Abstract
The yeast Saccharomyces cerevisiae senses and responds to nutrients by adapting its growth rate and undergoing morphogenic transitions to ensure survival. The Tor pathway is a major integrator of nutrient-derived signals that in coordination with other signaling pathways orchestrates cell growth. Recent advances have identified novel Tor kinase substrates and established the protein trafficking membranous network and the nucleus as platforms for Tor signaling. These and other recent findings delineate distinct signaling branches emanating from membrane associated Tor complexes to control cell growth.
Introduction
All living organisms sense and respond to nutrient-derived signals to adapt their physiology and adopt appropriate developmental decisions to promote survival. In eukaryotic organisms ranging from yeasts to humans, the Tor signaling pathway is a global regulator that controls cell growth. The central components of this signaling cascade are the Tor protein kinases, which were first identified in yeast as targets of the antifungal and immunosuppressive agent rapamycin. Treatment of cells with rapamycin results in dramatic physiological changes including: G1 cell cycle arrest, protein synthesis inhibition, glycogen accumulation and autophagy, which closely resemble those observed in cells deprived of nutrients. These and other findings support the view that Tor is activated by amino acid-derived signals to positively govern a myriad of anabolic processes, including translation, transcription, ribosome biogenesis and actin deposition to sites of active cell growth, while negatively regulating catabolic processes, such as protein degradation, mRNA destabilization, and autophagy [1*].
The versatility of the Tor kinases in impacting this wide range of cellular responses stems from their ability to physically associate with other proteins and thereby functionally coordinate diverse signaling pathways. The Tor proteins form two distinct, evolutionary conserved multimeric protein complexes known as TORC1 and TORC2 [2-6*]. Recent studies reveal these Tor complexes associate with internal membranes of the protein secretory pathway as well as with the nucleus, and these subcellular localizations are critical for Tor function [7*,8**]. The Tor pathway often works in parallel with the cAMP-PKA cascade to control common targets and also intersects with other signaling networks, such as the general amino acid control (GAAC) response, [9-13*]. Several newly identified Tor substrates, Sch9, Ypk1, and Slm1,2, further link Tor function to ammonium sensing, actin organization, control of cell integrity, and stress response [14*-20*]. Here we discuss the latest developments in the mechanisms of signaling by the Tor kinase cascade in the budding yeast S. cerevisiae.
Rapamycin sensitive and insensitive Tor complexes
In S. cerevisiae, the highly homologous Tor1 and Tor2 proteins associate with Kog1, Tco89, and Lst8 in the protein complex TORC1. A separate pool of Tor2 associates with Lst8, Avo1, Avo2, Avo3, Bit61, and Bit2 in a distinct complex called TORC2 [2-6*]. It has generally been accepted that each complex mediates distinct physiological processes in response to nutrient cues. TORC1, which is sensitive to rapamycin, regulates temporal processes of growth while the rapamycin-insensitive TORC2 is thought to control spatial aspects of growth such as actin polarization [1*].
While this notion has prevailed to date, three independent reports suggest the division of labor associated with each Tor complex is not as clearly delineated as previously thought. A comprehensive study has provided evidence for genetic interactions between TORC1 and a network of genes involved in actin polarization and cell wall integrity [7*], processes which were thought to be regulated by TORC2. Such evidence supports previous reports of actin polarization regulation by Kog1, an exclusive TORC1 component, as well as rapamycin-induced changes in actin organization [6*,21*].
Recent studies demonstrated that the AGC kinase Ypk2 is a direct substrate of TORC2 (20*). Both TORC2 and Ypk2 regulate actin polarization and are thought to act upstream of the Rho1/ Pkc1/ MAPK cell integrity pathway. This provides a likely route by which TORC2 controls actin cytoskeleton dynamics. Together, these findings show further evidence of functional overlap between TORC1 and TORC2, blurring the distinction between rapamycin-sensitive and insensitive pathways.
Signaling branches downstream of TORC1
The advent of genome-wide transcriptional analysis, as well as classical genetic approaches, revealed a remarkably robust transcriptional profile of TORC1 controlled genes [22]. This, in turn, identified a number of transcriptional regulators that act at the promoters of rapamycin-sensitive genes. Subsequent work pinpointed the nuclear exclusion of relevant transcription factors as a common theme in the regulation of TORC1 sensitive genes [23]. A continuing challenge is the elucidation of the mechanisms by which signals emanating from TORC1 exert appropriate transcriptional responses.
