Abstract
Vaccination leads to dramatic perturbations of the environment of parasite populations and this can have both demographic and evolutionary consequences. We present a theoretical framework for modelling the short- and long-term epidemiological and evolutionary consequences of vaccination. This framework integrates previous theoretical studies of vaccine-induced parasite evolution, and it allows one to make some useful qualitative predictions regarding the outcome of the competition between different types of vaccine-favoured variants. It can also be used to make quantitative predictions about the speed of such evolutionary processes. This work may help define the relevant parameters that need to be measured in specific parasite populations in order to evaluate the potential evolutionary consequences of vaccination. In particular, we argue that more work should be done evaluating the nature and magnitude of parasite fitness costs associated with adaptation to vaccinated hosts.
Keywords: evolution, epidemiology, vaccination, virulence
1. Introduction
The use of vaccines results in a profound alteration of the environment in which parasites live. Indeed, the goal of vaccination is to protect individual hosts and consequently decrease parasite prevalence. Ultimately, this may even lead to the eradication of the disease (Fenner et al. 1988). These epidemiological consequences of vaccination have received a considerable amount of attention, both from an empirical and a theoretical standpoint (Anderson & May 1991; McLean & Blower 1995; Earn et al. 2000; Rohani et al. 2000; Tildesley et al. 2006). One of the most significant conceptual developments from this research is the finding that there is a critical vaccination coverage above which a parasite can be driven to extinction, and that this coverage is typically less than 100% (i.e. herd immunity; Anderson & May 1991). Evolutionary ecologists would not find it surprising that the large epidemiological perturbations caused by vaccination also result in considerable changes in the way that natural selection acts on parasite populations. Nevertheless, studies of these effects have not yet been integrated into a coherent theoretical context. In fact, there are two largely separate bodies of research on this question (e.g. van Boven et al. 2005; Restif & Grenfell 2006), and each addresses different aspects of how vaccination affects parasite, and thus disease, evolution.
The first line of enquiry focuses on the so-called ‘escape’ mutants and is directed towards understanding how vaccination selects for parasite strains that are able to evade the protective effects of the vaccine (McLean 1995a, 1999; Wilson et al. 1998, 1999; Scherer & McLean 2002; van Boven et al. 2005). Since there is often substantial genetic variation in antigenic reactivity among parasite strains (Frank 2002), vaccination will select for those strains that are able to evade the vaccine-induced immunological response mounted by hosts. The primary interest of this research is in designing vaccines and vaccination protocols that minimize the risk of spread of such escape mutants.
The second line of enquiry focuses on the so-called ‘virulence’ or ‘life-history’ mutants and is directed towards understanding how vaccination causes evolutionary changes in the extent to which a parasite harms its host (i.e. evolutionary changes in its virulence; Gandon et al. 2001, 2003; van Boven et al. 2005; André & Gandon 2006; Ganusov & Antia 2006). The central premise behind this research is that virulence evolves as a result of constraints among parasite life-history characteristics, and that vaccination can alter the form of these constraints, thereby causing evolutionary changes in virulence.
Aside from the biological dichotomy set-up in the two above-mentioned bodies of research, there have also been important differences in the nature of the theoretical analyses used to address these two questions. Studies of escape mutants typically focus on short-term evolutionary change (McLean 1995a, 1999; Wilson et al. 1998, 1999; Scherer & McLean 2002; van Boven et al. 2005). This approach assumes that only two parasite strains are competing: the wild-type strain and the escape mutant. The objective is then to analyse the conditions under which the escape mutant can invade a wild-type population. Indeed, once the mutant is able to invade, this approach usually assumes that it will spread towards fixation.
By contrast, studies of virulence mutants typically focus on long-term evolutionary outcomes and do not pay much attention to short-term, transient, evolutionary dynamics (Gandon et al. 2001, 2003; André & Gandon 2006; Ganusov & Antia 2006). Most such studies allow for mutation to produce a whole continuum of different strain types, each of which is characterized by the disease life-history parameters that it induces upon infection (i.e. transmission rate, recovery rate and virulence). The objective is then to analyse the evolutionary equilibrium values of these quantitative traits, particularly those of virulence.
In this article, we attempt to place all of the above conceptual developments within a common theoretical framework, in order to better understand the connections among them, and to develop a more holistic perspective on the evolutionary epidemiology of vaccination. Our contention is that the above-mentioned dichotomy of escape versus virulence mutants is not the most fruitful categorization, and that all of these previous results are more profitably viewed as different aspects of the same epidemiological evolutionary process. We acknowledge that different types of genetic variants can arise and spread in the face of vaccination, but we suggest that, from an epidemiological standpoint, these are better classified by the disease life-history parameters that they induce (i.e. transmission rate, recovery rate and virulence; defined below), rather than classifying them as escape versus virulence mutants.
We begin by showing how previous theoretical studies of escape mutants and virulence evolution are related to one another, by placing both within a common epidemiological setting. Most of these previous studies have assumed that the evolutionary dynamics occur much more slowly than the epidemiological dynamics. This is probably often not the case for many parasites, however, and therefore we develop a more comprehensive theoretical framework for the evolutionary epidemiology of vaccination that relaxes this assumption. In §3, we use this framework to demonstrate that previous studies of both escape mutants and virulence evolution have missed some interesting and potentially important aspects of how vaccination affects parasite evolution, because they have ignored the possibility that evolutionary change occurs during transient epidemiological dynamics.
2. Theoretical development
Before getting into the formalization of vaccine-driven evolution, we lay out a clear and unambiguous language for discussing and comparing various examples. For all parasites considered in this article, we simplify matters by ignoring the possibility of multiple strains infecting the same host. First, this rules out the possibility of within-host evolution (although we discuss ways to introduce this complexity at the end of the article). Second, each parasite strain is characterized by three critical life-history characteristics that are displayed during an infection of either a naive or a vaccinated host: its transmission rate (i.e. the per capita rate at which infected individuals generate new infections per available susceptible host); its recovery rate (i.e. the per capita rate at which infected individuals recover from infection); and its virulence (i.e. the per capita rate at which infected individuals die from infection). For the sake of simplicity, we assume these three traits to be constant throughout the infectious period. How relaxing this assumption may affect the evolutionary outcome is discussed in Day (2003).
