Abstract
The puzzling linkage between genetic hemochromatosis and histocompatibility loci became even more so when the gene involved, HFE, was identified. Indeed, within the well defined, mainly peptide-binding, MHC class I family of molecules, HFE seems to perform an unusual yet essential function. As yet, our understanding of HFE function in iron homeostasis is only partial; an even more open question is its possible role in the immune system. To advance on both of these avenues, we report the deletion of HFE α1 and α2 putative ligand binding domains in vivo. HFE-deficient animals were analyzed for a comprehensive set of metabolic and immune parameters. Faithfully mimicking human hemochromatosis, mice homozygous for this deletion develop iron overload, characterized by a higher plasma iron content and a raised transferrin saturation as well as an elevated hepatic iron load. The primary defect could, indeed, be traced to an augmented duodenal iron absorption. In parallel, measurement of the gut mucosal iron content as well as iron regulatory proteins allows a more informed evaluation of various hypotheses regarding the precise role of HFE in iron homeostasis. Finally, an extensive phenotyping of primary and secondary lymphoid organs including the gut provides no compelling evidence for an obvious immune-linked function for HFE.
Genetic hemochromatosis (HC) is a recessive Mendelian disorder affecting ≈5–8/1,000 of Caucasoid individuals (heterozygous carriers are estimated at a frequency of 1/12) and represents, therefore, one of, if not the, most frequent hereditary anomalies (ref. 1; http://www.ncbi.nlm.nih.gov/htbin-post/Omim/dispmim?235200). The disease is characterized by gradual iron overload within parenchymal cells of several organs, principally liver, pancreas, and heart, leading to clinical manifestation in early adulthood and, if left untreated, to major complications: i.e., cirrhosis, hepatocellular carcinoma, diabetes, and chronic heart failure. Early diagnosis through biochemical assays of iron metabolism and ultimately liver biopsy is the indication for repeated phlebotomy ensuring the patient with a normal life span.
The long-sought basis for the linkage between iron overload and histocompatibility loci remained a mystery for over 20 years. First recognized by Simon and colleagues (2) through serological HLA testing, the association of HLA-A3, -B14 carrying haplotypes and inherited hemochromatosis led to a long and confusing period of candidate gene(s) identification with candidates swinging from one locus to another, none of which ultimately stood the test of genetics. In retrospect, it is now obvious that the work of various investigators was hampered by the formidable linkage disequilibrium surrounding, and extending far beyond, the telomeric segment of the HLA-A locus. The chapter was, however, successfully closed by a landmark reverse genetics and high throughput genomic sequencing strategy that led Feder and colleagues to the identification of the latest member of the mammalian major histocompatibility complex (MHC) class I (MHC-I) gene family (3). Initial as well as several subsequent large scale population studies confirmed that the locus first described as HLA-H, and shortly after renamed HFE, was responsible for the vast majority of hereditary hemochromatosis in Caucasian populations (ref. 3; http://www.ncbi.nlm.nih.gov/htbin-post/Omim/dispmim?235200) (despite the existence of authentic exceptions). Indeed, HC patients harbor at homozygosity a unique point mutation, C282Y, which disrupts the disulfide link between a pair of critical cysteine residues, precluding the association with β2 microglobulin (β2m) and hence cell surface expression (4). The pathological relevance of a second mutation/polymorphism, H63D, is still in doubt.
HFE is a bona fide MHC-I molecule by several key criteria. The human 9.5-kilobase (kb) gene (chromosome 6p21.3–22.1) and the mouse 6.7-kb gene (chromosome 13) have a typical class I architecture in which each domain is encoded by a separate exon: i.e., leader peptide, α1, α2, and α3 as well as the transmembrane and cytoplasmic-3′ untranslated region (3, 5). The gene gives rise to a 2.7/1.5-kb (human/mouse) transcript predominantly expressed within epithelial cells of various tissues (5). Once translated, the 48-kDa HFE heavy chain noncovalently engages the 12-kDa β2m light chain. Preliminary immunochemical analysis has revealed, in accord with the previously documented epithelial pattern of transcription, an apparent restriction of HFE polypeptide to the perinuclear area of crypt cells within the small intestine and the apical membrane of the syncytiotrophoblasts, both major sites of iron absorption, as well as various endothelia (6, 7). Crystallographic studies of a recombinant soluble HFE molecule divulged a tertiary structure closely resembling an authentic MHC-I fold: a heterodimer in which the membrane-proximal Ig domains (β2m and HFE α3 loop) support a distal putative ligand binding groove (8). However, despite numerous similarities, closer examination reveals that HFE diverges in a number of important points from canonical MHC-I molecules. These include a slightly longer gene essentially attributable to an unusually large first intron (several kilobases instead of 100 bp in class Ia glycoproteins), a cell-type specific pattern of expression (within epithelia) versus the almost ubiquitous pattern characteristic of classical MHC-I genes, and, finally, a definitive closure of the antigen binding cleft that prevents peptide binding and presentation. Unexpected, despite one prescient prediction (9), was the demonstration by Parkkila and colleagues of an HFE interaction with the transferrin receptor (TfR) (10). This puts HFE directly in the midst of iron metabolism because TfR is responsible for cellular uptake of iron bound transferrin. Indeed, gene transfer experiments, comparing mutant and wild-type HFE cDNAs, demonstrated that the mutant fails to interact with β2m and TfR and, as noted above, does not reach the cell surface. This defect appears to disturb the feedback between the body’s state of iron repletion and the regulation of intestinal iron absorption. Indeed, we and others have hypothesized that the HFE mutation reduces the iron supply to immature enterocytes in the duodenal crypts (11–13); hence, once these have reached the villi tips, they absorb iron excessively. A second piece of physical evidence hinting at this scenario of HFE involvement in iron homeostasis is the apparent antithetic regulation of HFE transcript levels with respect to that of a likely key player in iron physiology, the transmembrane iron transporter DMT1 (12).