The first link between TORC1 signaling and a downstream component, the PP2A-like phosphatase Sit4 and its regulatory subunit Tap42 derived from studies with tap42 alleles that blunted many rapamycin-induced phenotypes [24]. Subsequently, Tap42 was shown to be a direct target of Tor phosphorylation [25]. Analysis of additional tap42 rapamycin-insensitive alleles and sit4 mutants demonstrated that Tap42 represents a major branch of TORC1 signaling that is responsible for repression of stress regulated (STRE), nitrogen catabolite repressed (NCR), and retrogade signaling (RTG) genes, as well as for crosstalk between TORC1 and the GAAC response [26-28]. Remarkably, the effects of TORC1 signaling on RP (ribosomal protein) and Ribi (ribosome biogenesis) regulons are not Tap42-mediated. For some targets (NCR and RTG genes) Tap42 functions in concert with PP2A, whereas for other targets (STRE genes) Tap42 inhibits PP2A [26,29]. Likewise, Tip41, a Tap42-interacting protein that regulates Tap42-phosphatase interactions, appears to play both negative and positive roles in Tap42 signaling [30,29]. Interestingly, a pool of Tap42 is localized to membranes in complex with TORC1 in actively growing cells (Figure 1A) [31*]. In response to TORC1 inactivation, Tap42 is released from membranes coincident with Tap42-dependent activation of target genes (Figure 1B). In addition, rapamycin-resistant mutants of Tap42 largely fail to dissociate from TORC1 associated membranes. Whether this mode of regulation governs all TORC1-Tap42 signaling, or if Tip41 plays a role in this process, is unknown.
Figure 1. TORC1 dependent signaling events regulating synthesis of ribosome components.
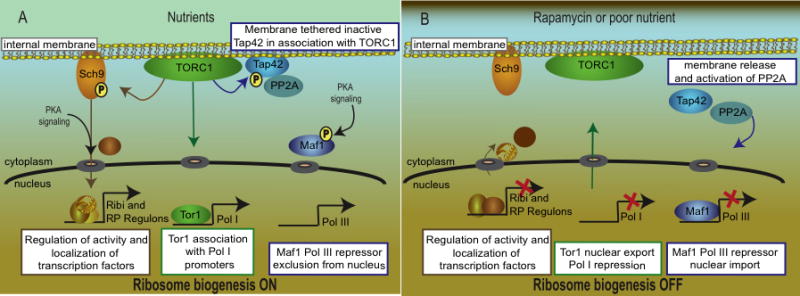
A, when nutrients are abundant Tap42 is localized to the membrane and is associated with TORC1. Under these conditions, TORC1 phosphorylates Sch9, which promotes expression of the Ribi and RP genes required for ribosome biogenesis. The RP and Ribi genes are controlled by the localization and activity of specific transactivators (including Abf1, Rap1, Sfp1, Ifh1, Fhl1, Hmo1, and Crf1) as well as the histone acetylase and deacetylase Esa1 and Rpd3, respectively. The precise mechanisms that regulate the activities and localization of these factors, as well as the signaling events that link Sch9 to RP and Ribi promoters, remain to be elucidated. Tor1 translocates to the nucleus in nutrient replete conditions and associates with Pol I and Pol III promoters. B, in response to nutrient deprivation or rapamycin treatment, Sch9 phosphorylation is inhibited and ribosome biogenesis is turned off. Tor1 is exported from the nucleus and Tap42 is displaced from the membrane and, in concert with the catalytic subunits of PP2A (and PP2A-like phosphatases), dephosphorylates specific targets, such as the Pol III repressor Maf1. The signaling events that govern Tor1 nuclear shuttling, and whether these are influenced by the activities of Tap42 or Sch9, are unknown.
Gln3 regulation underscores the complexity in the control of TORC1-regulated phosphatases. During growth in preferred nitrogen sources, Gln3 is cytoplasmic and translocates to the nucleus when cells are shifted to poor nitrogen sources or upon rapamycin treatment. Gln3 nuclear import induced by rapamycin is prevented by inactivation of Tap42, or deletion of Sit4 [23]. However, the nature of the signal generated by nitrogen quality may be different from that of rapamycin-induced Sit4 activity since nitrogen quality still controls Gln3 localization in sit4 deleted cells [32].