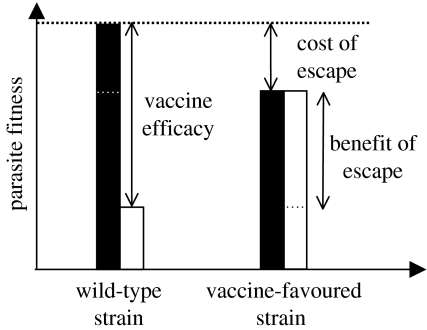
We refer to the predominant strain (or strains) present prior to vaccination as the ‘wild-type’ strain and to strains that are selectively favoured in vaccinated hosts as ‘vaccine-favoured variants’ (figure 1). The selective advantage of a vaccine-favoured variant arises from the differences in one or more of its three life-history parameters compared with the wild-type, when measured in a vaccinated host (i.e. its transmission rate, recovery rate and/or virulence in a vaccinated host). All else equal, increased transmission rate, reduced recovery rate or reduced virulence in the vaccinated host might confer the selective advantage to the vaccine-favoured variant. For example, in the dichotomy mentioned above, escape mutants might be viewed as variants that have reduced recovery rates in the vaccinated hosts (relative to the wild-type) owing to the changes in epitope. Similarly, life-history or virulence mutants might be viewed as vaccine-favoured variants that have an increased transmission in vaccinated hosts owing to an increased rate of replication. More generally, however, vaccine-favoured variants can differ from the wild-type in all three life-history parameters, and these parameters need not all differ in a direction that increases the variant's fitness (pathogen's fitness is defined in §2.2 and §2.3). All that is required is that the combined effect of the changes results in higher fitness in vaccinated hosts than the wild-type. We will also typically suppose that vaccine-favoured variants suffer some fitness cost in naive hosts (figure 1), since otherwise they would probably already have reached appreciable frequencies in the absence of vaccination. As with the benefit enjoyed by a vaccine-favoured variant, the cost paid in naive hosts also arises from differences in one or more of its three life-history components compared with the wild-type, when measured in naive hosts.
Figure 1.
Schematic of the comparison between a wild-type strain and a vaccine-favoured variant. The lifetime reproductive success (a relevant measure of fitness at endemic equilibrium) is plotted in naive hosts (black) and in vaccinated hosts (white) for both strains. The efficacy of the vaccine can be evaluated by the reduced performance of wild-type strain in vaccinated hosts. The cost of adaptation to vaccination is evaluated in naive hosts, while its benefit is evaluated in vaccinated hosts (in both cases, relative to the performance of the wild-type strain). When the population is away from the endemic equilibrium, one may use the selection coefficients (see equations (2.11a) and (2.11b)) for each strain to obtain relevant measures of fitness.
We now develop a mathematical framework that allows us to integrate previous theoretical studies of the epidemiological and evolutionary effects of vaccination. Our goal with this framework is not to treat the specifics of any particular disease, but rather to develop some simple ‘toy models’ that allow us to clearly draw out the conceptual connections among these previous lines of enquiry. Our rationale is that these same conceptual issues and connections will then arise in other theoretical treatments that are more specifically tailored to particular diseases.
We start with an analysis of a simple epidemiological model to illustrate the potential transitory epidemiological dynamics following the start of a vaccination campaign. Then, we use this epidemiological model to illustrate the connection between previous studies of escape mutants and virulence evolution in response to vaccination. As mentioned in the introduction, these previous studies have relied on a classical separation of time-scales approach that decouples epidemiological and evolutionary dynamics. In the latter part of this article, we then go on to develop a more general theoretical framework for the evolutionary epidemiology of vaccination that encompasses both types of questions that have been addressed previously, and that also tracks both the epidemiological and evolutionary dynamics simultaneously.
2.1 Epidemiological dynamics
We use a classical SIR model, modified to include imperfect vaccination (Scherer & McLean 2002). The host can be either susceptible (S), infected (I) or recovered (R). Vaccination adds another level of heterogeneity among the hosts. Susceptible and infected hosts can be either naive or vaccinated (indicated with a subscript N or V, respectively). All recovered hosts (vaccinated or not) are assumed to be fully immune to reinfection, and therefore the different types of recovered hosts (vaccinated or not) are pooled in a single host class. In this development of the epidemiological model, we assume the parasite population to be monomorphic (evolution of polymorphic parasite populations will be analysed in the following subsections). This yields the following set of differential equations:
| (2.1) |
The rate of arrival of new susceptible hosts in the population (immigrants and newborns) is λ. Among those individuals, a proportion p is vaccinated. Susceptible hosts become infected with rates and when they are naive or vaccinated, respectively. The rates of infection depend on the densities of infected hosts (IN and IV) and on the parasite transmission rates βij from host i to host j (where i and j can be either naive, N, or vaccinated, V). Uninfected hosts have a mortality rate δ, and infected hosts suffer extra mortality due to the presence of the parasite (i.e. parasite virulence). Parasite virulence may differ between naive and vaccinated hosts (αN and αV, respectively). Recovery rates may also differ between these two hosts (γN and γV, respectively).
A trivial equilibrium of equations (2.1) is the case where the parasite is absent (i.e. ) and where , (the ‘hat’ refers to parasite-free equilibrium). The stability of this equilibrium depends on the ability of a parasite to invade a fully susceptible population, which is given by the basic reproduction ratio of the parasite (McLean & Blower 1995; Dushoff 1996; Gandon et al. 2003)
| (2.2) |
where and are the basic reproduction ratios in unvaccinated and 100% vaccinated host populations, respectively. Parasite invasion of a fully susceptible host population will occur whenever R0>1. This condition can also be used to derive the critical vaccination coverage leading to disease eradication
| (2.3) |
When the vaccination coverage is above this threshold, the parasite cannot survive in the host population and is driven to extinction.
Alternatively, the above model can be used to study the dynamics of a population where the parasite has reached an endemic equilibrium. Before vaccination starts, if , the parasite will spread and reach an endemic and stable equilibrium (the ‘overbar’ refers to the endemic equilibrium)
Let us now assume that a vaccine becomes available and is used in this population. If vaccination coverage is above pc, then the parasite will be driven to extinction. For example, the intense vaccination campaign launched by the World Health Organization in 1967 led to the global eradication of smallpox virus Variola major in 1977 (Fenner et al. 1988) by this process. However, equation (2.3) can be used to see that, when parasites have a large basic reproduction ratio and when available vaccines function poorly (i.e. when is also relatively large), eradication is not feasible (because pc is very large). In all these cases, when p<pc, vaccination leads to a new endemic equilibrium with a reduced incidence of the disease (figure 2). Note, however, that in more complex models where the risk of severe complications after infection depends on the age at infection (e.g. rubella, poliomyelitis), increasing vaccination coverage can increase the incidence of severe cases (Anderson & May 1983, 1991).
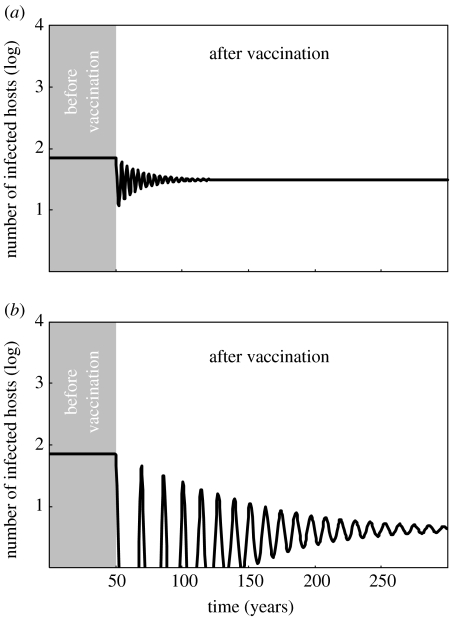
Figure 2.
