Abstract
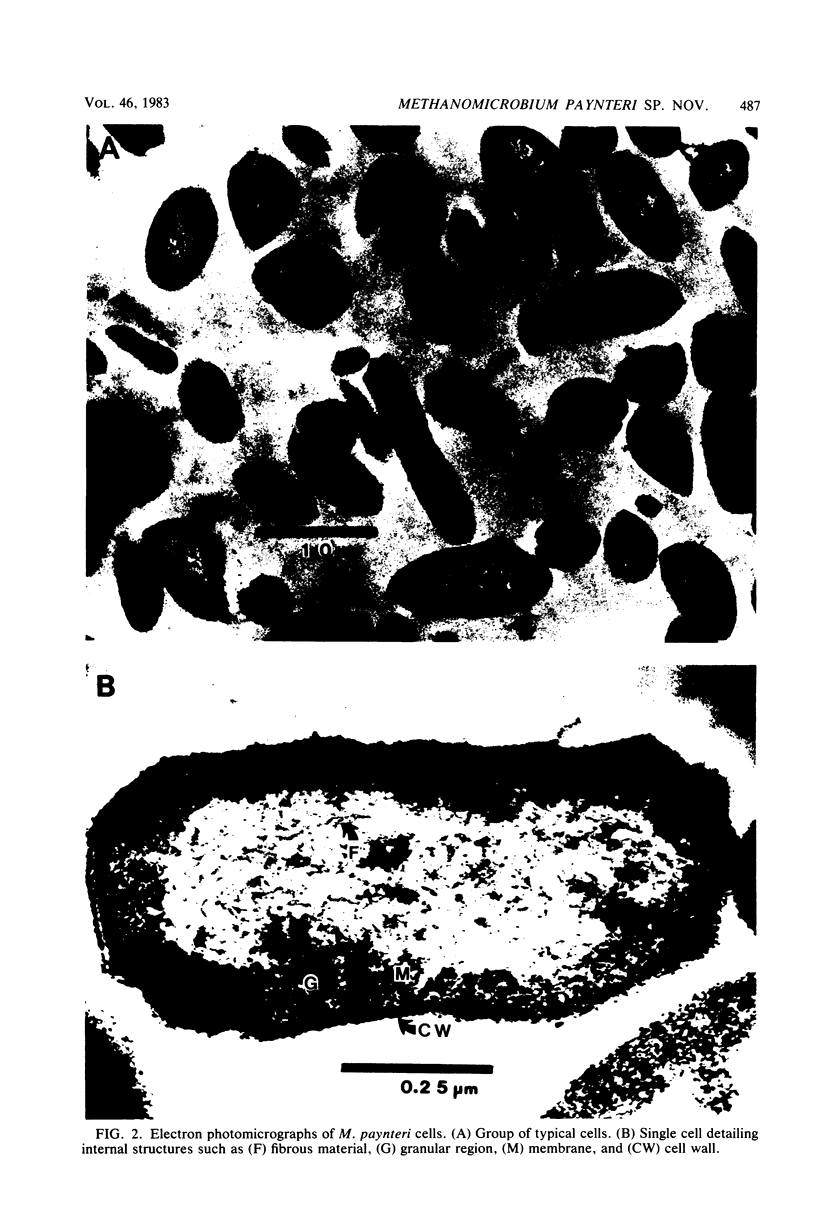
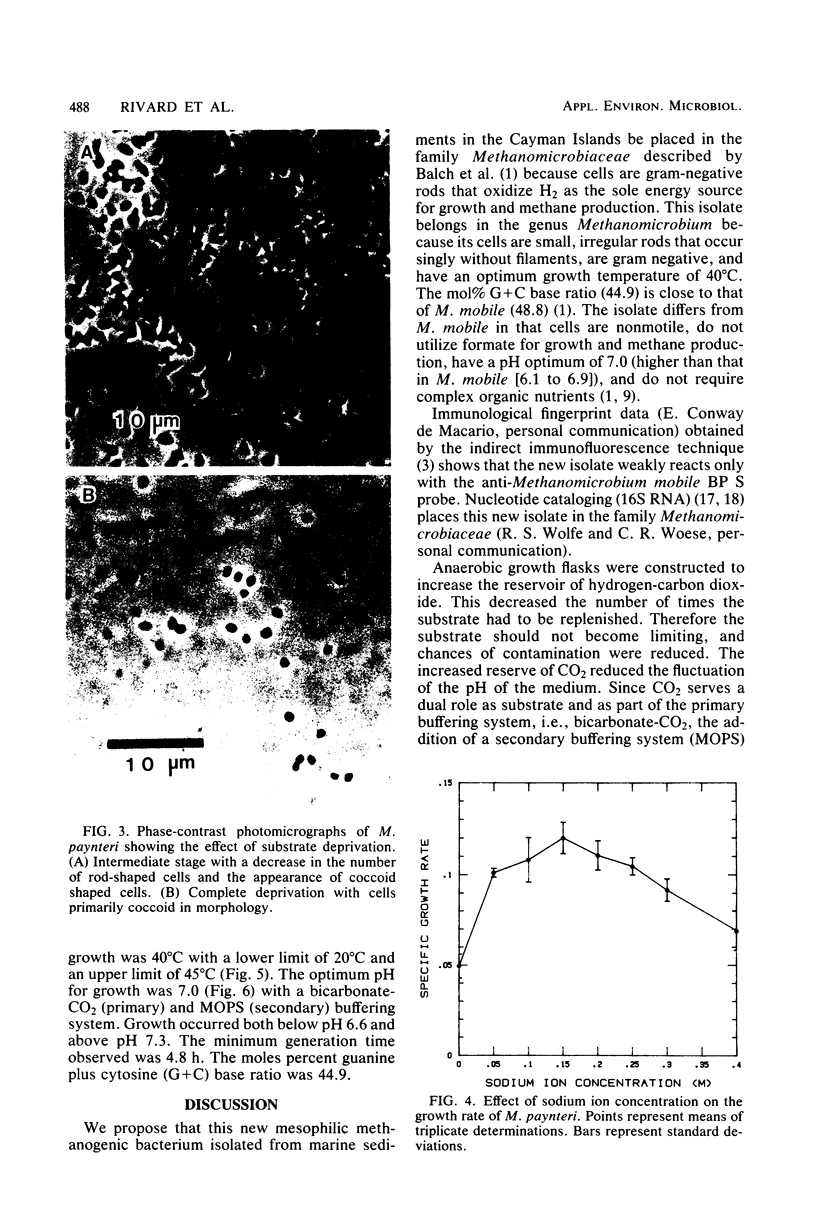
A new mesophilic methanogenic bacterial species isolated from marine sediments collected in the Cayman Islands is described. Cells are small rods occuring singly without filaments, are not motile, and do not possess flagella. Colonies are semitransparent and off-white in color. After 2 weeks of incubation at 37°C colonies are 1 to 2 mm in size, circular, and have entire edges. Only hydrogen-carbon dioxide is a substrate for growth and methane formation. Cells can tolerate a variety of organic secondary buffers (bicarbonate-CO2 being the primary buffer). Cells do not require yeast extract or Trypticase, but do require acetate, for growth. The optimum growth temperature is 40°C. The optimum sodium concentration is 0.15 M. The optimum pH for growth is 7.0. The minimum generation time is 4.8 h. The DNA base composition is 44.9 mol% guanine plus cytosine. The name Methanomicrobium paynteri is proposed in honor of M. J. B. Paynter. The type strain is G-2000 (=ATCC 33997, =DSM 2545).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol. 1976 Dec;32(6):781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E., Macario A. J., Wolin M. J. Specific antisera and immunological procedures for characterization of methanogenic bacteria. J Bacteriol. 1982 Jan;149(1):320–328. doi: 10.1128/jb.149.1.320-328.