Another target for PP2A signaling is Maf1, a regulator of ribosome biogenesis that inhibits Pol III in response to nutrient depletion or rapamycin treatment. Maf1 nuclear localization is prevented by PKA-dependent phosphorylation and translocation to the nucleus is triggered by rapamycin treatment [33]. A recent report demonstrated that rapid dephosphorylation of Maf1 in response to unfavorable environmental conditions is mediated by PP2A signaling; however; the involvement of Tap42 in this process has not been examined (Figure 1B) [34].
Many Tap42-independent TORC1 signaling events can be explained by the recent identification of the AGC kinase Sch9 as a direct TORC1 kinase substrate. Earlier, a genome-wide screen for new regulators of start uncovered a strong connection between ribosome synthesis and cell size [35]. Deletions of SFP1 or SCH9 were identified as confering dramatic small cell size phenotypes. Sfp1 controls the Ribi regulon and the RP genes. Artificial activation of Sfp1 or Sch9 upregulates the Ribi and RP genes resulting in a large cell phenotype. In addition, carbon starvation or rapamycin treatment leads to dephosphorylation and inactivation of Sch9 [36]. Either rapamycin treatment or sch9 mutation triggers nuclear localization of the Rim15 kinase, which in turn regulates a transcriptional program for entry into G0 and increased chronologic lifespan [37,38].
These and other effects indicated a close relationship between Tor and Sch9 signaling. The TORC1-sensitive phosphorylation sites on Sch9 were identified and mutation of these sites to non-phosphorylatable amino acids was shown to block all known functions of Sch9 with little effect on TORC1-regulated programs of NCR and RTG signaling [14*]. In mammalian cells mTor directly phosphorylates and activates the AGC kinase S6 kinase [39]. These results establish a signaling branch downstream of TORC1 that is distinct from the Tap42-mediated branch (Figure 1), and this brings TORC1 signaling in line with cell growth regulation in mammalian cells with Sch9 fulfilling a role analogous to S6 kinase. Importantly, some TORC1 controlled effects, most notably Msn2 and Msn4 localization, appear to be regulated by both branches [26,29,14*]. The Tap42 and Sch9 branches of TORC1 signaling explain most TORC1 cellular roles. Finally, Tor1 itself has been shown to translocate to the nucleus where directly regulates Pol I and Pol III transcription (discussed below) (Figure 1A) [8*]. Whether Tor1 nuclear localization is subject to control by Sch9 and/or Tap42 or represents a third independent TORC1 signaling branch is currently unknown.
Tor signals from internal membranes
Recent studies have forged an intimate relationship between Tor signaling and internal membranes of the secretory pathway. Several components of TORC1, including Tor1, Tor2, Lst8, Kog1, and Tco89, have been localized to endosomal, Golgi, prevacuolar, and vacuolar compartments [4,40,41,3,6*,5]. Similarly, the Gse/Ego complex, which control plasma membrane targeting of the general amino acid permease Gap1 in response to amino acids, resides in prevacuolar compartments [42**]. TORC1 and the Gse/Ego complex are also necessary for vacuolar membrane recycling via a process known as microautophagy, which occurs during recovery from rapamycin-induced growth arrest [43**]. Based on these findings an exciting role has been hypothesized for the Gse/Ego complex as a functional component of the mechanism involved in relaying amino acid signals from the vacuole to TORC1 [1*]. This model is particularly attractive given that the vacuole is a major cellular reservoir for amino acids.
Three current studies have indicated that the TORC1 link to intracellular membranes is not just physical but also functional. First, a novel role for the Golgi Ca2+/Mn2+ ATPase (Pmr1) in negatively regulating TORC1 signaling has been proposed [44*]. The relevant activity of Pmr1 was determined to be its ability to transport Mn2+ (rather than Ca2+) and Mn2+ supplemented growth medium restores rapamycin sensitivity to pmr1 cells [45]. The role of manganese remains unclear but it has been suggested that Mn2+ is required for mannosylation of proteins and lipids required for proper protein trafficking.