Epidemiological dynamics (incidence through time, in years) of a typical childhood disease (e.g. measles in an industrialized country) after the start of vaccination at t=50. Numerical simulations of equation (2.1) with the following parameter values: βij=29.10−4 (where i and j can be either naive, N, or vaccinated, V); δ=0.02; γN=γV=26; and αN=αV=0. In this model, we assume the total host population size to be constant (i.e. the flux of new hosts, λ, exactly balances the death rate) and equal to N=105. These parameter values yield a basic reproduction ratio of R0=11.15. Here, we assume the vaccine to be perfect. The critical vaccination coverage is thus (see equation (2.3) in the main text) pc=0.91. In (a) p=0.5 and in (b) p=0.85.
Before reaching the new endemic equilibrium, the disease incidence will oscillate during a transient phase (figure 2). Interestingly, when coverage is close to the vaccination threshold, the system may rapidly reach very low incidence without leading the parasite towards extinction (Anderson & May 1982; McLean & Anderson 1988; Scherer & McLean 2002). After this ‘honeymoon period’, several epidemics will occur (damped oscillations) before the system settles to the new endemic equilibrium (figure 2b). These transient dynamics have obvious epidemiological consequences, and this deserves more detailed analysis with specific models in order to identify vaccination strategies that minimize the risk of post-honeymoon epidemics (Rouderfer et al. 1994; McLean 1995b). As we will demonstrate later, such transient dynamics can have important evolutionary implications as well.
2.2 Previous analyses of the evolutionary consequences of vaccination
Most studies of the spread of escape mutants or the evolution of virulence in response to vaccination have assumed that epidemiological dynamics are very fast relative to evolution. This separation of time-scales derives from explicit models of population dynamics and population genetics (Beck 1984; Andreasen & Christiansen 1995; Andreasen 2002) where the phenotypic variation among the different genotypes is small (i.e. small mutation steps). It is also at the heart of game-theoretic analyses and adaptive dynamics (Geritz et al. 1998; Waxman & Gavrilets 2005).
To conduct the analysis, the parasite population is assumed to reach an endemic epidemiological equilibrium before mutation introduces genetic variation (new mutant strains with potentially different phenotypic traits). At this endemic equilibrium, the expected number of new infections produced by a randomly chosen infected host during its total infectious period (its lifetime reproductive success) is, under the simplifying assumption that βN=βNN=βNV and βV=βVV=βVN (Gandon 2004),
| (2.4) |
Since the number of infected hosts remains constant at endemic equilibrium, this necessarily yields R=1. Then, one may formalize evolution by focusing on the competition between a rare mutant strain (with different phenotypic traits ) and the resident strain at the endemic equilibrium set by the resident. The mutant will initially spread if and only if its lifetime reproductive success, R*, is higher than 1 (i.e. the lifetime reproductive success of the resident). Therefore, a condition for the mutant to invade the population is
| (2.5) |
Whether the mutant replaces the resident strain depends on the fate of the resident when it becomes rare. This can be determined by looking at the resident's lifetime reproductive success at the endemic equilibrium set by the mutant strain (denoted below by the symbol |*)
| (2.6) |
If R*>1 and R|*>1, then the mutant will initially increase in frequency but will not go to fixation (because the fitness of the resident increases when the mutant becomes frequent). Consequently, the mutant and the resident will coexist in the population (Porco & Blower 1998, 2000; André & Gandon 2006; Restif & Grenfell 2006). In contrast, when R*>1 and R|*<1, the mutant will replace the resident strain.
The above description of evolutionary dynamics has been used to describe the spread of escape mutants after a vaccination campaign (McLean 1995a, 1999; Scherer & McLean 2002). The approach is to begin by supposing that the wild-type is still present after the introduction of the vaccine, and then to suppose that a mutant arises with reduced recovery rates and/or increased transmission rate (owing to its ability to evade the vaccine-induced immunological response of the host). Parasite strains are characterized by their reproduction ratios, and one then asks whether or not the vaccine-favoured mutant will spread, as a function of the costs incurred by the mutant in naive hosts, as well as the level of vaccine coverage in the population. If such a vaccine-favoured mutant does spread, then one can use the epidemiological model to determine how the critical vaccine coverage required is thereby affected.
The above description of evolutionary dynamics has also been used to characterize virulence evolution as a result of vaccination (Gandon et al. 2001, 2003; André & Gandon 2006; Ganusov & Antia 2006). Again the approach is to begin by supposing that the wild-type is still present after the introduction of the vaccine and that a mutant arises with altered life-history components. Typically, it is supposed that the mutant's life-history components satisfy some constraint (e.g. a positive relationship between transmission rate and virulence). Again one then asks whether or not such mutants can spread. The ultimate question in this type of analysis, however, is not whether a particular mutant can spread, but rather, whether or not there is a vaccine-favoured strain that, once present in the population, can resist invasion by all other possible mutant strains. Indeed, since the direction of evolution is given by the sign of R*−R, a strain that maximizes R* is evolutionarily stable (because no other strategy can invade it). The goal then is to characterize the life-history parameters (e.g. transmission rate, recovery rate and virulence) that maximize R* in the presence of vaccination, and to compare these to the life-history parameters of the wild-type. For example, Gandon et al. (2001, 2003) used this approach to show that vaccines limiting transmission have little effect on evolution, while vaccines reducing the within-host growth rate of the parasites may favour more virulent strains (see also §3.2).
The above epidemiological model illustrates the conceptual connections between theoretical studies of escape mutants and of virulence/life-history evolution. In both situations, the analysis relies on the use of the lifetime reproductive success (i.e. R*) as a measure of parasite fitness. This is based on the assumption that the system reaches an epidemiological equilibrium after vaccination before any of the mutant strains of the parasite arise. This is clearly unlikely when the transient dynamic following the start of the vaccination is long (figure 2b) and/or for parasites with high mutation rates. Therefore, it is of interest to know how relaxing this assumption affects the evolutionary predictions. In §2.3, we develop a more general theoretical framework that addresses both escape and virulence evolution in response to vaccination, and that allows evolutionary change to occur on any time-scale relative to that of the epidemiological dynamics.
2.3 A general framework for the evolutionary epidemiology of vaccination
In the framework developed below, the parasite population may be polymorphic at any given time. In particular, we assume that n different parasite strains are competing within a heterogeneous host populations composed of naive and vaccinated individuals (recall that, for the sake of simplicity, multiple infections are not allowed; see Day & Proulx 2004 for extensions to such cases). We use and to denote the frequencies of parasite strain i infecting naive and vaccinated hosts, respectively (where the superscripts N and V refer to the different types of hosts). We also assume that mutation occurs with rate μ in all strains. A strain j mutates to a strain i with probability mji. These strains may differ in one or several life-history traits (e.g. transmission, virulence or recovery) and, consequently, their relative growth rates in the population.
