Abstract
Microarray is a powerful tool to compare the gene expression of different tumour specimens and cell lines simultaneously and quantitatively. To get a better insight into genes that are involved in uveal melanoma tumorigenesis, we compared the gene expression profiles of 12 different uveal melanoma cell lines with three melanocyte cell cultures obtained from healthy donor eyes. Gene expression profiles were obtained by nylon filter arrays, containing 1176 gene spots related to cancer development. The expression levels of selected genes were validated on cell lines and primary uveal melanomas by real time RT–PCR, and were subsequently included in cluster analysis. Four candidate tumour markers, Laminin Receptor 1, Endothelin 2, Von Hippel Lindau Binding protein 1 and Cullin 2, have been selected from genes that were differentially expressed in the uveal melanoma cell lines compared to the normal uveal melanocytes. In primary uveal melanomas, these four markers could discriminate between two classes of uveal melanoma, which may be indicative of a differential disease process.
Keywords: uveal melanoma, gene expression, microarray
Up- and downregulated genes can be important determinants of tumour development. To understand the biological behaviour of tumour growth and progression, it is important to know how genes are expressed in malignancies, compared to normal tissue.
Uveal melanoma is the most common primary intraocular tumour of the eye in adults. The incidence ranges from six to eight cases per one million subjects per year among Caucasians. Approximately, 50% of patients with large uveal melanoma die from metastases (Diener-West et al, 1992).
The well-known prognostic parameters for uveal melanoma are clinico-histopathological parameters cell type, tumour diameter, tumour localisation, mitotic rate and the presence of vascular networks (Mooy and de Jong, 1996). Other defined factors are, for instance, the expression of the epidermal growth factor receptor (Hurks et al, 2000) and the reduced CDKN2A expression caused by promoter hypermethylation (van der Velden et al, 2001). In spite of these markers, metastases can still not be detected in an early stage. The mortality rates, after metastasis has occurred, are very high and hardly any treatment is effective for uveal melanoma metastases. Therefore, better insight into tumour progression and tumour dissemination is still of utmost importance.
Information regarding genes that are involved in the tumour growth of uveal melanoma and in the development of metastases is frequently derived from studies that relate biological parameters of tumour progression to prognostic outcome. For instance, loss of chromosome 3 and gain of the q arm of chromosome 8 are significantly associated with poor survival and loss of 1p is a specific hallmark of primary uveal melanoma metastases (White et al, 1998; Aalto et al, 2001).
Recently, cDNA microarray studies have become a powerful tool to detect differences in gene expression in different kinds of tumours (Ross et al, 2000), and have subsequently demonstrated that expression profiling can serve as a prognostic tool (Alizadeh et al, 2001; van de Vijver et al, 2002).
In this study, we used filter arrays with more than a thousand cDNAs related to cancer development, to reveal genes that are differentially expressed in uveal melanoma cell lines compared to cultured normal human melanocytes. In order to validate potential progression markers, real-time quantitative RT–PCR analysis on differentially expressed genes was used in cell lines and in fresh frozen primary uveal melanoma specimens.
An important role in uveal melanoma development is now suggested for Laminin Receptor 1 (LAMR1), Endothelin 2 (ET2), Von Hippel Lindau Binding Protein 1 (VBP1) and Cullin 2 (CUL2) genes, since their expression levels discriminate between two classes of uveal melanoma.
MATERIALS AND METHODS
Cell lines and tumour characteristics
A total of 15 cell lines were included in this study. Nine cell lines were derived from primary choroidal uveal melanomas (Mel202, Mel285, Mel270, Mel290, 92.1, 92.2, OCM1, OCM3 and OCM8), three cell lines were derived from uveal melanoma metastases (Omm1.3 and Omm1.5 are liver metastases from Mel270, and OMM1 was obtained from a subcutaneous metastasis) (Luyten et al, 1996). The cell lines OCM1, OCM3 and OCM8 were kindly provided by Dr Kan-Mitchell (Kan-Mitchell et al, 1989). Cell lines Mel202, Mel285, Mel290, Mel270 and the two cell lines derived from liver metastases were provided by Professor BR Ksander (Schepens Eye Institute, Boston, MA, USA). Cell lines 92.1 and 92.2 were derived from the same primary tumour and were established in our own laboratory (Waard-Siebinga et al, 1995).