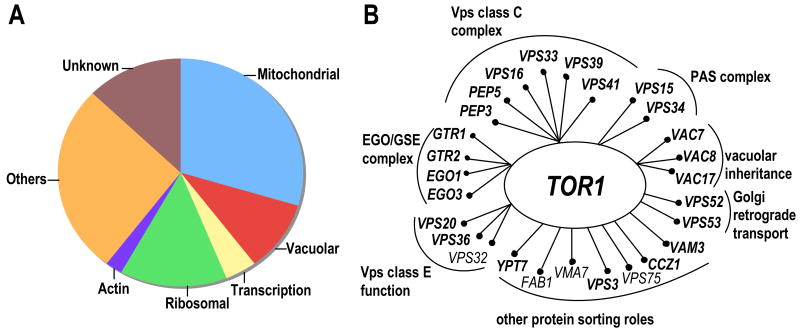
Second, Tor complexes are bound to detergent-resistant membranes (DRMs) and proteomic analysis reveal that several proteins involved in actin organization are colocalized with Tor1 and Tor2 there [4,7*]. Interestingly, when any of nine genes encoding these proteins are mutated in cells lacking the TORC1 non-essential components Tor1 and Tco89, synthetic lethality or reduced fitness defects are observed, indicating the function of these gene products may be linked to a TORC1-related role (see below). Third, a systematic genome-wide screen yielded over 200 genes that, when mutated in combination with the tor1 mutation, result in synthetically lethal or reduced fitness phenotypes [46*]. These genes represent diverse functional categories (Figure 2A), illustrating further the complexity of TORC1 signaling. In particular, a striking link was evident between TORC1 and the class C Vps, the Ego/Gse, and the preautophagosome (PAS) complexes which function in vesicle docking and fusion, protein sorting, and autophagy, as well as with other genes involved in vacuolar segregation (Figure 2B). A previous study that examined the rapamycin sensitivity of the yeast gene deletion collection also reported a class C Vps complex connection unique to Tor1 and not shared with Tor2 [47]. These findings were surprising since it had been thought that Tor2 was capable of providing all of the cellular functions associated with Tor signaling. Instead we now appreciate that there are Tor1/2-shared functions as well as both Tor1- and Tor2-unique functions.
Figure 2. Genetic synthetic interaction network of Tor1.
A, graphical representation of genes (grouped in functional categories) that when mutated in combination with tor1 result in synthetic lethal or reduced fitness defects. B, schematic representation of the distinct functional categories involved in protein sorting and vacuolar function, that exhibit synthetic interactions with tor1. Mutation of the genes shown in bold confer rapamycin hypersensitivity as demonstrated by two recent studies [46*,47].
These results prompted studies to assess in greater detail the impact of TORC1 signaling in protein sorting. Mutation of Tor1 or rapamycin exposure did not have a detectable effect on maturation of vacuolar hydrolases sorted via the endosomal, the Cvt, or the non-endosomal pathways. Similarly, experiments examining alpha factor processing revealed that forward and retrograde transport between the Golgi complex and endosomal compartments is not affected by loss of Tor1 function [46*]. In contrast, while TORC1 activity was dispensable for receptor mediated endocytosis or endocytosis of the Mep2 ammonium permease, rapamycin treatment caused a modest delay in a late step of fluid phase endocytosis [48,46*,7*]. Whether this delay is of sufficient magnitude to be physiologically relevant remains to be explored and one possibility is that it is a secondary consequence of the actin polarization defect caused by rapamycin.
Remarkably, a role for the class C Vps complex function in mediating amino acid homeostasis for proper Tor signaling was also revealed and suggested that Tor1 is more efficient than Tor2 in supporting growth under conditions of amino acid limitation [46*]. Collectively, these findings suggest a model whereby localization of TORC1 to membranes of the protein transport apparatus is important for reception of amino acid-derived signals and, in turn, to relay these signals to TORC1 effectors. These findings further underscore the role of the protein transport membranous network and its components as a prominent and strategic platform that functionally influences a growing number of signaling processes, including the Rim101-mediated pH-response, MAP kinase activation via the Gα subunit Gpa1 and the PI-3 kinase Vps34, and (as discussed here) nutrient sensing [49,50].