Despite this wealth of information gained in less than 3 years since the identification of the molecule, the precise nature of HFE intervention in iron homeostasis remains elusive, and several important questions are presently unanswered (11). Among these are: (i) What is the physiological relevance of HFE-TfR interaction, given that the duodenal mucosa, the site of regulated iron absorption, has a surprisingly low iron content in HC and most organs with large iron deposits lack HFE expression? (ii) What is the evolutionary advantage of using an MHC class I structure, apparently primarily evolved for presenting peptide antigens through a membrane distal platform to the αβ T-cell receptor and to engage the CD8 coreceptor via a membrane proximal Ig-loop (14), to regulate iron levels? The reciprocal question is equally interesting: Does HFE have any, yet undefined, immunological functions, which would link the immune system to iron physiology?
To answer some of these questions, we report the generation, as well as in-depth analysis, of an HFE mutant mouse strain. Our model, in addition to mimicking the human disorder, provides a tool for various physiological and immunological assays impossible by nature to realize in humans and valuable for comparison with other models of hemochromatosis (15, 16). Metabolic and histological parameters were followed in animals subjected to different levels of iron intake. Intestinal 59Fe absorption rates were determined in the duodenum, as was the activity of the iron regulatory protein (IRP-1) and the endogenous iron content. Finally, a comprehensive phenotyping of primary and secondary lymphoid organs provides a detailed picture of the immune system in the absence of HFE.
Materials and Methods
Gene Targeting.
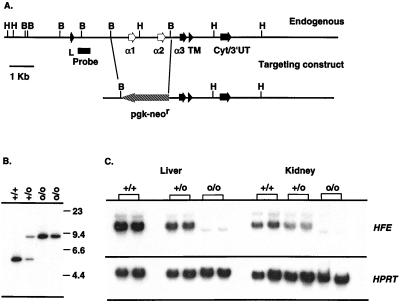
Cloning and sequence analysis of the murine HFE gene from a 129/SvJ library has been previously reported (5). A 2.5-kb BglII fragment encompassing the second and third exons (α1 and α2 domains) was replaced with a 2-kb pgk-neor gene flanked by loxP sites (gift of P. Kastner, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) (Fig. 1A). The targeting fragment was excised from the vector, was purified, and was electroporated into E14 ES cells (subclone kindly provided by T. Miyazaki, Basel Institute for Immunology, Basel). Correctly targeted HFE alleles were detected by Southern blot analysis in 6 of 240 G418-resistant clones, two of which (with normal karyotypes as assessed by Genome Systems, St. Louis) were subsequently injected into C57BL/6 blastocysts. Chimeric, heterozygous, and homozygous mice were generated according to standard procedures and were obtained in Mendelian ratios. Transmission of the mutation was assessed by either genomic Southern blot analysis (Fig. 1B) or PCR using the following oligonucleotides: 5′-GAATTAACAGGCCGTTTCTAAAG-3′, 5′-CTTGGAGTAGTGGCTCACACT-3′ (HFE forward and reverse respectively), and 5′-GAGATCAGCAGCCTCTGTTCC-3′ (pgk-neor). Northern blots prepared with total RNA isolated from kidney and liver tissue confirmed the absence of full length HFE mRNA (Fig. 1C) in HFEo/o mice. The faint, shorter transcript visible in o/o animals is the product of direct splicing of exons 1–4 (leader-α3 domain) as ascertained by reverse transcription–PCR (data not shown). Mice used for these analysis were produced by intercrossing heterozygous animals (with 129/Ola–C57BL/6 mixed backgrounds).
Figure 1.
Generation of an HFE deficient mouse strain. (A) Structure of the endogenous locus and the targeting construct. HFE exons are noted, as is the probe used for screening. As indicated, exons encoding the α1 and α2 domains are replaced by pgk-neor in the mutant animals. H, HindIII; B, BglII. (B) Representative Southern blot of DNA from mutant and control mice. Genomic DNA was digested with HindIII, and the blot was hybridized with the 5′ probe indicated in A; the genotypes of the animals are shown above. (C) Northern blot analysis of kidney and liver RNA from +/+, +/o, and o/o mice. Twenty micrograms of total RNA was run in each lane, and filters were hybridized with an HFE cDNA (spanning exons 2–5). The filter was stripped and hybridized with a hypozanthine phosphoribosyl transferase probe as a loading control.
Animal Care and Diets.