Three melanocyte cell cultures were grown from three normal human donor eyes of two different individuals (Melcyt1A, Melcyt1B and Melcyt2). After the corneae of these eyes were prepared for donor purposes, the remaining tissues were kindly provided by the Netherlands Ophthalmic Research Institute (Eye Bank, IOI, Amsterdam). The freshly obtained eyes were rinsed with PBS and superfluous tissue was removed. The eye was cut into two parts, and the part containing the iris and lens was discarded. The vitreous body and retina of the remaining part were removed. After incubation in 0.125% trypsin (Gibco, Paisley, Scotland) in 0.02% EDTA during 1 h at 37°C the retinal pigment epithelium was removed. The choroid was peeled off the sclera and incubated in 0.25% trypsin during 18 h at 4°C. After the addition of culture medium, the cells were collected by centrifugation and plated. Geneticin (Gibco) was added to the culture medium to a final concentration of 100 μg ml−1 to eliminate contaminating cells, like fibroblasts. The cell cultures were characterised by immunohistochemistry for S100 and NKI/beteb (Vennegoor et al, 1988).
The melanoma cell lines were cultured in RPMI 1640 (Gibco) medium and supplemented with 3 mM L-glutamine (Gibco), 2% penicillin/streptomycin and 10% FBS (Hyclone, UT). The three melanocyte cell lines were grown in F12 medium (Gibco), following the directions of Hu et al (1993). All cell cultures were incubated at 37°C in a humidified 5% CO2 atmosphere.
The fresh frozen uveal melanoma samples were derived from 19 different primary choroidal melanomas. The research protocol followed the Tenets of the Declaration of Helsinki. Histopathological analysis was performed by an ocular pathologist. Table 1 presents the clinical and pathological characteristics of the fresh frozen uveal melanoma samples. We are limited in survival analysis due to follow-up data of three years.
Table 1. Clinical and pathological characteristics of 19 fresh frozen primary uveal melanoma samples.
| Tumour sample | Cell type | Prominence (mm) | Diameter (mm) | Pigment | Mitosisa | Intravascular ingrowth | Vascular loops | Metastases | Location |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Epithelioid | 8 | 14 | >50% | 3 | Yes | Yes | No | Chorb |
| 2 | Spindle | 10 | 12 | >50% | 2 | No | Yes | No | Chor |
| 3 | Mixed | 7 | 15 | >50% | 3 | No | Yes | Yes | Cbd-chorc |
| 4 | Spindle | 11 | 11 | <50% | 5 | No | Yes | No | Cbd-chor |
| 5 | Mixed | 9 | 14 | >50% | 5 | Yes | No | No | Cbd-chor |
| 6 | Mixed | 12 | 17 | <50% | 8 | No | Yes | No | Chor |
| 7 | Spindle | 9 | 16 | <50% | 2 | No | Yes | No | Cbd-chor |
| 8 | Mixed | 8 | 14 | Severe | NAd | No | Yes | No | Chor |
| 9 | Mixed | 8 | 17 | <50% | 5 | Yes | Yes | No | Cbd-chor |
| 10 | Mixed | 9 | 16 | >50% | 4 | No | Yes | No | Cbd-chor |
| 11 | Mixed | 13 | 20 | >50% | 7 | No | Yes | No | Cbd |
| 12 | Spindle | 11 | 20 | >50% | 20 | No | Yes | No | Cbd |
| 13 | Mixed | 13 | 18 | 50% | 21 | Yes | Yes | No | Cbd-chor |
| 14 | Mixed | 13 | 20 | <50% | 20 | No | NA | No | Chor |
| 15 | Mixed | 6 | 18 | >50% | 1 | No | No | No | Cbd-chor |
| 16 | Mixed | 11 | 20 | 50% | 21 | Yes | No | No | Cbd-chor |
| 17 | Mixed | 8 | 21 | <50% | 6 | No | Yes | No | Chor |
| 18 | Mixed | 8 | 20 | Severe | 9 | Yes | Yes | No | Cbd-chor |
| 19 | Mixed | 12 | 20 | 50% | 10 | No | Yes | No | Cbd-chor |
Number of mitoses per 15 fields of microscopic examination at a magnification of × 320.
Chor=choroidea.
Cbd=ciliar body.
NA=not available.
Microarray cDNA hybridisation and analysis
Cells from early passages were grown to subconfluency and the total RNA of each cell line was isolated with the RNeasy kit, following the directions of the manufacturer (Qiagen, Hilden, Germany). Biotin-labelled oligo-dT and streptavidin-labelled magnetic beads were used to isolate the mRNA from the total RNA. Probe synthesis and hybridisation were performed according to the protocol of the manufacturer of the Human Cancer Atlas 1.2 filters (Clontech, Palo Alto, CA, USA). The cDNA was labelled with α32P-dATP. Each cell line was hybridised to two separate filters in order to obtain a technical replicate. In this study, the melanocyte cell lines were used as references and controls. Cell lines Melcyt1A and Melcyt1B are derived from two different eyes of one person in order to create an internal biological control.