Tor signaling in the nucleus
One major role of Tor signaling is the control of ribosome biogenesis, which requires the coordinated action of the three nuclear RNA polymerases: Pol I, Pol II, and Pol III, resulting in transcription of 35S rRNA, RP, and 5S rRNA and tRNA genes.
An interesting aspect in Tor control of ribosome biogenesis was recently brought to light by the discovery that a significant fraction of Tor1 localizes to the nucleus (Figure 1A) [8**]. This nuclear localization is dynamic and can be prevented by either nutrient starvation or rapamycin exposure (Figure 1B). Tor1 nucleocytoplasmic shuttling is assisted by the alternate action of the importin Srp1 and the exportin Crm1 and by nuclear localization (NLS) and nuclear export sequences (NES) within the Tor1 protein. Nuclear Tor1 was found to bind the 35S rDNA promoter via a helix turn helix motif (HTH). Either mutation of the Tor1 NLS or HTH motifs, which prevent nuclear entry and promoter binding respectively, impaired the ability of Tor1 to regulate Pol I-directed expression of 35S rRNA and Pol III-driven expression of 5S rRNA. These mutations however, had no affect on Pol II-regulated expression of the NCR genes or the RP genes. This later event is tightly coordinated with rRNA expression to ensure a balanced supply of building blocks for ribosome biogenesis. In contrast, the Tor1 NES mutation, which prevented nuclear export, did not impair TORC1 control, thought to occur in the cytoplasm, of Pol II-driven NCR and RP gene expression. Interestingly, mTor has also been previously reported to shuttle between the nucleus and the cytoplasm, and thus this may be an evolutionary conserved feature of the Tor kinases [51].
These exciting findings challenge current models that TORC1 regulation of Pol II-directed genes is initiated in the cytoplasm by promoting the nuclear translocation of the transcriptional activators. However, several outstanding questions remain to be addressed. First, which are the direct targets of TORC1 at the rRNA promoters and within the nucleocytoplasmic trafficking machinery? An obvious candidate for a nuclear TORC1 target is the ubiquitously conserved Pol I transcriptional activator Rrn3/TIF-1A, which recruits Pol I to the 35S rDNA promoters. In human cells TIF-1A is phosphorylated and thereby activated by TORC1 [52]. Although this phosphorylation has not as yet been demonstrated in yeast cells, TORC1 signaling does positively regulate Rrn3 interaction with RNA Pol I (53). Other possible Tor substrates are the Rpd3 histone deacetylase, which is thought to be recruited to the rDNA repeats in a TORC1-dependent fashion under conditions of active transcription, and the RNA Pol III repressor Maf1 (discussed above) [54,34]. Because Zheng and coworkers showed that TORC1 regulates its own nuclear–cytoplasmic shuttling, karyopherins and accessory factors could also be potential targets for TORC1 action. Second, by which mechanisms is TORC1 recruited to the rDNA promoters? While it is possible that Tor1 might directly bind the rDNA promoters via its HTH motif, this remains to be tested.
The answers to these questions should clarify the extent of the impact of TORC1 nuclear signaling and will thereby advance our understanding of ribosome biogenesis and cell growth control.
Integration of Tor with other pathways
In response to nutrient cues the TOR pathway intersects with other signaling cascades to orchestrate cell growth (summarized in Table 1). Both Tor/Sch9 and the cAMP-PKA pathways often function in parallel to regulate common targets, including expression of the RP, Ribi and STRE genes as well as genes required for entry into the G0 phase regulated by Rim15, and converge on downstream effectors (Table 1) [10-14*].
Table 1. Summary of the major pathways that interact with Tor signaling.