All animal experiments were performed in accordance with institutional and governmental guidelines. Iron deficiency and overload were induced by feeding animals for 21 days with a diet containing 6 mg Fe/kg (C1038, Altromin, Lage, Germany) or during 14 days with the same diet supplemented with 2.5% (wt/wt) carbonyl iron (Sigma). Control animals received an iron-balanced diet of the same composition (C1000, Altromin) but containing 180 mg Fe/kg.
Hematology, Metabolism, and Histology.
Plasma iron concentrations were determined photometrically by the bathophenantroline method (Merckotest No. 3317, Merck). Plasma transferrin was measured by immunodiffusion technique as described (17). For conversion to total iron binding capacity (micrograms of Fe/100 ml), plasma transferrin concentrations (milligrams of protein/milliliter) were multiplied by 140, assuming a molecular mass of 80 and 56 Da for transferrin and iron, respectively (18). Photometrically determined total iron binding capacity values agreed well with immunologically determined transferrin concentrations. Iron content was measured in liver, duodenal mucosa, spleen, and kidney by using ICP-AE spectrometry (Thermo Jarrell-Ash, Atom Comp Series 800, Franklin, MA) after digestion of the tissue in nitric acid (suprapur, Merck) under pressure (170°C, 2.5 h; Model 14571, Fa. Berghof, Ening, Germany). For histological analysis, small liver specimens were fixed in formalin. Paraffin sections (4 μm) were stained with hematoxylin and eosin, periodic acid Schiff reagent, and trichrome. Prussian blue was used for histochemical determination of iron storage. All reagents were of analytical purity and were purchased from Merck and Sigma. 59Fe was supplied by NEN.
In Vivo 59Fe Absorption Studies.
In situ tied-off duodenal segments were used to determine 59Fe absorption as described earlier (19, 20). A duodenal segment (2–3 cm) was filled after overnight fasting with 50–100 μl of physiological medium at pH 7.4, containing 59Fe3+ chelated with nitrilotriacetate (1:2). After 15 min, the ligated segment was removed, and 59Fe activity in the carcass was assayed in a whole body counter for small animals (ARMAC Model 446, Packard). Mucosal transport was expressed as pmol 59Fe/cm segment length/min. The results obtained in wild-type mice were in good agreement with findings by others (19, 20).
IRP Activity.
Mucosa samples were scraped with a glass slide from the duodenum. Tissue samples were kept frozen at −70°C. By using a small tissue grinder, fragments of frozen tissue were solubilized in Eppendorf tubes in 100 μl of Munro buffer (21), which is composed of 10 mM Hepes (pH 7.6), 3 mM MgCl2, 40 mM KCl, 1 mM PMSF (Sigma), and 0.1% Triton X-100. Samples were centrifuged at 12,000 × g, and the protein concentration of the supernatant was determined by using the Bio-Rad protein assay. Protein (0.5 μg) in presence or absence of 2-mercaptoethanol was incubated with a molar excess of 32P-labeled iron responsive element of human ferritin (transcribed from pSPT-fer) essentially as described (22). In most experiments, the iron responsive element was gel-purified. Addition of heparin and RNase T1 treatment was omitted. RNA–protein complexes were resolved in 6% polyacrylamide gels and were quantified by a phosphorimager (Fuji Medical Systems, Stamford, CT).
Phenotype Analysis of Various T-Cell Subpopulations.
Thymi and lymph nodes were removed and gently disrupted with frosted glass microscope slides to make single cell suspensions. Littermates or age-matched pairs were analyzed at 5 weeks–1 year of age, and similar numbers of thymocytes and lymph node cells were recovered in each case. Intestinal epithelial lymphocytes (IELs) were isolated essentially as described (23). In brief, small intestines were removed and flushed with 0.15 M NaCl. After dissecting away the Peyer’s patches, intestines were opened longitudinally, were gently washed in 0.15 M NaCl, and were cut into 5-mm pieces; these were stirred vigorously in 100 ml of Ca2+ Mg2+-free Hank’s balanced salt solution supplemented with 4 mM NaHCO3, 25 mM Hepes, and 2 mM dithiotreitol for 30 min at 37°C. Tissue fragments were allowed to settle, and the cell suspension (epithelial cells and IELs) was decanted through gauze; a portion of this mixture was analyzed to estimate total IEL numbers in each intestine. After washing in RPMI 1640 medium/5% FCS, IELs were further purified by discontinuous Percoll density gradient centrifugation. Cells were resuspended in 10 ml of Percoll (ρ= 1.062), were underlayed with 2 ml of Percoll (ρ = 1.109), and were centrifuged for 12,850 g⋅min. Cells at the ρ = 1.062/1.109 interface were harvested, were washed three times, and then were stained for analysis. Littermates or age-matched pairs were analyzed from 11 weeks–one year of age; IEL recovery was variable, with 1–5 × 106 IELs isolated from each mouse. For cytofluorometric analysis, 0.25–1 × 106 cells were incubated with saturating amounts of antibody in PBS, 2% FCS, and 0.1% NaN3 on ice for 20 min. Biotinylated antibodies were revealed with avidin-FITC (PharMingen). Anti-γδ (GL3)-FITC and-PE, anti-TCRβ (H57–597)-PE and -FITC, anti-CD4 (GK1.5)-PE, anti-CD8α (53–6.7)-Cy-Chrome, anti-Vδ4 (GL2)-bio, anti-CD3ɛ (145–2C11)-FITC, anti-CD8β (53–5.8)-FITC, were obtained from PharMingen (San Diego, CA). Cells were passed on a FACSCalibur flow cytometer, and data were analyzed by using cellquest software (Becton Dickinson).
