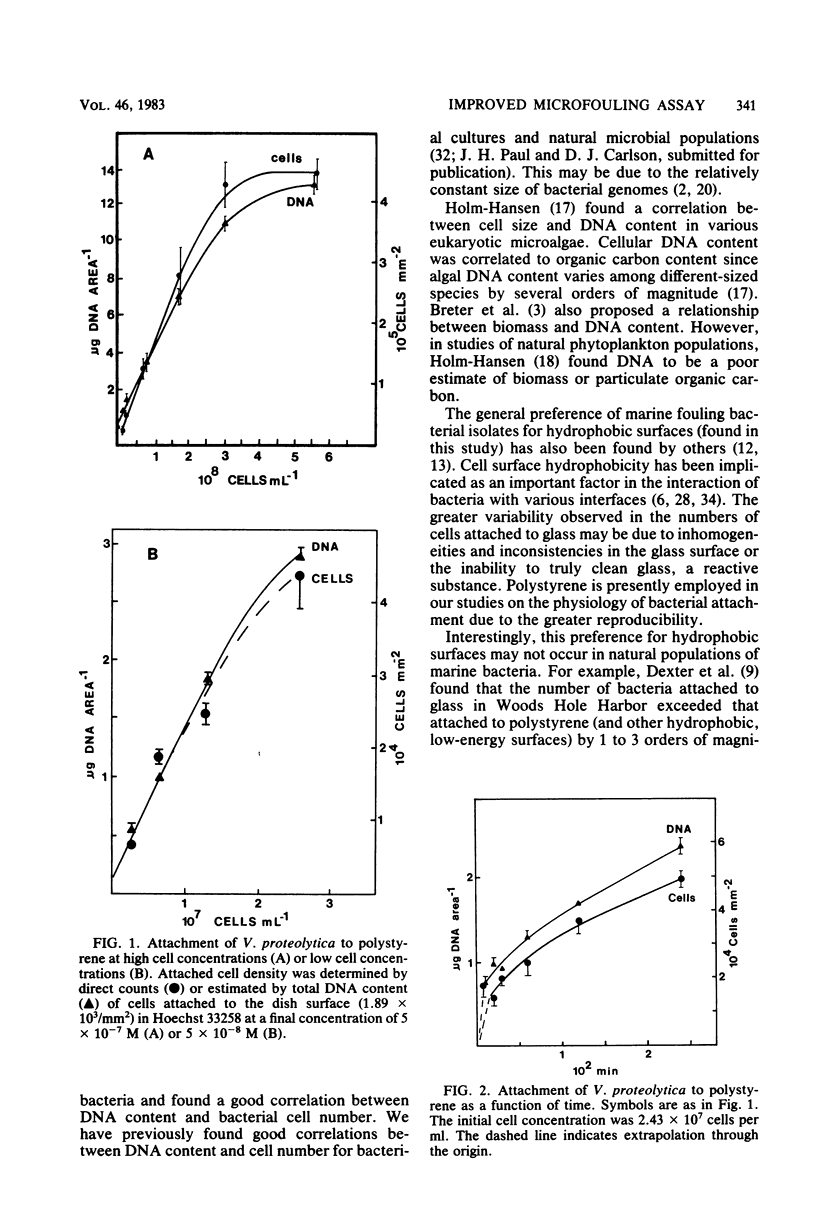
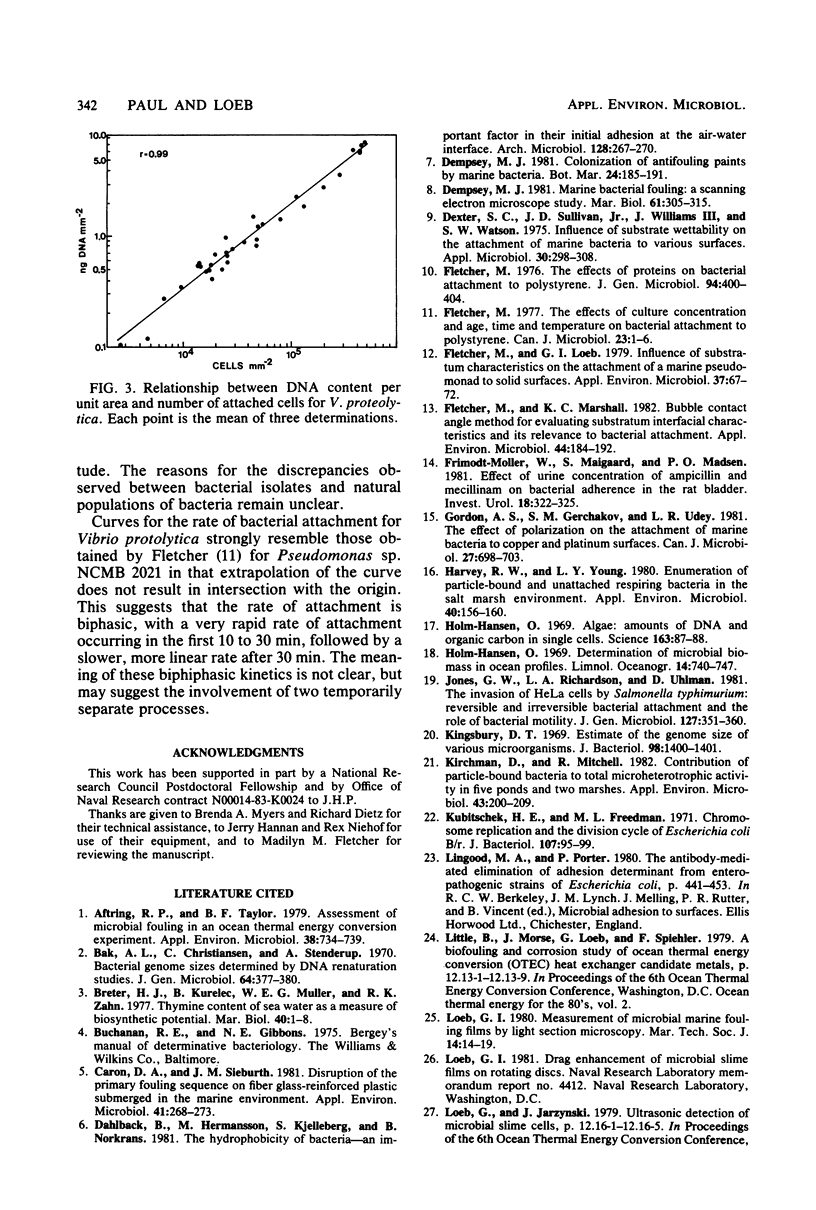
With a direct count assay, 10 fouling bacterial isolates have been characterized for their ability to adhere to glass cover slips and polystyrene dishes. Although most adhered in greater numbers to polystyrene, the preference was statistically significant for only seven isolates at the 95% confidence level, due in part to the greater variability in cell attachment to glass (coefficient of variation, 32.3% for glass compared with 10.0% for polystyrene). Employing polystyrene dishes, a novel microfouling assay was developed, based on the extraction and fluorometric determination of DNA. The assay was rapid, enabled the detection of as little as 0.15 μg of DNA per dish (∼5,000 cells per mm2), and showed good agreement with the direct count assay. The DNA method resulted in less variability among three replicates (average coefficient of variation, 7.06%) and allowed for estimation of bacterial density over a larger surface area per sample (1.89 × 103 mm2) than was feasible with epifluorescence microscopy (0.06 to 0.1 mm2).