A nylon Human Cancer Atlas filter consists of 1176 different cDNA spots designed to reflect molecular pathways in oncogenesis and several house-keeping genes.
To analyse and quantify the spots obtained by the phosphorimager scans, we used the GenePix® Pro analyzing computer program (Axon Instruments, Inc., Union City, CA, USA). Each cell line was normalised to the value of the uveal melanocyte, Melcyt1A. The obtained ratios were analysed by a hierarchical clustering method (Cluster) to compare the expression data. In this type of clustering method, the data points are forced into a strict hierarchy of nested subsets, resulting in a phylogenetic tree with branch lengths that indicate the degree of similarity between the samples (Tamayo et al, 1999). The data obtained by these analysis programs were visualised by TreeView (Eisen et al, 1998; Tavazoie et al, 1999).
Real-time quantitative RT–PCR analysis
To validate the results of the microarray, we performed real-time quantitative RT–PCR analyses on the cell lines and primary frozen tumour samples (Heid et al, 1996). The same method and protocol of the RNeasy kit was applied to isolate total RNA from sections of the fresh frozen melanoma samples. In all 1 μg of total RNA of all cell lines and tumour samples was reverse-transcribed with M-MLV reverse transcriptase (Gibco-BRL, Eggenstein, Germany) in a 20 μl reaction volume. The cDNA synthesis was performed in the presence of random hexamere primers at a temperature of 42°C for 50 min (Promega, San Louis Obispo, CA, USA). We applied the real-time PCR method using the SYBR® Green PCR Master Mix as supplied by the manufacturer (PE Applied Biosystems, Inc., Foster City, CA, USA). The SYBR® Green I dye detects double-stranded DNA, not requiring specific probes. The Mix includes a passive reference, which is required for signal normalisation. To minimise nonspecific product formation, the reaction Master Mix contains the enzyme AmpliTaq Gold® DNA polymerase. An ABI PRISM 7700 Sequence Detection System monitors the fluorescence signal during the PCR reaction. It registers the number of cycles at which the signal reaches a certain threshold level. The differences between the threshold value of the target transcripts and the endogenous references are used to calculate the relative expression.
The primers for the four genes under study, ET2, LAMR1, VBP1 and CUL2, and the two endogenous controls U1A and RPS11 were designed with the Primer Express software (PE Applied Biosystems) (Table 2). All reactions were repeated twice with two different endogenous controls to ensure reproducibility of the results. For comparing the efficiency of amplification of the gene targets and the controls, a serial dilution was made of each different cDNA sample. All experiments revealed a nearly optimal efficiency close to 100%.
Table 2. Primer sequences of the genes validated in the real-time quantitative RT–PCR assays.
| Primer | Primer (forward) | Primer (reverse) |
|---|---|---|
| U1A | 5′-gca gct tat gcc agg aca gat-3′ | 5′-ttg gtg agg aac aag atg tga ttc-3′ |
| RPS11 | 5′-aag cag ccg acc atc ttt ca-3′ | 5′-cgg gag ctt ctc ctt gcc-3′ |
| ET2 | 5′-tgg agg ctg cgt cct ga-3′ | 5′-ctg agg cat gcg tgt tcg t-3′ |
| LAMR1 | 5′-cca ctg gag cca ctc caa tt-3′ | 5′-ggc tgc ctg gat ctg gtt agt-3′ |
| VBP-1 | 5′-gac cag att ctt gct ggc aga t-3′ | 5′-agc ccc caa cca cag aca c-3′ |
| CUL-2 | 5′-gtt cgt atc atg aaa gca cga aaa-3′ | 5′-tta aac cta gct ctt gac tgg cta atc-3′ |
Statistical analysis
Correlations between clinico-histopathological parameters and the expression level of the four genes were determined using the Pearson correlation test.
RESULTS
Microarray analysis
In this study, the expression profile of 12 different human uveal melanoma cell lines was compared to three uveal melanocyte cultures. The mRNA of all cell cultures was hybridised in replicate to filter arrays containing 1176 cDNAs related to tumorigenesis. The duplicate experiments revealed nearly identical hybridisations that clearly showed the fingerprints of each cell line. Nevertheless, a small percentage of genes were differentially expressed between the duplicates. Only the few genes with opposite results were discarded from analysis. The gene expression of each cell line was normalised to one melanocyte cell culture (Melcyt1A). A total of 449 genes revealed no expression in all 15 cell lines, that is, their expression was not significantly higher than the local background. Only 52 genes were expressed in all cell cultures, that is, melanomas and melanocytes. In total, 54 genes were expressed only in melanoma cell lines and 187 genes exclusively in the melanocytes.