Under these assumptions, the epidemiological dynamics depends on the mean parasite trait values in the two types of hosts
| (2.7) |
where, for instance, the mean levels of virulence are and .
Equation (2.1) can be viewed as a special case of equations (2.7) when there is no variance among strains in their transmission, virulence or recovery. Following Day & Gandon (2006, eqns 2.7 and 2.10), it is also possible to derive the rate of change in frequency of a given strain i in both types of hosts (see also appendix A)
| (2.8a) |
The quantities rAB(i) in equations (2.8a) link these evolutionary equations to the epidemiological equations (2.7) and are given by
| (2.8b) |
where the subscripts refer to the route of transmission. For example, rNV(i) is the per capita rate of production of new infections of strain i in vaccinated individuals, by naive hosts infected with strain i. Note also that in equations (2.7), (2.8a) and (2.8b), the averages and are calculated over the distribution .
Equations (2.8a) and (2.8b) can be used to better understand the three different forces acting on the evolution of parasite strain frequencies in both types of hosts (selection, mutation and migration). For example, let us focus on the evolution of the parasite population infecting the subpopulation of naive hosts (the first equation in equation (2.8a) and (2.8b)). The first term, , refers to the action of natural selection in naive hosts. For example, strain i will increase in frequency in naive hosts if it has a higher growth rate than the average in the naive host subpopulation. The second term, , refers to the action of mutation. Whether this will result in an increase or a decrease of strain i frequency depends on all strain frequencies, as well as on the mutation model. The third term, , refers to the effect of immigration (i.e. transmission) of parasites from the subpopulation of vaccinated hosts into naive hosts. This effect is weighted by the relative size of the two subpopulations of hosts (because the effect of immigration on strain frequency depends on the relative size of the two subpopulations) and depends on the difference between the frequency of strain i in the two subpopulations. For example, if the frequency of the focal strain is higher in parasites infecting vaccinated hosts, then immigration (transmission) from this subpopulation to naive hosts will increase the frequency of this strain in naive hosts. The fourth term, , is also due to immigration from the other subpopulation and illustrates that such transmission can have evolutionary consequences even if the frequency of strain i does not differ between the two subpopulations. Indeed, this fourth term expresses the fact that the frequency of strain i in naive hosts may increase if this strain is overrepresented among immigrants from the population of parasites infecting the vaccinated hosts (Day & Gandon 2006). For example, if strain i is more transmissible than other strains, the immigration (i.e. transmission) from vaccinated to naive hosts will increase the frequency of strain i in naive hosts even if the two subpopulations do not initially differ in strain frequencies.
The above equations will now be useful to study the two evolutionary questions addressed in the previous analyses of vaccination. First, if we are particularly interested in a given parasite strain (e.g. one vaccine-favoured variant or quasispecies with one strain—the master sequence—being much fitter than the others; see Day & Gandon 2006), then we can use these equations to track the change in frequency of this focal parasite strain. Second, if we are interested in the evolution of life-history components of the parasite, such as the level of transmission, recovery or virulence, then we can use these equations to track the evolution of the mean value of these phenotypic traits in the parasite population, allowing for any number of strains to be present at any given time.
2.3.1 Tracking the frequency of a focal strain
We use the above model to track the frequency of a focal strain (among the n possible strains) which is , where and are the frequencies of the focal strain in the two types of hosts and I=IN+IV is the total number of infected hosts. In the following, refers to the per capita rates of production of new infections by the focal strain. The average per capita rate of production of new infections by the remaining strains is . In particular, note that when the focal strain is competing against a unique resident strain, this simply equals the per capita rate of production of new infections induced by the resident strain . We will further assume that μ* is the rate of mutation of the focal strain (towards remaining strains) and μ is the rate of mutation of the remaining strains towards the focal strain.
Equations (2.8a) and (2.8b) can then be used to describe the change in frequency of the focal strain in the different types of hosts
| (2.9a) |
where and are the variances in the focal strain frequency in the parasite populations infecting naive and vaccinated hosts, respectively, and
| (2.9b) |
The dynamics of the global frequency of the focal strain (i.e. averaged over both naive and vaccinated hosts) is thus
| (2.10) |
where fN=IN/I and fV=IV/I are the proportions of the parasite that are in naive and vaccinated hosts, respectively.
The first two terms in equation (2.10) simply express the fact that the focal strain frequency may change owing to a change in its frequencies in each of the two host types. The third term in equation (2.10) shows that, even if the focal strain frequency does not change, a change in the proportion of the infections in each of the two types of hosts may also affect its total frequency, provided its frequencies differ between host types. Consider, for example, the situation where vaccinated hosts are only infected by the focal strain (qV=1), while none of the naive hosts are infected by this strain (qN=0). In this situation, if there is a relative increase in the prevalence of naive hosts, the total frequency of the focal strain will decrease.
To complete the model, we can combine equations (2.9a), (2.9b) and (2.10) to obtain
| (2.11a) |
where
| (2.11b) |
In other words, sN and sV measure the intensity of selection on the focal strain in naive and vaccinated hosts, respectively. Not surprisingly, the first two terms in equations (2.11a) and (2.11b) show that the speed of evolution depends on the intensity of selection and on the variance in frequency of the focal strain. The following two terms describe the effect of mutation. The final term expresses the potential impact of migration between the two types of hosts, but also the influence of epidemiological dynamics through . Equation (2.7) can then be used to expand this final component. In §3, we will demonstrate how this general framework can provide new insights into the spread of vaccine-favoured variants as a result of vaccination.
2.3.2 Phenotypic evolution
Instead of focusing on a particular strain frequency, one may be interested in the evolution of the mean value of some phenotypic trait. Each of the competing strains may have different life-history traits, xi (where x may refer to transmission, virulence or recovery) and we are interested in the evolution of the mean life-history trait and of the parasite infecting these two types of hosts.
In each generation, mutations push the average value of the trait in host A towards . But selection within the different types of hosts and migration between types of hosts also affect the evolution of average phenotypic values. Following Day & Gandon (2006, eqns 2.9 and 2.10), we can derive equations for the overall evolutionary dynamics as
| (2.12) |
Equation (2.12) is a multiple habitat version of the Price equation (Price 1970; Day & Gandon 2006). It summarizes the interplay between the main factors (already identified in equation (2.8a) and (2.8b)) acting on phenotypic evolution: selection (the first term); mutation (the second term); and migration and selection within the migrant pool (the final term). We can also average over the two types of host to focus on the evolution of the overall mean trait
| (2.13) |
where
where is the phenotypic variance in the population infecting host type A, while is the regression coefficient between the per capita growth rate rAB and the phenotype of individuals infecting host type A. The new variables and are thus analogous to σNsN and σVsV, respectively, in equation (2.11a) and measure the variance of the phenotypic trait times the intensity of selection, in the different types of habitats (i.e. hosts).