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aftring R. P., Taylor B. F. Assessment of microbial fouling in an ocean thermal energy conversion experiment. Appl Environ Microbiol. 1979 Oct;38(4):734–739. doi: 10.1128/aem.38.4.734-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Caron D. A., Sieburth J. M. Disruption of the primary fouling sequence on fiber glass-reinforced plastic submerged in the marine environment. Appl Environ Microbiol. 1981 Jan;41(1):268–273. doi: 10.1128/aem.41.1.268-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlbäck B., Hermansson M., Kjelleberg S., Norkrans B. The hydrophobicity of bacteria - an important factor in their initial adhesion at the air-water interface. Arch Microbiol. 1981 Jan;128(3):267–270. doi: 10.1007/BF00422527. [DOI] [PubMed] [Google Scholar]
- Dexter S. C., Sullivan J. D., Williams J., Watson S. W. Influence of substrate wettability on the attachment of marine bacteria to various surfaces. Appl Microbiol. 1975 Aug;30(2):298–308. doi: 10.1128/am.30.2.298-308.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M., Loeb G. I. Influence of substratum characteristics on the attachment of a marine pseudomonad to solid surfaces. Appl Environ Microbiol. 1979 Jan;37(1):67–72. doi: 10.1128/aem.37.1.67-72.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M., Marshall K. C. Bubble contact angle method for evaluating substratum interfacial characteristics and its relevance to bacterial attachment. Appl Environ Microbiol. 1982 Jul;44(1):184–192. doi: 10.1128/aem.44.1.184-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M. The effects of proteins on bacterial attachment to polystyrene. J Gen Microbiol. 1976 Jun;94(2):400–404. doi: 10.1099/00221287-94-2-400. [DOI] [PubMed] [Google Scholar]
- Frimodt-Møller N., Maigaard S., Madsen P. O. Effect of urine concentration versus tissue concentration of ampicillin and mecillinam on bacterial adherence in the rat bladder. Invest Urol. 1981 Mar;18(5):322–325. [PubMed] [Google Scholar]
- Gordon A. S., Gerchakov S. M., Udey L. R. The effect of polarization on the attachment of marine bacteria to copper and platinum surfaces. Can J Microbiol. 1981 Jul;27(7):698–703. doi: 10.1139/m81-108. [DOI] [PubMed] [Google Scholar]
- Harvey R. W., Young L. Y. Enumeration of particle-bound and unattached respiring bacteria in the salt marsh environment. Appl Environ Microbiol. 1980 Jul;40(1):156–160. doi: 10.1128/aem.40.1.156-160.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm-Hansen O. Algae: amounts of DNA and organic carbon in single cells. Science. 1969 Jan 3;163(3862):87–88. doi: 10.1126/science.163.3862.87. [DOI] [PubMed] [Google Scholar]
- Jones G. W., Richardson L. A., Uhlman D. The invasion of HeLa cells by Salmonella typhimurium: reversible and irreversible bacterial attachment and the role of bacterial motility. J Gen Microbiol. 1981 Dec;127(2):351–360. doi: 10.1099/00221287-127-2-351. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T. Estimate of the genome size of various microorganisms. J Bacteriol. 1969 Jun;98(3):1400–1401. doi: 10.1128/jb.98.3.1400-1401.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D., Mitchell R. Contribution of particle-bound bacteria to total microheterotrophic activity in five ponds and two marshes. Appl Environ Microbiol. 1982 Jan;43(1):200–209. doi: 10.1128/aem.43.1.200-209.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitschek H. E., Freedman M. L. Chromosome replication and the division cycle of Escherichia coli B-r. J Bacteriol. 1971 Jul;107(1):95–99. doi: 10.1128/jb.107.1.95-99.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall K. C., Cruickshank R. H. Cell surface hydrophobicity and the orientation of certain bacteria at interfaces. Arch Mikrobiol. 1973 Apr 8;91(1):29–40. doi: 10.1007/BF00409536. [DOI] [PubMed] [Google Scholar]
- Nickels J. S., Bobbie R. J., Lott D. F., Martz R. F., Benson P. H., White D. C. Effect of manual brush cleaning on biomass and community structure of microfouling film formed on aluminum and titanium surfaces exposed to rapidly flowing seawater. Appl Environ Microbiol. 1981 Jun;41(6):1442–1453. doi: 10.1128/aem.41.6.1442-1453.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. Method for studying microbial biofilms in flowing-water systems. Appl Environ Microbiol. 1982 Jan;43(1):6–13. doi: 10.1128/aem.43.1.6-13.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. Bacterial adherence to polystyrene: a replica method of screening for bacterial hydrophobicity. Appl Environ Microbiol. 1981 Aug;42(2):375–377. doi: 10.1128/aem.42.2.375-377.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]