Statistics.
Values of the different groups were presented as means ± SD. Significant differences between more than two corresponding groups were analyzed by one-way ANOVA (5% level). When significant differences were found, the least-significant-difference test was used (5% level) to test which means were significantly different from each other and which formed homogenous groups. Differences between two corresponding parameters were assessed by use of the unpaired Student’s t test (P < 0.05).
Results and Discussion
The “business-end” of an MHC class I molecule is the α1–2 ligand binding superdomain (14). For classical MHC-I molecules, this involves the well understood tight interaction with a nonameric peptide cargo, presented to the αβ T-cell receptor. With respect to HFE, despite the apparent absence of peptide within a narrowed groove, the most distinct biological function of the molecule identified so far—interaction with the transferrin receptor—has been mapped precisely within this same structure, although to a distinct site (the C terminus of the α1 domain α helice) (24). Hence, to rationally disrupt HFE function, we decided to remove these two critical exons from the mouse germline by using a targeting vector in which they were replaced with the neomycin resistance gene driven by the phosphoglycerate kinase promoter (pgk-neor) (Fig. 1). Experiments were thereafter designed to critically evaluate the lymphoid and metabolic status of these animals.
HFEo/o mice were born in Mendelian ratios from heterozygous parents (of 474 offspring, 127 were o/o, 219 o/+, and 128 +/+). They developed and bred normally and were morphologically indistinguishable from heterozygous as well as wild-type littermates (data not shown). Animals were assayed at 12 weeks of age because before this date, they witnessed a rapid growth [12× weight gain in 12 weeks, irrespective of genotype (data not shown)]. To achieve a thorough analysis of metabolic functions, the colony was subdivided into three groups, which were subjected to normo-, hypo-, and hyperferric regiments, respectively.
Table 1 depicts plasma iron concentration, as well as transferrin saturation in homozygous, heterozygous, and wild-type HFE animals. Closely paralleling the human disorder, both of these parameters were significantly higher in HFEo/o mice as compared with HFE+/+ controls, clearly documenting iron overload in these animals. Plasma transferrin concentration showed no significant differences due to genetic status or iron overload (data not shown). Table 2 clearly reports a significant (3–4×) rise in hepatic iron concentrations of HFEo/o mice compared with wild-type and heterozygous littermates under all dietary iron regiments. These results are in accordance with those obtained in the β2m knockout mice as well as Zhou and colleagues’ HFE-deficient strain (12, 15, 16). Differences in absolute values for this parameter seem related to differences in age and growth rate (15, 16). Iron-deficient feeding reduces the hepatic iron load, although, again, distinctly higher values were found in HFEo/o animals in comparison with HFE+/+ littermates. In contrast to the liver, the iron content in duodenal mucosa was not significantly different in HFEo/o mice and HFE+/+ controls. As in humans (25), the duodenal iron content in HFEo/o mice increased only slightly in spite of excessive hepatic iron load. Finally, iron overload increased the iron content in the spleen whereas iron deficiency caused a decrease, again to lower values in HFE+/+ than in HFEo/o mice. The iron content in the kidney was influenced by neither the genotype nor the state of iron repletion.
Table 1.
Hematological parameters
| HFE genotype | +/+ | +/o | o/o |
|---|---|---|---|
| Plasma iron, μg/100 ml | |||
| Iron adequate | (7) 134 ± 60 A | (8) 117 ± 37 A | (7) 195 ± 64 B |
| Iron overload | (8) 171 ± 46 A | (5) 162 ± 49 A | (10) 276 ± 61 B* |
| Transferrin saturation, % | |||
| Iron adequate | (7) 41 ± 18 A | (8) 39 ± 19 A | (7) 59 ± 24 A |
| Iron overload | (8) 58 ± 17 A | (5) 52 ± 17 A | (10) 84 ± 12 B |
Numbers in parentheses correspond to number of animals assayed or number of determinations performed. Groups in the same row that have no capital letter in common are significantly different from each other (ANOVA; P < 0.05). See Materials and Methods for details.
Significant differences between iron-adequate and iron-overloaded segments (P < 0.05, Student t-test).
Table 2.