Note that equation (2.13) can be used to track the transient evolutionary dynamics of traits, and it can also be used to find the evolutionarily stable trait values. Indeed, at an evolutionary equilibrium, we necessarily have . For example, if we assume that the mutation rate is low and, consequently, the difference in mean trait values between the two types of hosts is low, then we get the following condition at evolutionary equilibrium: . This condition means that, at evolutionary equilibrium, selection acting on one habitat is perfectly balanced by selection on the other habitat.
3. Examples
In this section, we illustrate the potential utility of the above general framework for addressing questions about the spread of specific vaccine-favoured variants, as well as about the evolution of virulence in response to vaccination. In both the cases, we use the same underlying epidemiological model (equation (2.1)). In the first evolutionary scenario, mutation introduces three vaccine-favoured variants that have relatively large differences in their life histories (in terms of virulence, transmission or recovery). These are meant to represent different escape strategies, and we study which of these is most likely to emerge (§3.1). In the second evolutionary scenario, again mutation introduces strains with different life-history components, but here we suppose that the differences are relatively small. Furthermore, in this case, a larger number of different strains are allowed to coexist (i.e. many more than the three above vaccine-favoured variants). In this situation, we focus on the evolutionary dynamics of virulence when averaged over the distribution of parasite strains (§3.2).
3.1 Qualitative evolution
We assume that four different types of strains exist (table 1). First, the wild-type strain (with trait values α, β and γ) is characterized by the highest basic reproduction ratio in the absence of vaccination. Therefore, before the vaccination campaign, it will be the most prevalent one (the other strains will only be maintained through recurrent mutation from the wild-type). We further assume that the vaccine prevents wild-type infections with an efficacy of 99%. The three other strains represent different types of vaccine-favoured variants. They can be viewed as generalist strains because they do as well in naive as in vaccinated hosts (figure 1). This resistance, however, is assumed to be costly, and what distinguishes these three different strains is their different costs of resistance (when measured in naive hosts in comparison with the wild-type strain): strain 1 has a higher virulence; strain 2 has a lower transmission rate; and strain 3 is cleared more rapidly. Under the above assumptions, it is clear that, if vaccination coverage is large, escape strains will spread and replace the wild-type. However, which of the three escape mutants will replace the wild-type?
Table 1.
Qualitative evolution after vaccination. The first column presents the four possible types of parasites. The wild-type is the fittest before vaccination but its infectiousness is 99% lower on vaccinated hosts. The three remaining strains are resistant to the vaccine. In contrast with the wild-type, the resistant strains are equally fit on naive and vaccinated hosts. The second column gives the basic reproduction ratio of the different strains after vaccination, R*(see equation (2.5)). Here, it is assumed that the population has reached the post-vaccination endemic equilibrium set by the wild-type strain. The third column gives an approximation of selection intensity just after the start of vaccination (far from the post-vaccination endemic equilibrium, when the wild-type is the most prevalent strain). The third column thus gives a measure of the fitness of vaccine-favoured variants relative to the wild-type (figure 4b).
| parasite strains | basic reproduction ratio after vaccination, | intensity of selection |
|---|---|---|
| wild-type: α, β, γ | 0 | |
| strain 1: α*, β, γ (with α*>α) | 0.99βSV−(α*−α) | |
| strain 2: α, β*, γ (with β*<β) | 0.99β*SV−(β−β*)SN | |
| strain 3: α, β, γ* (with γ*>γ) | 0.99βSV−(γ*−γ) |
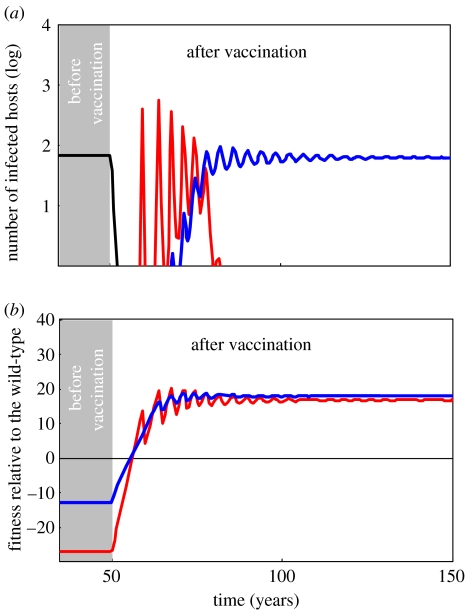
The classical evolutionary analysis (see §2.2) shows that the evolution of escape strategies can be predicted by the relative basic reproduction ratios of the different strains. The escape mutant with the highest reproductive ratio should win the competition. Figure 4 presents the results of a simulation where, for illustrative purposes, only three strains are competing (the wild-type, strain 1 and strain 2; figure 3). Mutation may occur among these strains with the same probabilities. In other words, mutation introduces some genetic variance but does not favour one strain over the other. In the long term, the basic reproduction ratio does predict the evolutionary outcome: before vaccination the wild-type wins and after vaccination strain 2 wins (because parameter values are such that the basic reproduction ratio of strain 2 is higher than strain 1). However, just after the start of the vaccination, the winner (strain 1) is not the mutant with the highest reproductive ratio.
Figure 4.
Qualitative evolution after the start of vaccination with a coverage p=0.85 at t=50 (compare with figure 2b in the absence of evolution). In (a) we plot the density (note the logarithmic scale) of the three different parasite variants, and in (b) we plot the fitness of different vaccine-favoured variants relative to the wild-type (given in the third column of table 1). In this scenario, we allow three strains to emerge by mutation (μ=10−5) in the parasite population (figure 3): (i) the wild-type (black) with α=0, β=29.10−4 and γ=26 on naive hosts (on vaccinated hosts β=29.10−6, thus the vaccine as a 99% efficacy on the infectivity of the wild-type), (ii) strain 1 (red) with α=27, β=29.10−4 and γ=26 on both naive and vaccinated hosts, and (iii) strain 2 (blue) with α=0, β=29.10−4/2 and γ=26 on both naive and vaccinated hosts. Other parameter values as in figure 1b.
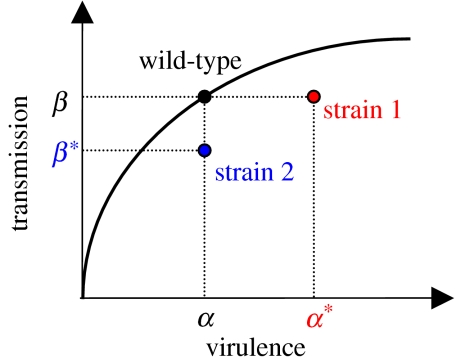
Figure 3.