In at least eight melanoma cell lines, 29 genes were expressed that were not expressed in any of the three normal melanocyte cultures (not significantly higher than the local background). Fifteen of these genes were highly differentially expressed, that is, we considered a gene to be highly expressed if the expression ratio of an uveal melanoma cell line was at least 1.5-fold higher compared to the normal uveal melanocytes Melcyt 1B and Melcyt 2 (Table 3). From this list, we selected four genes revealing the highest percentage of uveal melanoma cell lines displaying an increase in the expression ratio compared to the controls (Table 3).
Table 3. Microarray results: selection of genes revealing high differential expression ratios in uveal melanoma cell lines normalised to uveal melanocyte 1A (Melcyt1A).
| Name | Melcyt1B | Melcyt2 | Mel270 | Omm1.3 | Omm1.5 | Mel202 | Mel285 | Mel290 | 92.1 | 92.2 | Ocm1 | Ocm3 | Ocm8 | Omm1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAMR1 | 0.87 | 1.11 | 3.62 | 6.66 | 4.72 | 2.15 | 2.40 | 2.28 | 7.24 | 6.33 | 6.30 | 3.04 | 3.21 | 4.47 |
| ET2 | 1.18 | 1.39 | 26.68 | 34.92 | 42.63 | 1.18 | 0.92 | 1.34 | 3.67 | 3.47 | 50.11 | 8.32 | 4.02 | 36.48 |
| CUL2 | 1.16 | 1.15 | 1.05 | 1.14 | 1.17 | 1.69 | 1.81 | 3.69 | 3.82 | 4.35 | 10.27 | 6.53 | 1.47 | 6.90 |
| VBP1 | 1.09 | 1.42 | 0.89 | 0.94 | 1.01 | 2.19 | 3.52 | 9.64 | 6.52 | 4.75 | 3.87 | 3.23 | 2.02 | 2.64 |
| RHOC | 1.13 | 1.28 | 2.66 | 3.77 | 3.27 | 1.47 | 1.48 | 1.62 | 2.25 | 2.41 | 4.43 | 3.08 | 3.58 | 3.18 |
| PKCI1 | 1.21 | 1.46 | 5.1 | 3.28 | 1.71 | 2.41 | 3.34 | 5.16 | 8.3 | 5.71 | 4.32 | 3.43 | 3.55 | 4.85 |
| TIMP1 | 0.83 | 0.96 | 3.03 | 9.90 | 15.24 | 1.07 | 2.20 | 1.80 | 2.01 | 1.85 | 4.05 | 2.96 | 2.82 | 4.06 |
| SOD1 | 0.89 | 1.06 | 2.21 | 0.84 | 0.65 | 1.94 | 3.30 | 2.48 | 5.40 | 5.73 | 3.16 | 1.91 | 2.94 | 2.01 |
| HMGIY | 0.89 | 0.78 | 3.93 | 1.95 | 0.81 | 1.62 | 2.92 | 1.18 | 2.47 | 2.04 | 2.27 | 1.35 | 1.52 | 1.79 |
| RPS5 | 0.76 | 0.80 | 3.18 | 6.01 | 3.51 | 1.58 | 1.77 | 1.09 | 4.50 | 2.79 | 2.13 | 1.44 | 2.33 | 1.22 |
| CCND1 | 0.89 | 1.06 | 1.39 | 0.85 | 1.07 | 2.35 | 3.52 | 1.69 | 2.37 | 1.84 | 2.12 | 3.40 | 3.35 | 1.69 |
| IL13 | 1.19 | 1.17 | 5.19 | 2.70 | 2.92 | 1.22 | 0.99 | 0.93 | 3.01 | 4.08 | 177.83 | 17.15 | 6.01 | 24.37 |
| EIF3-beta; EIF3B | 0.99 | 0.73 | 2.71 | 1.15 | 1.06 | 2.80 | 2.87 | 1.55 | 1.85 | 5.01 | 1.60 | 1.27 | 1.83 | 1.14 |
| ETV4; E1A-F | 1.17 | 0.82 | 2.20 | 1.25 | 1.17 | 1.54 | 0.92 | 1.00 | 1.94 | 2.21 | 34.47 | 7.70 | 24.75 | 5.99 |
| CDC37 homolog | 0.99 | 0.92 | 2.91 | 1.38 | 1.16 | 2.01 | 2.05 | 1.39 | 3.81 | 4.35 | 2.98 | 1.56 | 1.53 | 1.44 |
Furthermore, the common origin of the primary tumour cell line Mel 270 and its two liver metastatic cell lines Omm1.3 and Omm1.5 was perfectly demonstrated by their similar expression profiles (Table 3). Hierarchical cluster analysis revealed their close relationship in a phylogenetic cluster tree. The similarity could be clearly recognised in the tree by the short branches reflecting their degree of correlation. The latter was also apparent for cell lines 92.1 and 92.2, both derived from the same primary tumour. The genetic origin of these cell lines appears to be the major determinant of expression, since the lines resemble each other more than any of the other cell lines, indicating that these cell lines have kept their identity during in vitro culturing and their reliability as a model for tumour research. This importance of genetics in gene expression was also demonstrated by Cheung et al (2003).