Tissue iron content (μg/g dry weight; n = 4–7)
| HFE genotype | +/+ | +/o | o/o |
|---|---|---|---|
| Liver | |||
| Iron deficient | a 193 ± 21 A | a 949 ± 368 B | |
| Iron adequate | a 423 ± 114 A | a 417 ± 105 A | a 1656 ± 420 B |
| Iron overload | b 1143 ± 264 A | b 1428 ± 468 A | b 3657 ± 778 B |
| Duodenal mucosa | |||
| Iron deficient | a 132 ± 46 A | a 143 ± 44 A | |
| Iron adequate | a 142 ± 14 A | a 143 ± 27 A | a 176 ± 44 A |
| Iron overload | b 470 ± 53 A | b 472 ± 127 A | b 621 ± 126 A |
| Spleen | |||
| Iron deficient | a 935 ± 211 A | a 1919 ± 438 B | |
| Iron adequate | b 2132 ± 365 A | a 2214 ± 533 A | a 2337 ± 484 A |
| Iron overload | c 3813 ± 291 A | b 3108 ± 373 A | b 3260 ± 631 A |
| Kidney | |||
| Iron deficient | a 292 ± 21 A | a 344 ± 28 A | |
| Iron adequate | a 387 ± 120 A | a 330 ± 77 A | a 356 ± 63 A |
| Iron overload | a 378 ± 93 A | a 386 ± 53 A | a 394 ± 71 A |
Numbers in parentheses correspond to number of animals assayed or number of determinations performed. Groups in the same row that have no capital letter in common are significantly different from each other, as are lowercase letters, which indicate significant differences between various states of iron repletion (ANOVA; P < 0.05). See Materials and Methods for details.
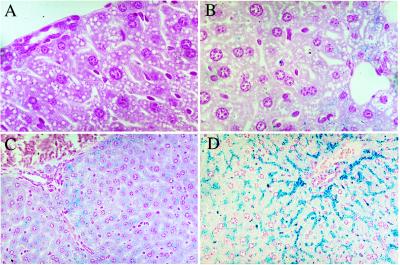
Iron staining predominantly in periportal hepatocytes is a typical histological feature in early stages of human HC (Fig. 2). Its increase in HFEo/o mice as well as the restriction of iron accumulation in Kupffer cells to iron-overloaded animals confirms earlier results in other animal models (15, 16). These findings emphasize the validity of our model in mimicking the hepatic iron distribution pattern of human HC.
Figure 2.
Iron storage in hepatic cells from +/+ and o/o mice fed on different diets. Light micrographs of a periportal liver section from +/+ controls on a standard diet (A) and after 2 weeks of iron-rich feeding (B). The white areas are peripheral vessels. Although no iron staining is visible in A, parenchymal iron accumulation in (B) is represented by small blue dots in the cells around the periportal vessel. C and D show periportal sections from o/o animals on iron-adequate and iron-rich diets, respectively. Pearl’s staining shows marked parenchymal iron accumulation in C and to an even higher extent in D. In all cases, iron accumulation decreases with increasing distance from the periportal area. Pearl’s Prussian blue staining. (A and B, ×1,000; C and D, ×400).
As previously mentioned, the Cys282Tyr mutation renders HFE incapable of interacting with β2m and TfR; consequently, HFE is no longer expressed at the cell surface (4). This defect in the HFE protein appears to disturb the feedback between the body’s state of iron repletion and the regulation of intestinal iron absorption. The HFE mutation is assumed to reduce the iron supply to immature enterocytes in the duodenal crypts (11, 12, 13). However, to date, analysis of HFE knockout mice has not included measurements of the total duodenal iron content, on its available low molecular iron fraction (IRP activation) as well as on intestinal iron absorption, all of which are essential to investigate the mechanistic link between HFE mutation and increase iron absorption. Hence, we set out to critically evaluate these issues. Table 3 clearly demonstrates a 2.6× increase in 59Fe absorption in normally fed HFEo/o vs. HFE+/+ and HFE+/o animals. This roughly parallels the increase in hepatic iron load on a control diet (Table 2) and can be reconciled with the 7.7-fold increase in mRNA of the potential duodenal iron transporter DMT1 observed in another murine HFEo/o model (12). Subjected to 2 weeks of an iron-rich diet, 59Fe absorption decreased significantly in all genotypes alike, although it remained significantly higher in HFEo/o as compared with the HFE+/+ animals. Finally, in HFE+/+ animals, iron-deficient feeding increased duodenal iron absorption more than 6-fold whereas in HFEo/o mice it doubled (Table 3). Within the duodenal mucosa, the intracellular sensing of iron levels is achieved by the two critical iron regulatory proteins, IRP-1 and IRP-2 (26). Upon a decrease in intracellular iron concentration, IRPs bind to the so-called iron response elements in the 5′ or 3′ end of various mediators. The regulatory iron pool in the duodenal mucosa as estimated by determination of IRP-1 activity showed no significant differences between iron adequately fed HFE+/+ and HFEo/o animals (Table 4), which is in agreement with observations in HC (25). In iron deficiency, however, IRP activity increased markedly in both genotypes (Table 4), and intestinal iron absorption increased simultaneously (Table 3). However, there is no indication for a decreased functional iron pool in the duodenal mucosa of iron adequately fed HFEo/o mice (Table 4). Hence, even if the interaction between the defective HFE-gene product and the functional β2m and TfR at the basolateral membrane site is probably the mechanism for decreasing the iron supply to crypt enterocytes (12, 15, 16), increased iron absorption in HC seems to be influenced by other mechanisms than those acting in iron deficiency. However, our experiments did not determine IRP-1 activity in crypt and villus cells separately. Absorbed as well as endogenous iron have access to cytosolic IRP-1 and modulate its activity (13). Thus, an impact of the very high food iron content on IRP-1 activity at the villus tips cannot be excluded, although the animals were starved for ≈18 h before the experiments. Finally, no measurable IRP-2 activity was detected (data not shown).