| Pathway | Cellular cue | Major outputs | Intersection point(s) with TOR signaling |
|---|---|---|---|
| Calcineurin | Ca2+ | Negative regulation of STRE genes, Ion homeostasis | Slm1, Slm2, Fpr1 |
| GCN | Amino acid deprivation (uncharged tRNA) | Upregulation of amino acid anabolism and scavenging | Gcn2, eIF2α |
| Snf1 | Glucose depletion | Activation of alternative carbon source utilization | Snf1, Gln3 |
| Nitrogen discrimination | Quality of N source | Activation of non-preferred nitrogen source utilization | Ure2, Gln3, Gat1 |
| Retrograde | Mitochondrial dysfunction | Replenishing of TCA cycle components | Lst8, Mks1, Rtg1, Rtg2, Rtg3 |
| PKC | Osmotic homeostasis | Cell wall integrity | Rom2 |
| PKA | Fermentable carbon source | Promotion of ribosome biogenesis, repression of stress response and G0 entrance | Fhl1, Ihf1, Sfp1, Maf1, Yak1, Msn2, Msn4, Rim15, |
| PHG (diploid specific) | Poor N source | Foraging behavior-filamentous growth | Snf1, Mep2 |
| Autophagy | N starvation | Autophagy | Apg1, Apg13 |
Similarly, the NCR pathway that controls cellular responses to nitrogen quality and the retrograde pathway that responds to mitochondrial dysfunction are intimately related to TORC1 activity. This relation is underscored by the fact that Lst8, a regulator of retrograde signaling, is a component of both Tor complexes [55]. Tor also negatively regulates two signaling programs that are activated by nitrogen source limitation: the GAAC response and autophagy [27,28,56,57].
Finally, two branches downstream of TORC2 act on signaling programs that promote polarization of the actin cytoskeleton. In one branch, TORC2 phosphorylates Ypk2 to activate Rho1-PKC, as discussed above. In the second branch, the newly identified TORC2 effectors Slm1 and Slm2 (PH domain proteins subject to control by PI4, 5P (2) and sphingolipid signaling) drive actin polarization and are required to cope with heat and oxidative stress [15*-19]. TORC2-Slm signaling antagonizes the Ca2+ and calmodulin-dependent phosphatase calcineurin; Slm1 and Slm2 negatively influence calcineurin signaling and Slm1,2-depleted or slm mutant cells exhibit actin depolarization and stress hypersensitivity phenotypes, which are suppressed by calcineurin defects [19,18*].
Concluding remarks
Recent years have witnessed considerable advances in the understanding of nutrient sensing and control of cell growth via Tor signaling in S. cerevisiae. In this review we sought to take a “reductionist view” of these advances to draw attention to three important, emerging themes. First, signaling events downstream of Tor complexes occur in separate branches. Second, nutrient sensing and Tor signaling are intimately linked with internal membranes. Third, TORC1 responsive phenomenon involves direct nuclear shuttling of Tor1 itself to Pol I and Pol III promoters. Whether this represents a third TORC1 branch or is subject to control by one or both of the other TORC1 signaling branches is unknown.
The challenges ahead are to elucidate the mechanisms by which nutrient and stress signals are transmitted to Tor, to identify the remaining Tor substrates, and to understand the precise mechanisms by which Tor and its substrates orchestrate cell growth.
Acknowledgments
We thank Joseph Heitman for critical reading of the manuscript and insightful discussions with members of Mike Tyers laboratory. This work was supported by R01 CA114107 from the National Cancer Institute (to Maria E. Cardenas).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of special interest, published within the last two years reviewed, have been highlighted as:
* of especial interest
** of outstanding interest
- 1*.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]; An extensive and excellent review on Tor signaling in yeast.
- 2.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen EJ, Kaiser CA. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedaman KP, Reinke A, Anderson S, Yates J, 3rd, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 6*.Araki T, Uesono Y, Oguchi T, Toh EA. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]; This study, together with references 7 and 20, provides evidence for the overlapping regulation of the actin cytoskeleton by TORC1 components.
- 7*.Aronova S, Wedaman K, Anderson S, Yates J, 3rd, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive study that demonstrates that Tor complexes are bound to internal detergent resistant membranes and colocalize with proteins involved in actin polarization.
- 8**.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]; This study shows that an important fraction of Tor1 localizes to the nucleus where it binds to 35S and 5S rDNA promoters to control expression of rRNA in response to nutrients.
- 9.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 10.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'hea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, Thevelein JM, De Virgilio C, De Moor B, Winderickx J. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol. 2005;55:862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 12*.Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study, together with studies reported in reference 13, demonstrate that the Tor and cAMP-PKA cascades function in parallel pathways to regulate several common functions.
- 13*.Chen JC, Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr Genet. 2006;49:281–293. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]; See reference 12.
- 14*.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]; This work demonstrates that TORC1 can directly phosphorylate the AGC kinase Sch9 which, in turn, is responsible for many TORC1-dependent processes that regulate growth.