Real-time quantitative PCR analysis
Cell lines
ET2, LAMR1, VBP1 and CUL2 belong to the group of genes exhibiting the most discriminating differential expression ratio in uveal melanomas cell lines compared to the expression ratio of the normal melanocytes (Table 3). We validated the differential expression of these genes by real-time RT–PCR. Similar expression of the two endogenous controls U1A (small nuclear protein) and RPS11 (ribosomal protein) excluded possible influences of regulation.
The absolute expression levels of the four genes were markedly different. The average expression of CUL2 was 64 times higher compared to the average expression level of ET2, VBP1 showed 512 times higher and LAMR1 4096 times higher than ET2.
Despite differences in absolute expression levels obtained by filter array and real time RT–PCR, both assays separately confirmed the importance of these genes.
Comparing the expression data between the filter array and the real-time PCR, real time RT–PCR confirmed a high expression level of ET2 in two of the 12 cell lines. The VBP1 gene revealed a high expression in nine out of 12 cell lines vs seven out of 12 cell lines in the PCR assay, with corresponding results between both assays in the case of four cell lines (Mel285, Ocm3, Ocm8 and 92.1). The same trend was observed in the expression pattern of CUL2; in the microarray eight out of 12 cell lines revealed a high expression compared to normal melanocyte expression, whereas in the PCR assay nine out of 12 cell lines fulfilled this criterion.
LAMR1 expression in the microarray was very consistent with its expression measured by real-time PCR. In both assays we observed low expression in the melanocytes and cell lines Ocm8, Mel202 and Mel290, whereas the cell lines 92.1, Ocm1, Omm1 and Omm1.3 revealed high expression levels. This is of great interest, since two of these cell lines with a high expression level of LAMR1 are derived from metastases.
Fresh frozen primary uveal melanoma samples
In order to validate the candidate tumour markers obtained in cell lines, we performed real-time quantitative PCR analysis on 19 fresh frozen uveal melanoma samples.
The genes LAMR1 and especially ET2 revealed highly differential expression levels, whereas the expression levels of the genes CUL2 and VBP1 were not markedly highly differential.
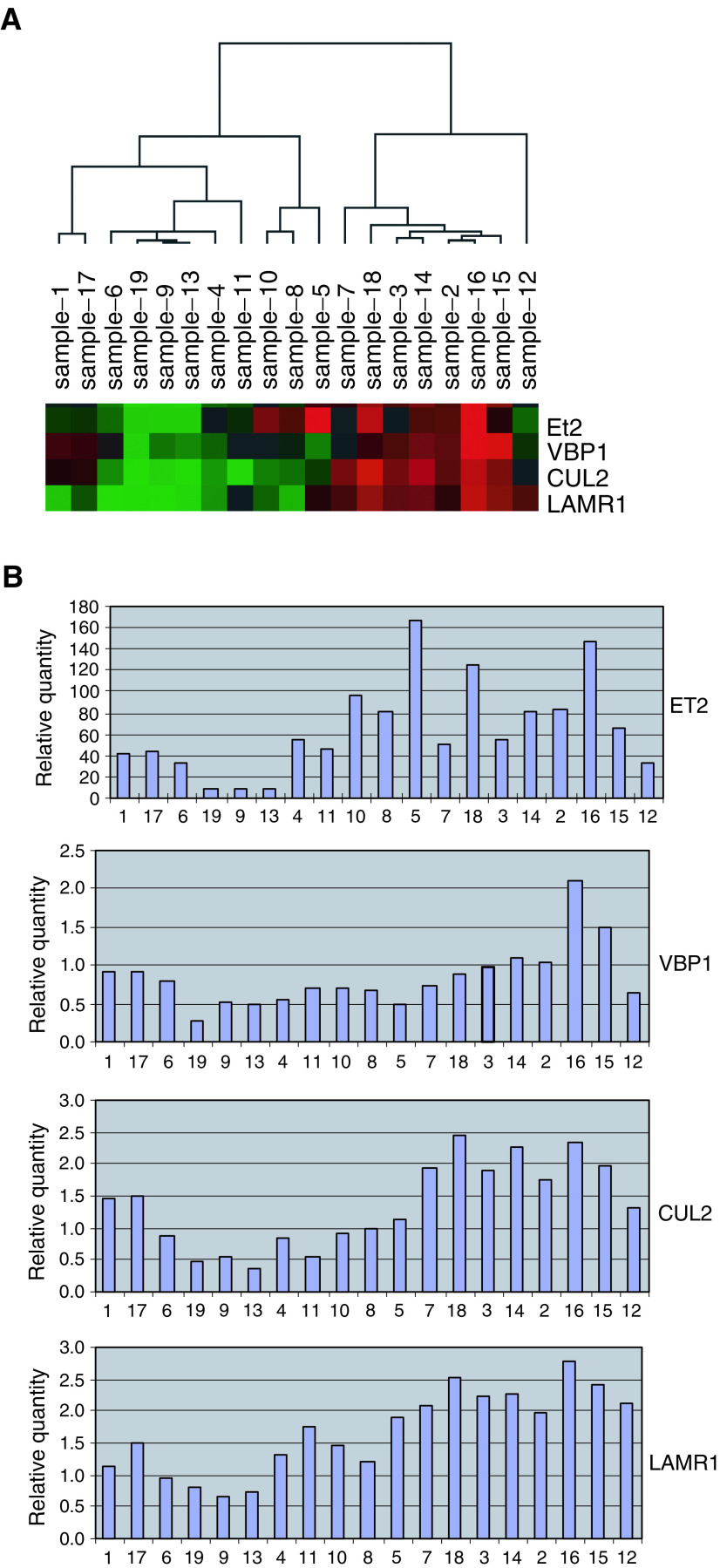
Cluster analysis of the real-time PCR results of the genes under study revealed two distinct clusters in the primary tumour samples (Figure 1). The first cluster was formed by eight samples revealing a relatively high expression of the genes ET2, LAMR1, CUL2 and VBP1. The relatively low expression levels of all these genes were observed in a second cluster of 11 tumour samples.
Figure 1.