Table 3.
Duodenal 59Fe absorption data
| HFE genotype | +/+ | +/o | o/o |
|---|---|---|---|
| Iron deficient | (4) a 17.762 ± 2.611 A | (5) a 13.849 ± 3.172 A | |
| Iron adequate | (5) b 2.790 ± 0.867 A | (6) a 3.460 ± 0.980 A | (6) b 7.249 ± 4.087 B |
| Iron overload | (5) c 0.743 ± 0.181 A | (5) b 0.917 ± 0.407 AB | (6) c 1.199 ± 0.393 B |
Numbers in parentheses correspond to number of animals assayed or number of determinations performed. Groups in the same row that have no capital letter in common are significantly different from each other, as are lowercase letters, which indicate significant differences between various states of iron repletion (ANOVA; P < 0.05). Total body 59Fe uptake was determined and converted to pmol Fe/cm/min. See Materials and Methods for details.
Table 4.
IRP-1 activity in duodenal mucosa
| HFE genotype | +/+ | +/o | o/o |
|---|---|---|---|
| Iron deficient | (3) a 60 ± 12 A | (5) b 60 ± 9 A | |
| Iron adequate | (8) a 44 ± 13 A | (8) a 33 ± 9 A | (7) a 41 ± 10 A |
| Iron overload | (6) a 40 ± 24 A | (6) a 42 ± 16 A | (7) a 41 ± 11 A |
Numbers in parentheses correspond to number of animals assayed or number of determinations performed. Groups in the same row that have no capital letter in common are significantly different from each other, as are lowercase letters, which indicate significant differences between various states of iron repletion (ANOVA; P < 0.05). Values correspond to the percentage of active IRP-1 measured without 2-mercaptoethanol compared to total IRP-1 activity after addition of 2-mercaptoethanol. See Materials and Methods for details.
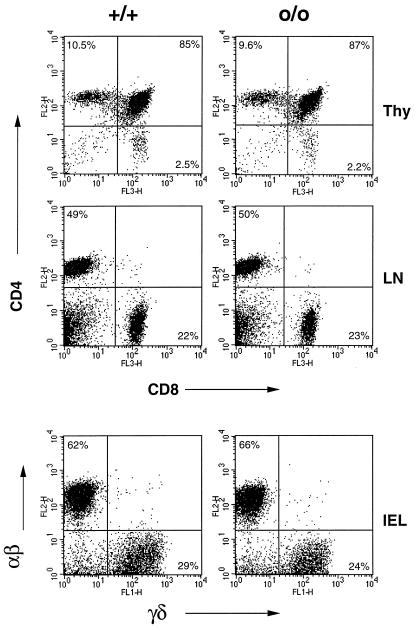
The identification of the HLA-linked hemochromatosis locus as an MHC class I molecule took both the “iron overload community” as well as immunologists off guard. First, there was no apparent need for such a structure within the well defined transferrin receptor absorption pathway, and, second, ferric homeostasis was not regarded as an immunologically relevant problem, despite pioneering work by de Sousa and colleagues who first recognized iron overload in β2m deficient mice and subsequently reported quantitative anomalies in peripheral CD8+ T cells in some hemochromatotic patients (27, 28). The availability of HFEo/o mice provides the ultimate tool to analyze a putative role for HFE in immunity, impossible to perform with β2m-deficient animals because these fail to produce any (or very few) CD8+ T cells due to their lack of classical MHC class I molecules or obviously in human HC patients (beyond peripheral blood analysis). We initially did a general assessment of T and B cell populations in the bone marrow, thymus, spleen, and lymph nodes in HFEo/o and HFE+/+ littermates and found no major perturbations (data not shown). Because HFE is a MHC class I molecule, we then focused on T cells; an extensive phenotyping of various T-cell compartments is summarized in Table 5, and representative FACS data are reported in Fig. 3. Within the thymus, the number and percent of the four major populations, double negative (CD4−CD8−), double positive (CD4+CD8+) and single positive (CD4+ and CD8+), were comparable in HFE-deficient and wild-type animals (5–22 weeks of age). In addition, thorough analysis of the double negative population revealed no differences in the early stages of T cell development (data not shown). A decrease in peripheral CD8+ T cell number was not observed in the HFEo/o mice; in the lymph nodes, there were equal numbers of CD4+ and CD8+ T cells in HFEo/o, HFE+/o, and HFE+/+ animals. Perhaps the most relevant anatomical site with regard to HFE function is the gut epithelium because this is where HFE is mainly expressed; in the proximal intestine, this seems to be the location where iron absorption is geared to the demand (12, 13, 17). As the digestive tract carries a unique population of intraepithelial lymphocytes (IELs), mainly T cells (29), alterations in these populations could have been possible in HFEo/o mice, a prospect not previously explored (16). Although the vast majority of T cells in other lymphoid organs bear the αβ TCR, up to half of all gut T cells express the γδ TCR, known to interact with other atypical MHC-I molecules. Given this, we decided to take a close look at the distribution of these cells within the small intestine (Table 5 and Fig. 3). Clearly, the IEL populations residing in the HFE-deficient gut are quite similar to those in HFEo/+ and HFE+/+ controls with respect to αβ, γδ T cells. In fact, HFE-deficient and control mice harbor similar numbers of all lymphocyte populations characteristic of the gut epithelium, including an enrichment of T cells bearing the CD8αα homodimer. In sum, the immune system does not show, at least phenotypically, any abnormality after HFE deletion. This was also true of HFEo/o and HFE+/+ mice at 1 year of age (data not shown). These observations suggest that HFE has no obvious immune-related function. Moreover, as noted above, we saw no evidence for anomalies in CD8+ T cell number, as has previously been reported for HC patients (30). This suggests that these peculiarities observed in HC patients may, at least in part, be posttherapeutic or perhaps be secondary manifestations of a more severe iron overload.