Schematic description of the different strains in competition in figure 4. The figure presents the life-history traits of three parasite strains (wild-type, strain 1 and strain 2) when measured in naive hosts (table 1). In vaccinated hosts, strains 1 and 2 keep the same life-history traits, but the wild-type is assumed to have a 99% reduction in transmission (table 1).
The discrepancy between simulation results and the classical long-term evolutionary analysis can be explained using equations (2.11a) and (2.11b). The final column of table 1 gives an approximation of the intensity of selection (as defined in equation (2.11b)) for the different strains when the wild-type is the most common strain (valid just after the start of the vaccination). If we neglect the presence of the other strains, the coefficient of selection of the wild-type is 0. As for the escape mutants, it is important to note that their coefficients of selection depend on the availability of susceptible hosts and, in particular, on vaccinated hosts. Just after the start of a vaccination, there are a large number of susceptible vaccinated hosts, and this favours strategies that do not pay the cost in terms of transmission (strains 1 and 3; figure 4b). In contrast, the relative values of basic reproduction ratios of the different strains are independent of the number of susceptible hosts. This explains the short-term success of strain 1 in figure 4, despite having a lower basic reproduction ratio. Figure 4b plots the approximation of the selection coefficients of the two vaccine-favoured mutants (given in the last column of table 1) and shows that strain 1 has a selective advantage over strain 2 only in the transitory phase after vaccination. Interestingly, figure 4a may be viewed as an ecological succession. After a perturbation of the environment (vaccination), the habitat is invaded by better colonizers (strain 1) with large fecundity (transmission) and low survival (lower duration of the infection owing to higher virulence). In the long term, however, the environment reaches a new steady state where the strategies that allow more resource in survival and less in fecundity (strain 2) win.
The transitory emergence of strain 1 would not have been predicted from a classical evolutionary analysis based on R0 (with a separation of time-scales), and this type of finding regarding the transient evolutionary dynamics has important implications. With a case mortality of more than 50%, strain 1 is indeed much more virulent than the wild-type or the other escape strategy (strain 2; both of which are harmless). Furthermore, strain 1 has a higher transmission rate than strain 2, thereby causing it to spread much more quickly. Given that it might take a substantial amount of time after a vaccination programme has been introduced before the epidemiological dynamics reach a new equilibrium (if they ever, in fact, do), it seems very desirable to use a broader theoretical framework such as equations (2.11a) and (2.11b) to capture the important transient or non-equilibrium evolutionary dynamics that might occur.
3.2 Quantitative evolution
In this second evolutionary scenario, we focus on the evolution of virulence (measured in naive hosts). We assume mutation rates to be small, and therefore using equations (2.8a), (2.8b) and (2.12) yields
| (3.1) |
with (i.e. the mean virulence of parasites sampled in host type B and measured in host type A). Equation (3.1) can be used to gain some insight regarding the direction of evolution just after the start of a vaccination. In this situation, there are very few infections in vaccinated hosts (i.e. ) and the difference in the mean virulence between naive and vaccinated hosts is low (i.e. ). This results in the following approximations:
| (3.2) |
Furthermore, if the parasite population is at an evolutionary equilibrium before vaccination (i.e. before vaccination), then the following must also hold: , where is the endemic equilibrium number of susceptible hosts before vaccination.
Just after the start of the vaccination , and therefore, using the above approximations, equation (3.1) becomes
| (3.3) |
If we further assume that there is a positive covariance between virulence and transmission (a common assumption in the virulence evolution literature), the above equations show that virulence initially evolves in opposite directions in the different types of hosts. Just after the start of the vaccination, virulence decreases in naive hosts and increases in vaccinated ones. To understand this result, one needs to see the difference between the dynamics of infection in naive and vaccinated hosts. Just after the start of the vaccination campaign, most infected hosts will be from the naive subpopulation. This changes the balance between the cost and the benefits of virulence. In naive hosts, a lower number of susceptible hosts select for lower virulence (see Lenski & May 1994; Frank 1996; Day & Proulx 2004). In contrast, in vaccinated hosts, the new infections are produced by the most transmissible strains. Since we further assume a positive covariance between transmission and virulence, selection for higher transmission favours the evolution of higher virulence among vaccinated hosts.
To explore this qualitative prediction more quantitatively, we need to specify the relationships among the various traits, measured in the same or in different hosts. Following the assumptions used by Gandon et al. (2001, 2003), it is possible to run numerical simulations (figure 5) and derive approximations for the evolutionary dynamics (appendix A). Numerical simulations confirm that the direction of evolution may differ in the different types of hosts just after the start of the vaccination. This effect, however, depends on the amount of genetic variance for virulence. When it is low, this effect is relatively weak and disappears rapidly (figure 5). Some estimations of the additive genetic variance before vaccination are thus required to make short-term predictions about virulence evolution for specific infectious diseases.
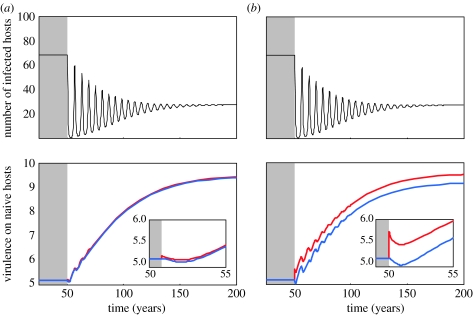
Figure 5.
Quantitative evolution after the start of vaccination with a coverage p=0.75 at t=50. In (a) and (b), we show epidemiological dynamics (number of infected hosts). In (c) and (d), we show both long- and short-term evolutionary dynamics (insert graphs show evolution 5 years after the start of vaccination) of virulence (when measured in naive hosts) of parasites infecting naive (blue) and vaccinated hosts (red). In (a) the genetic variance in virulence is 2, and in (b) the genetic variance is 20. Parameter values (see also appendix A for the notations and the functional relationships between virulence and transmission in the different types of hosts): ν1=0.9 and ν2=0.9. Other parameter values are as in figure 2b.
It is also possible to use the above framework to obtain a more comprehensive understanding of how various properties of vaccines affect the evolutionary dynamics of virulence. Gandon et al. (2001, 2003) explored this question using the classical separation of time-scales approach, and found that vaccines acting solely to diminish the mortality caused by parasites result in a higher evolutionary equilibrium level of virulence (measured in naive hosts), whereas those that act solely to diminish transmissibility result in no change.
Using ν1 to denote the proportion by which the vaccine decreases transmissibility, and ν2 to denote the proportion by which the vaccine decreases parasite-induced mortality (ν1 and ν2 are equivalent to parameters r1 and r2 in Gandon et al. 2001, 2003), the dynamics of virulence just after vaccination can be approximated as (see appendix A)
| (3.4) |
where
which equals 1 when ν2=0, and is larger than 1 otherwise.