(A) Cluster figure (Treeview) of the real-time RT–PCR results of fresh frozen uveal melanoma samples. Red indicates a relatively high expression level and green indicates a relative low expression level. (B) Column graphs presenting the relatively quantity of expression per gene. The samples are set in the same order as in the cluster figure. The differences in the relative expression levels between the two clusters of the uveal melanoma samples are demonstrated in these graphs as well.
Pearson test identified one significant correlation between gene expression and histopathological parameters of the tumours; ET2, LAMR1 and VBP1 were positively correlated with PAS-positive loops (P=0.005, 0.028 and 0.01, respectively).
We were not able to perform a solid survival analysis, since all tumours were collected recently.
DISCUSSION
We used microarray technology and real-time RT–PCR to compare gene expression profiles of uveal melanoma cell lines and normal melanocytes, in order to identify potential tumour progression markers.
Based on the type and purpose of our array technique, we validated a selection of four genes, that is, LAMR1, ET2, VBP1 and CUL2, with high differential expression by quantitative real-time RT–PCR. In addition, we performed real-time RT–PCR of these four genes in a panel of primary uveal melanomas. Hierarchical clustering of these data resulted in two distinct subgroups with differences in the expression level.
Between the two assays, there is a difference in expression level of the selected genes. The higher the average expression level, the stronger the correlation with the microarray results. The filter technique applied in this study was not able to detect genes with a low expression level securely enough to obtain a valid estimate of the expression differences; nevertheless, it appeared to be a useful method to identify a combination of genes that might play a role in uveal melanoma development.
The expression patterns of the genes VBP1 and CUL2 exhibited a small overlap in both assays and revealed no elevated expression levels in the primary tumour samples. Both genes are Von Hippel-Lindau (VHL)-related genes. VHL regulates transcription elongation and is known to act as a tumour suppressor protein. CUL2 is a member of the cullin family of proteins and binds to the VHL/elongin B/elongin C complex (VBC). The members are believed to target certain cellular proteins for ubiquitination and degradation and are in some cases involved in cell cycle regulation (Pause et al, 1997; Clifford et al, 1999). VBP1 forms complexes with the VHL gene product. VHL acts as a molecular chaperone that carries VBP1 from perinuclear granules to the nucleus or cytoplasm (Tsuchiya et al, 1996). In spite of the highly differential expression ratios of these two genes in the microarray, the expression levels in the primary uveal melanomas were not high. Nevertheless, they seem to have a discriminating role in the primary uveal melanomas.