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink R. W., Dugan P. R. Tentative identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 Mar;33(3):713–717. doi: 10.1128/aem.33.3.713-717.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J. F., Boone D. R. Analytical determination of the buoyant density of DNA in acrylamide gels after preparative CsCl gradient centrifugation. FEBS Lett. 1973 Dec 1;37(2):321–324. doi: 10.1016/0014-5793(73)80487-9. [DOI] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- STADTMAN T. C., BARKER H. A. Studies on the methane fermentation. X. A new formate-decomposing bacterium, Methanococcus vannielii. J Bacteriol. 1951 Sep;62(3):269–280. doi: 10.1128/jb.62.3.269-280.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Uchida T., Bonen L., Schaup H. W., Lewis B. J., Zablen L., Woese C. The use of ribonuclease U2 in RNA sequence determination. Some corrections in the catalog of oligomers produced by ribonuclease T1 digestion of Escherichia coli 16S ribosomal RNA. J Mol Evol. 1974 Feb 28;3(1):63–77. doi: 10.1007/BF01795977. [DOI] [PubMed] [Google Scholar]
- Woese C., Sogin M., Stahl D., Lewis B. J., Bonen L. A comparison of the 16S ribosomal RNAs from mesophilic and thermophilic bacilli: some modifications in the Sanger method for RNA sequencing. J Mol Evol. 1976 Apr 9;7(3):197–213. doi: 10.1007/BF01731489. [DOI] [PubMed] [Google Scholar]