Equation (3.4) reveals that higher values of ν2 select for higher virulence just as found in Gandon et al. (2001, 2003). It also shows, however, that higher values of ν1 select for lower virulence. This result is at odds with those of Gandon et al. (2003), and it arises as an initial transient evolutionary response to vaccination. Essentially, at the start of the vaccination campaign, the vaccine reduces the effective availability of susceptible hosts. This reduces the advantage of high transmissibility and thereby allows virulence (which is coupled to transmission) to evolve to lower levels (see also Day & Proulx 2004). Eventually, once the post-vaccination epidemiological equilibrium is reached, however, the average level of virulence will have turned around and evolved back up to its initial value prior to vaccination.
Finally, although the above framework has been developed in the context of the evolution of a single life-history component, this approach is readily extended to allow the evolution of multivariate phenotypes as well (e.g. the simultaneous evolution of virulence, transmission and/or recovery as independent, potentially genetically correlated traits; Day & Gandon 2006; appendix B).
4. Discussion
In this article, we briefly review previous attempts to model parasite evolution after a vaccination campaign, and we provide a single theoretical framework in which the different models that have been used can be analysed. Our overall contention is that it is most fruitful to focus on the life-history parameters of different strains in both naive and vaccinated hosts. Indeed, two different escape strategies may have the same basic reproduction ratio (the classical measure of fitness used in previous evolutionary studies) but very different life-history traits. We have shown that these differences can have major implications for evolution during the transient phase, after the start of the vaccination. In particular, we show that very virulent strains (with lower basic reproduction ratio) may rapidly spread after vaccination even if, in the long term, these strains can be out competed by avirulent strains with higher basic reproduction ratios (figure 4).
This example also illustrates the possibility of making predictions about short-term evolution. This contrasts with most other evolutionary analyses of the consequences of vaccination. The two case studies presented here illustrate the relevance of short-term evolutionary dynamics, since they may conflict with the long-term evolutionary outcomes. This framework may thus represent a better tool to evaluate the short-term epidemiological and evolutionary impact of vaccination.
To predict the direction and the speed of evolution, however, one needs a good measure of the diversity of life-history traits. This means having a good quantitative assessment of all three life-history parameters in both vaccinated and naive hosts (i.e. an assessment of the costs and the benefits faced by each strain). Figure 3 illustrates a hypothetical example for two of the life-history parameters as measured in naive hosts. More generally, we need this type of cost–benefit information for all three parameters (i.e. recovery rate as well) in both types of host. Ideally, this information would be obtained for as many strains as possible. One way to summarize this information on strain diversity is via the variance and the covariance between parasite life-history traits. This is analogous to estimating the G matrix in quantitative genetics studies (Lande 1982; Blows 2006), and the framework presented here clearly illustrates how this summary of the cost–benefit information can then be used to make evolutionary and epidemiological predictions (e.g. see appendix B). Without this information to feed the models, it will be difficult to evaluate the potential risks associated with vaccine-induced evolution. Besides, it is possible that even more efficient escape strategies (i.e. lower costs in naive hosts and/or higher adaptation on vaccinated hosts) will emerge after the start of the vaccination. This situation could still be analysed within this framework by allowing the G matrix to evolve. In other words, this requires pre- and post-vaccination monitoring of the parasite population.
4.1 Some limitations of the model
Despite the generality of the approach presented, it nevertheless has some important limitations. First, the model ignores the complication of within-host evolution. This is clearly unrealistic for RNA viruses like HIV and hepatitis C virus (HCV) where there is strong evidence that an arms race is going on between the virus population and the adaptive immune system of the host (Shimizu et al. 1994; Goulder & Watkins 2004). Here, the within-host evolution of the parasite is fuelled by the antigenic diversity produced by the typically high mutation rates of such RNA viruses. Within-host evolution may also occur if multiple infections are frequent, and this may be the case in malaria (de Roode et al. 2005 and references therein) and potentially many other infectious diseases. These complexities, however, could be integrated within the present framework. Day & Proulx (2004) present a way to do this in a homogeneous population (e.g. see eqn A12 in Day & Proulx 2004): , where M is the mean change of within-host virulence in a small interval of time (these changes could be either due to mutation or due to secondary infections). This model could be further extended to heterogeneous host populations and include the selective pressure imposed by the immune system on within-host evolution.
A second limitation of the present model is that it does not deal with situations where the initial population is already antigenically variable. All the theoretical case studies that we analysed assumed no pre-existing strain structure and, in other words, that the parasite population was dominated by a unique wild-type strain. However, pre-existing strain structure does occur in hepatitis B (Hsu et al. 1999), Streptococcus pneumoniae (Smith et al. 1993) and also in human malaria (Gupta 2002). To model these situations, one must first understand the emergence of strain structure before vaccination. In particular, it might be necessary to start from a host population that is already genetically or immunologically heterogeneous. Indeed, cross-specific immunity has been shown to allow the coexistence of antigenically variable strains (Gupta 2002). Vaccination could then act through the addition of another level of heterogeneity, i.e. immunological heterogeneity. Doing so would require allowing strain-specific immunity in the theory, and this represents an interesting avenue for future research.
A third limitation relates to the genetic determination of virulence and other disease characteristics in bacterial pathogens. Many of the phenotypic attributes of pathogenic bacteria, including the ability to colonize certain host tissues, the ability to evade an immune response and the ability to induce deleterious effects on the hosts, can be transmitted horizontally by bacteriophages (Wagner & Waldor 2002; Brüssow et al. 2004). For example, the toxin produced by diphtheria is genetically encoded by a temperate phage, and therefore this pathogenicity can be transmitted vertically via the prophage during bacterial replication, as well as horizontally through lysis (Pappenheimer & Murphy 1983). In fact, there is a growing realization that such horizontal gene movement probably plays a central role in the evolution of many other pathogens, including Vibrio cholera, Staphylococcus aureus, Streptococcus A, Neisseria meningitis, Pseudomonas aeruginosa and many of the bacterial pathogens from the family Enterobacteriaceae (Joklik et al. 1988).
The existence of bacteriophage-mediated horizontal transfer of pathogenicity genes brings with it at least two complications for the theoretical framework described here. First, as shown by Anderson & Cowles (1958) for the case of diphtheria toxin, the phage itself can be antigenic. The antiphage sera of infected hosts may thus block the horizontal transmission of the phage within an infected host, and because vertical transmission of the phage is imperfect (Crowell 1926; Anderson & Cowles 1958), the lysogenic bacteria can be completely lost from an infection. This dynamic has been described both in vitro and in vivo for Diptheria (Anderson & Cowles 1958) and represents an additional selective pressure acting on the production of the toxin. These two modes of transmission require specific models to understand the epidemiological dynamics of both the bacteria and its phage (Moxon & Jansen 2005). Second, many of the pathogenic attributes of bacteria need not be selectively beneficial to the bacteria, but rather they might simply be incidental by-products of phage evolution. If this is true, we will need to broaden the way we model disease evolution, allowing for the existence of a third evolutionary player in the game (i.e. the phage), that has its own potential evolutionary trajectory.