The microarray data displayed differential expression of ET2 in nine of the 12 cell lines. However, real-time PCR confirmed an upregulated ET2 expression in only two of the 12 melanoma cell lines. On the other hand, all 19 tumour samples revealed a high expression of ET2. The validation of ET2 in the cell lines would not consider this gene to be of importance for uveal melanoma development. However, several studies provided abundant evidence for ET2 in cancer development. The NCI60 Cancer Microarray project described a high expression level of the endothelin receptor type B (ENRB) in skin melanoma (Ross et al, 2000), and Demunter et al (2001) reported evidence for its role as a tumour progression marker in this tumour type.
ENRB is a nonspecific receptor for all three isotypes of endothelin: ET1, ET2 and ET3 (Arai et al, 1990; Sakurai et al, 1990). ET2 can act as a mitogen in many cancer systems, and ETs in general stimulate melanocyte proliferation and differentiation (Reid et al, 1996). Recently, Smith et al (2002) associated decreased ENRB expression in large primary uveal melanoma with early clinical metastasis and short survival. These results, together with the high expression levels of ET2 in our tumour samples, might support investigation of the expression of endothelin receptors and their ligands in uveal melanoma.
LAMR1 revealed the most interesting results. Combining both assays we observed at least a 2.5-fold higher LAMR1 expression ratio in eight uveal melanoma cell lines (including all metastatic cell lines Omm1.3, Omm1.5 and Omm1) compared to the basic expression ratio of normal melanocytes. Furthermore, eight of the 19 primary tumour samples revealed relatively high expression levels of LAMR1, including the only patient in whom metastases had already developed. Interestingly, LAMR1 is significantly correlated with the PAS-positive loops, containing laminin (Thies et al, 2001). LAMR1 is known for its role in cancer proliferation, invasion and metastasis (Satoh et al, 1992; Castronovo, 1993). Hipfel et al (2000) observed a high expression of this laminin receptor in skin melanoma but not in naevi and normal skin. Despite the lack of survival data in our study, an interesting relationship between the level of LAMR1 expression and metastatic potential might be present, encouraging further exploration of the role of LAMR1 in the development of uveal melanoma metastases.
Validation of the four genes on the primary melanoma samples resulted in two distinct subgroups with specific expression profiles. Besides LAMR1, the histopathologic parameter PAS-positive loops revealed a significant association with the expression level of the genes ET2 and VBP1. The ability to form vascular loops is an important prognostic parameter in uveal melanoma metastases development (Folberg et al, 2001). Therefore, these two clusters within these samples might have a prognostic value in tumour dissemination.
In conclusion, filter microarray is a strong tool to compare the gene expression profiles of cell lines of different origin, but careful attention should be paid to the quantitative value of this filter method in the case of genes with low expression levels. By dint of this powerful technique, we identified discriminating fingerprints, and subsequently defined tumour markers possibly involved in uveal melanoma tumour development and progression, such as ET2 and LAMR1.
Acknowledgments
We thank Tonny CJM Kröse and Remco Dijkman for their technical assistance and helpful discussions. PA van der Velden is supported by the foundation ‘De drie Lichten’. W Zuidervaart is supported by ‘Rotterdamse Vereniging Blindenbelangen’ and ‘Landelijke Stichting voor Blinden en Slechtzienden’. FA Van Nieuwpoort is supported by ‘Netherlands Organisation and Scientific Research’.
References
- Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S (2001) Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci 42: 313–317 [PubMed] [Google Scholar]
- Alizadeh AA, Ross DT, Perou CM, van de RM (2001) Towards a novel classification of human malignancies based on gene expression patterns. J Pathol 195: 41–52 [DOI] [PubMed] [Google Scholar]
- Arai H, Hori S, Aramori I, Ohkubo H, Nakanishi S (1990) Cloning and expression of a cDNA encoding an endothelin receptor. Nature 348: 730–732 [DOI] [PubMed] [Google Scholar]
- Castronovo V (1993) Laminin receptors and laminin-binding proteins during tumor invasion and metastasis. Invasion Metastasis 13: 1–30 [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS (2003) Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33: 422–425 [DOI] [PubMed] [Google Scholar]
- Clifford SC, Walsh S, Hewson K, Green EK, Brinke A, Green PM, Gianelli F, Eng C, Maher ER (1999) Genomic organization and chromosomal localization of the human CUL2 gene and the role of von Hippel-Lindau tumor suppressor-binding protein (CUL2 and VBP1) mutation and loss in renal-cell carcinoma development. Genes Chromosomes Cancer 26: 20–28 [PubMed] [Google Scholar]