- 15*.Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004;23:3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with references 16, and 18, these studies demonstrate that two downstream effectors of TORC2, Slm1 and Slm2, act in concert with other signaling programs to polarize actin.
- 16*.Fadri M, Daquinag A, Wang S, Xue T, Kunz J. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell. 2005;16:1883–1900. doi: 10.1091/mbc.E04-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]; See reference 15.
- 17.Bultynck G, Heath VL, Majeed AP, Galan JM, Haguenauer-Tsapis R, Cyert MS. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol Cell Biol. 2006;26:4729–4745. doi: 10.1128/MCB.01973-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Mulet JM, Martin DE, Loewith R, Hall MN. Mutual antagonism of target of rapamycin and calcineurin signaling. J Biol Chem. 2006;281:33000–33007. doi: 10.1074/jbc.M604244200. [DOI] [PubMed] [Google Scholar]; See reference 15.
- 19.Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol Cell Biol. 2007;27:633–650. doi: 10.1128/MCB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report provides a genetic and biochemical demonstration that the AGC kinase Ypk2 is a direct target for TORC2 to regulate actin polarization and the cell integrity cascade.
- 21*.Wang H, Jiang Y. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol Cell Biol. 2003;23:3116–3125. doi: 10.1128/MCB.23.9.3116-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; See reference 6.
- 22.Rohde JR, Cardenas ME. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr Top Microbiol Immunol. 2004;279:53–72. doi: 10.1007/978-3-642-18930-2_4. [DOI] [PubMed] [Google Scholar]
- 23.Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 27.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohde JR, Campbell S, Zurita-Martinez SA, Cutler NS, Ashe M, Cardenas ME. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol Cell Biol. 2004;24:8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 31*.Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals that a major regulator of TORC1's downstream effects, Tap42, is associated with TORC1 in a membranous environment in a rapamycin-sensitive fashion.
- 32.Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameroni E, Hulo N, Roosen J, Winderickx J, De Virgilio C. The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle. 2004;3:462–468. [PubMed] [Google Scholar]
- 38.Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 41.Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]; This study, and reference 43, identified the GSE/EGO complex. Gao and Kaiser localized this complex to the prevacuolar compartment and demonstrated its involvement in regulating sorting of the amino acid permease Gap1 in response to amino acids. Duboloz et al. showed that, in concert with TORC1, the GSE/EGO complex promotes microatophagy following growth arrest induced by rapamycin.
- 43*.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]; See reference 42.
- 44*.Devasahayam G, Ritz D, Helliwell SB, Burke DJ, Sturgill TW. Pmr1, a Golgi Ca2+/Mn2+-ATPase, is a regulator of the target of rapamycin (TOR) signaling pathway in yeast. Proc Natl Acad Sci U S A. 2006;103:17840–17845. doi: 10.1073/pnas.0604303103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report proposes an interesting role for the Golgi Ca2+/Mn2+ ATPase (Pmr1) in negatively regulating TORC1 signaling and, together with studies shown in reference 45, demonstrates that the relevant function of Pmr1 in this process is the Mn2+ transport activity.
- 45.Devasahayam G, Burke DJ, Sturgill TW. Golgi manganese transport is required for rapamycin signaling in Saccharomyces cerevisiae. Genetics. 2007;177:231–238. doi: 10.1534/genetics.107.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systematic study that employed diploid synthetic lethal analysis by microarray to define the genetic synthetic lethal and reduced fitness interaction network of Tor1. This approach demonstrated a strong connection between TORC1 signaling and functions involved in protein trafficking and identified a role for the class C Vps complex in mediating amino acid homeostasis for efficient Tor signaling.
- 47.Xie MW, Jin F, Hwang H, Hwang S, Anand V, Duncan MC, Huang J. Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc Natl Acad Sci U S A. 2005;102:7215–7220. doi: 10.1073/pnas.0500297102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.deHart AK, Schnell JD, Allen DA, Tsai JY, Hicke L. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol Biol Cell. 2003;14:4676–4684. doi: 10.1091/mbc.E03-05-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 51.Kim JE, Chen J. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc Natl Acad Sci U S A. 2000;97:14340–14345. doi: 10.1073/pnas.011511898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 56.Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2alpha Kinase GCN2. J Biol Chem. 2003;278:20457–20460. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- 57.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]