Table 5.
Immunological phenotyping of HFE-deficient mice
| Thymus | |||
|---|---|---|---|
| +/+ (n = 11) | +/o (n = 6) | o/o (n = 2) | |
| CD4−CD8− | 2.4 ± 0.9 | 2.4 ± 0.4 | 2.2 ± 0.9 |
| CD4+CD8+ | 86.5 ± 2.2 | 85.9 ± 0.7 | 86.8 ± 2.2 |
| CD4+ | 9.0 ± 0.5 | 9.4 ± 0.6 | 9.3 ± 1.4 |
| CD8+ | 2.2 ± 0.5 | 2.3 ± 0.3 | 2.1 ± 0.5 |
| Lymph node | |||
| +/+ (n = 10) | +/o (n = 6) | o/o (n = 11) | |
| CD4+ | 41.6 ± 6.9 | 41.4 ± 5.9 | 45.9 ± 7 |
| CD8+ | 17.1 ± 5 | 17.1 ± 5.8 | 16.6 ± 4.7 |
| Gut intraepithelial lymphocytes | |||
| +/+ (n = 7) | +/o (n = 6) | o/o (n = 8) | |
| IELs, ×10−6 | 3.3 ± 2.2 | 2.7 ± 1.4 | 2.0 ± 1.3 |
| CD3+IELs, ×10−6 | 3.0 ± 2.0 | 2.3 ± 13 | 1.8 ± 1.2 |
| Percent of CD3+ | |||
| αβ TCR+ | 58.8 ± 14.3 | 56.9 ± 11.3 | 60.2 ± 12.6 |
| γδ TCR+ | 40.8 ± 13.0 | 39.9 ± 10.7 | 37.9 ± 17.0 (7) |
| Vδ4+ | 5.3 ± 1.7 (4) | Not determined | 6.8 ± 0.9 (4) |
| (Percent of γδ) | (15.5 ± 3.9) | (20.4 ± 4.3) | |
| CD8αα+ | 48.1 ± 16.1 | 49.4 ± 19.1 | 36.8 ± 14.0 |
| CD8αβ+ | 31.5 ± 17.5 | 32.1 ± 15.6 | 42.9 ± 15.5 |
| CD4+ | 5.3 ± 3.2 | 5.5 ± 2.2 | 7 ± 4.1 |
| CD4+CD8+ | 4.3 ± 3.5 | 4.8 ± 5.6 | 3.2 ± 2.1 |
Mice included in this table were 5–22 weeks of age. Data are expressed in percent unless otherwise noted.
Figure 3.
HFE-deficient mice harbor normal number of various T cell subpopulations. Shown is a representative FACS analysis of lymphocytes isolated from HFEo/o and HFE+/+ littermates (13 weeks of age). CD4/CD8 staining of thymocytes and lymph node cells is shown above and αβ (TCRβ)/γδTCR staining of IELs below. Total cell numbers isolated from each mouse were similar, and the percentages of relevant populations are noted.
In conclusion, careful and in-depth analysis of HFE deletion in vivo establishes an animal model faithfully mimicking human hemochromatosis in key diagnostic areas. These include an elevated plasma iron concentration and a raised transferrin saturation as well as increased iron accumulation in the liver. Moreover, the model allows measurements of a number of parameters obviously unachievable in humans and not previously performed in other animal models. Paramount among these is the clear augmentation of duodenal iron absorption. Finally, a comprehensive phenotyping of the immune system reveals no detectable anomalies within various T cell compartments. Enfin, backcrossing our mutation on several mice strains displaying distinct differences in iron metabolism should help decipher the genetic heterogeneity of HC and help identify non-MHC linked iron regulatory loci (31).
Acknowledgments
We thank Marco Colonna and Louis Du Pasquier for critical reading of this manuscript. We also thank E. Wagner, W. Metzger, and colleagues for superb mouse care; U. Mueller and C. Westphal for blastocyst injection; R. Ceredig for help with the initial FACS analysis; and W. Marston for technical assistance. This work was supported by the Association pour la Recherche sur le Cancer, Ligues Départementales (67-68) contre le Cancer, Action Concertée Incitative Blanche du Ministère de l’Éducation Nationale, de la Recherche et de la Technologie (S.B.), Grant 31-37729.93 of the Swiss National Foundation (L.K.), and the Basel Institute for Immunology, which was founded and is supported by F. Hoffmann-La Roche, Basel, Switzerland (S.G.).