- Demunter A, Wolf-Peeters C, Degreef H, Stas M, van den Oord JJ (2001) Expression of the endothelin-B receptor in pigment cell lesions of the skin. Evidence for its role as tumor progression marker in malignant melanoma. Virchows Arch 438: 485–491 [DOI] [PubMed] [Google Scholar]
- Diener-West M, Hawkins BS, Markowitz JA, Schachat AP (1992) A review of mortality from choroidal melanoma. II. A meta-analysis of 5-year mortality rates following enucleation, 1966 through 1988. Arch Ophthalmol 110: 245–250 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folberg R, Chen X, Boldt HC, Pe’er J, Brown CK, Woolson RF, Maniotis AJ (2001) Microcirculation patterns other than loops and networks in choroidal and ciliary body melanomas. Ophthalmology 108: 996–1001 [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6: 986–994 [DOI] [PubMed] [Google Scholar]
- Hipfel R, Schittek B, Bodingbauer Y, Garbe C (2000) Specifically regulated genes in malignant melanoma tissues identified by subtractive hybridization. Br J Cancer 82: 1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DN, McCormick SA, Ritch R, Pelton-Henrion K (1993) Studies of human uveal melanocytes in vitro: isolation, purification and cultivation of human uveal melanocytes. Invest Ophthalmol Vis Sci 34: 2210–2219 [PubMed] [Google Scholar]
- Hurks HM, Metzelaar-Blok JA, Barthen ER, Zwinderman AH, Wolff-Rouendaal D, Keunen JE, Jager MJ (2000) Expression of epidermal growth factor receptor: risk factor in uveal melanoma. Invest Ophthalmol Vis Sci 41: 2023–2027 [PubMed] [Google Scholar]
- Kan-Mitchell J, Mitchell MS, Rao N, Liggett PE (1989) Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci 30: 829–834 [PubMed] [Google Scholar]
- Luyten GP, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, van Drunen E, Vuzevski V, de Jong PT, Luider TM (1996) Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer 66: 380–387 [DOI] [PubMed] [Google Scholar]
- Mooy CM, de Jong PT (1996) Prognostic parameters in uveal melanoma: a review. Surv Ophthalmol 41: 215–228 [DOI] [PubMed] [Google Scholar]
- Pause A, Lee S, Worrell RA, Chen DY, Burgess WH, Linehan WM, Klausner RD (1997) The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA 94: 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K, Turnley AM, Maxwell GD, Kurihara Y, Kurihara H, Bartlett PF, Murphy M (1996) Multiple roles for endothelin in melanocyte development: regulation of progenitor number and stimulation of differentiation. Development 122: 3911–3919 [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, van de RM, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24: 227–235 [DOI] [PubMed] [Google Scholar]
- Sakurai T, Yanagisawa M, Takuwa Y, Miyazaki H, Kimura S, Goto K, Masaki T (1990) Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature 348: 732–735 [DOI] [PubMed] [Google Scholar]
- Satoh K, Narumi K, Sakai T, Abe T, Kikuchi T, Matsushima K, Sindoh S, Motomiya M (1992) Cloning of 67-kDa laminin receptor cDNA and gene expression in normal and malignant cell lines of the human lung. Cancer Lett 62: 199–203 [DOI] [PubMed] [Google Scholar]
- Smith SL, Damato BE, Scholes AG, Nunn J, Field JK, Heighway J (2002) Decreased endothelin receptor B expression in large primary uveal melanomas is associated with early clinical metastasis and short survival. Br J Cancer 87: 1308–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci 96: 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Thies A, Mangold U, Moll I, Schumacher U (2001) PAS-positive loops and networks as a prognostic indicator in cutaneous malignant melanoma. J Pathol 195: 537–542 [DOI] [PubMed] [Google Scholar]
- Tsuchiya H, Iseda T, Hino O (1996) Identification of a novel protein (VBP-1) binding to the von Hippel- Lindau (VHL) tumor suppressor gene product. Cancer Res 56: 2881–2885 [PubMed] [Google Scholar]
- van der Velden PA, Metzelaar-Blok JA, Bergman W, Monique H, Hurks H, Frants RR, Gruis NA, Jager MJ (2001) Promoter hypermethylation: a common cause of reduced p16(INK4a) expression in uveal melanoma. Cancer Res 61: 5303–5306 [PubMed] [Google Scholar]
- Vennegoor C, Hageman P, Van Nouhuijs H, Ruiter DJ, Calafat J, Ringens PJ, Rumke P (1988) A monoclonal antibody specific for cells of the melanocyte lineage. Am J Pathol 130: 179–192 [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil D, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Waard-Siebinga I, Blom DJ, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, Danen EH, Jager MJ (1995) Establishment and characterization of an uveal-melanoma cell line. Int J Cancer 62: 155–161 [DOI] [PubMed] [Google Scholar]
- White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE (1998) Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer 83: 354–359 [PubMed] [Google Scholar]