Abbreviations
- HC
hemochromatosis
- β2m
β2 microglobulin
- kb
kilobase
- TfR
transferrin receptor
- IRP
iron regulatory protein
- IEL
intraepithelial lymphocyte
References
- 1.Sheldon J H. Haemochromatosis. London: Oxford Univ. Press; 1935. [Google Scholar]
- 2.Simon M, Bourel M, Fauchet R, Genetet B. Gut. 1976;17:332–334. doi: 10.1136/gut.17.5.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 4.Feder J N, Tsuchihashi Z, Irrinki A, Lee V K, Mapa F A, Morikang E, Prass C E, Starnes S M, Wolff R K, Parkkila S, et al. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 5.Riegert P, Gilfillan S, Nanda I, Schmid M, Bahram S. Immunogenetics. 1998;47:174–177. doi: 10.1007/s002510050344. [DOI] [PubMed] [Google Scholar]
- 6.Parkkila S, Waheed A, Britton R S, Feder J N, Tsuchihashi Z, Schatzman R C, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:2534–2539. doi: 10.1073/pnas.94.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastin J M, Jones M, O’Callaghan C A, Schimanski L, Mason D Y, Townsend A R. Br J Haematol. 1998;103:931–941. doi: 10.1046/j.1365-2141.1998.01102.x. [DOI] [PubMed] [Google Scholar]
- 8.Lebron J A, Bennet M J, Vaughn D E, Chirino A J, Snow P M, Mintier G A, Feder J N, Bjorkman P J. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 9.Dorak M T, Burnett A K, Worwood M. Immunol Cell Biol. 1994;72:435–439. doi: 10.1038/icb.1994.64. [DOI] [PubMed] [Google Scholar]
- 10.Parkkila S, Waheed A, Britton R S, Bacon B R, Zhou X Y, Tomatsu S, Fleming R E, Sly W S. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kühn L C. Trends Biochem Sci. 1999;24:164–166. doi: 10.1016/s0968-0004(99)01386-9. [DOI] [PubMed] [Google Scholar]
- 12.Fleming R E, Migas M C, Zhou X, Jiang J, Britton R S, Brunt E M, Tomatsu S, Waheed A, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:3143–3148. doi: 10.1073/pnas.96.6.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schümann K, Moret R, Künzle H, Kühn L C. Eur J Biochem. 1999;260:362–372. doi: 10.1046/j.1432-1327.1999.00155.x. [DOI] [PubMed] [Google Scholar]
- 14.Garboczi D N, Biddison W E. Immunity. 1999;10:1–7. doi: 10.1016/s1074-7613(00)80001-1. [DOI] [PubMed] [Google Scholar]
- 15.Santos M, Schilham M W, Rademakers L H, Marx J J, de Sousa M, Clevers H. J Exp Med. 1996;184:1975–1985. doi: 10.1084/jem.184.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumann K, Elsenhans B, Forth W. Am J Physiol. 1999;276:G431–440. doi: 10.1152/ajpgi.1999.276.2.G431. [DOI] [PubMed] [Google Scholar]
- 18.El Guindi M, Skikne B S, Covell A M, Cook J D. Am J Clin Nutr. 1988;47:37–41. doi: 10.1093/ajcn/47.1.37. [DOI] [PubMed] [Google Scholar]
- 19.Raja K B, Simpson R J, Peters T J. Biochim Biophys Acta. 1987;901:52–60. doi: 10.1016/0005-2736(87)90255-0. [DOI] [PubMed] [Google Scholar]
- 20.Simpson R J. J Nutr. 1996;126:1858–1864. doi: 10.1093/jn/126.7.1858. [DOI] [PubMed] [Google Scholar]
- 21.Leibold E A, Munro H N. Proc Natl Acad Sci USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müllner E W, Neupert B, Kühn L C. Cell. 1989;58:373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- 23.Poussier P, Julius M H. In: Immunology Methods Manual: The Comprehensive Sourcebook of Techniques. Lefkovits I, editor. San Diego: Academic; 1997. pp. pp.1509–1513. [Google Scholar]
- 24.Lebron J A, Bjorkman P J. J Mol Biol. 1999;289:1109–1118. doi: 10.1006/jmbi.1999.2842. [DOI] [PubMed] [Google Scholar]
- 25.Flanagan P R, Hajdu A, Adams P C. Hepatology. 1995;22:828–832. [PubMed] [Google Scholar]
- 26.Hentze M W, Kühn L C. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Sousa M, Reimao R, Lacerda R, Hugo P, Kaufmann S H, Porto G. Immunol Lett. 1994;39:105–111. doi: 10.1016/0165-2478(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 28.De Sousa, M. (1992) Mem. Inst. Oswaldo Cruz87,S 5, 23–29. [DOI] [PubMed]
- 29.Lefrancois L. J Immunol. 1991;147:1746–1751. [PubMed] [Google Scholar]
- 30.Arosa F A, Oliveira L, Porto G, da Silva B M, Kruijer W, Veltman J, de Sousa M. Clin Exp Immunol. 1997;107:548–554. doi: 10.1046/j.1365-2249.1997.d01-967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonnell S M, Hover A, Gloe D, Ou C Y, Cogswell M E, Grummer-Strawn L. Am J Med. 1999;107:30–37. doi: 10.1016/s0002-9343(99)00163-1. [DOI] [PubMed] [Google Scholar]