Finally, the use of fully deterministic models prevents us to take into account the potential consequences of demographic stochasticity and genetic drift on the evolution of parasite populations. Genetic drift may often lead to the eradication of some parasite strains and thus differ with the dynamics of a continuous deterministic model (where, by definition, strains never go to extinction). Restif & Grenfell (2006) contrast the different evolutionary outcomes in a parasite population using an epidemiological model with or without stochasticity. However, Restif & Grenfell (2006) only considered situations where mutation is a limiting factor (i.e. a given parasite variant is introduced only once). We believe that if there are recurrent mutations (as it is the case in the above case studies; figures 4 and 5), strain extinction will become transitory and the deterministic model may still provide a good approximation of the evolutionary dynamics. Of course, this will depend on the relative amount of drift and mutation. In any case, the study of the evolutionary dynamics in finite population of parasites does deserve further theoretical investigation.
4.2 Implications for other areas of research
We believe that the generalized theoretical framework presented here could also be relevant to formalize other evolutionary questions. First, it could be used to model within-host evolution of chronic infections and the arms race going on between the parasite population and the adaptive immune response of the host. For example, in HIV and SIV, there is some evidence that different types of escape mutants emerge early or late after infection (Goulder & Watkins 2004). This is currently interpreted by the possibility that early mutants may carry lower costs than late mutants. But an alternative explanation is suggested by the first case study (figure 3). These different mutants may be selected at different times because the host is a variable environment. Early in the infection, a large number of susceptible cells are available. This may select life-history strategies that become poorly efficient in the longer term. To evaluate this hypothesis, one needs to evaluate the life-history traits (replication rates within cells, killing rate of cells, survival outside the cells, etc.) of these different escape mutants (not available yet).
Another potential application of this model is the analysis of antibiotic resistance. In this case, the perturbation is not induced by vaccination but by the use of therapeutic drugs. The present model could be used to follow the evolutionary and epidemiological implications of the use of antibiotics in the short and long terms. Similarly, the model could also be used to model the spread of escape strategies in plant pathogens following the introduction of resistant lines in cultures. This may help analyse the short-term evolutionary dynamics of escape strategies and therefore estimate the durability of different management strategies.
Finally, the present model may also find some potential application in the evolution of free-living organisms. Vaccination can be viewed as a durable perturbation of the environment. After the start of the vaccination, the parasite population is away from both its epidemiological and evolutionary equilibria. Analogous situations may also occur with free-living organisms. For example, it has been shown that climate change was responsible for dramatic shifts in the geographical ranges of several insect species (Thomas et al. 2001). Interestingly, the newly colonized populations are characterized by larger dispersal abilities and lower reproductive outputs in the speckled wood butterfly (Hughes et al. 2003) and in the wing-dimorphic bush cricket (Simmons & Thomas 2004). Similar results have been pointed out in metapopulations of butterflies (Hanski et al. 2006) and plants (Olivieri & Gouyon 1985; Peroni 1994; Cody & Overton 1996) where more recently colonized sites tend to host more dispersive individuals. This is analogous to the numerical result obtained in figure 4, where the mean transmission and virulence of parasites infecting vaccinated hosts increase just after the start of the vaccination. In all these cases, the underlying process leading to this pattern is selection for higher transmission abilities in the newly colonized sites and an associated decrease in longevity and/or fecundity due to negative genetic covariance between dispersal ability and these important fitness-related traits. These final examples illustrate the importance of the ecological and demographical context to understand evolution. Merging the gap between ecology and evolutionary theory for parasites, as for any other free-living organisms, requires better life-history descriptions (genetic variations and covariations). Recent studies provide interesting data in this direction (Davies et al. 2001; Mackinnon & Read 2003; Bull et al. 2004) and we hope that the present theoretical considerations will motivate further empirical research in this direction.
Acknowledgments
We would like to thank three anonymous reviewers, Maciek Boni, Margaret Mackinnon, Frits Mooi, Andrew Read and Ophelie Ronce for comments on an earlier draft of this article. S.G. was supported by the Centre National de la Recherche Scientifique, and T.D. was supported by a grant from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs program.
Appendix A. Numerical example of virulence evolution
The numerical example presented in figure 5 is based on the following assumptions about the transmission rate, recovery rate, virulence and their relationships with one another in both naive and vaccinated hosts
| (A1) |
With the above functional forms, it is possible to approximate the evolutionary dynamics after the start of vaccination. In particular, if the variances are relatively small, we can use the approximation , where refers to the derivative evaluated at the average value of αN in naive hosts (Day & Proulx 2004). This yields (using equation (2.12) under the assumption that mutation rate is negligible)
| (A2) |
Using equation (2.13), the evolutionary dynamics of virulence, averaged over the two types of hosts, is
| (A3) |
This expression can be further simplified if we assume (as in the derivation of equation (3.3)) that (i) just after the start of the vaccination, the mean virulence is close to its ES level before vaccination in fully naive population, meaning and (ii) the difference in mean virulence between parasites infecting naive and vaccinated hosts is negligible, , yielding
| (A4) |
where
Appendix B. The evolution of multivariate traits
The framework for studying the evolutionary epidemiology of vaccination presented in the text can also be readily extended to allow for the evolution of multivariate traits (e.g. the simultaneous evolution of transmission rate, recovery rate and virulence; Day & Gandon 2006). For example, the extension of equation (A 3) to the evolution of virulence, transmission and recovery in the different types of hosts is
| (A5) |
In this case, evolution (i.e. its direction and its speed) depends on the gradient of selection (which is determined by the epidemiological setting; the column vectors in expression A 4) and on the matrices of genetic covariances between the different traits in the different habitats (GN and GV in expression A 5; Day & Proulx 2004; Day & Gandon 2006). This is similar to the classical quantitative genetics framework of Lande (1976, 1982). Such a formalism may be more practical, because it distinguishes clearly the selection gradient (which can be derived from epidemiological dynamics) from the matrix of genetic covariance G (which could potentially be measured for any organism). This matrix summarizes the constraints on evolution, i.e. the genetic variance and correlation (trade-offs) among traits. In practice, the evaluation of G is a daunting task (it requires huge experiments). A few recent studies have attempted to measure G in schistosomes (Davies et al. 2001) and rodent malaria (Mackinnon & Read 2003). These two studies obtained qualitatively contrasting results, which may yield different evolutionary scenarios (Gandon 2004). Clearly, the estimation of G is critical to making short-term evolutionary predictions. Such an approach might also yield long-term predictions if the G matrix remains constant over time.
Footnotes
One contribution of 20 to a Theme Issue ‘Cross-scale influences on epidemiological dynamics: from genes to ecosystems’.
References
- Anderson P.S, Cowles P.B. Effect of antiphage serum on the virulence of Corynebacterium diphtheriae. J. Bacteriol. 1958;76:272–280. doi: 10.1128/jb.76.3.272-280.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Directly transmitted diseases: control by vaccination. Science. 1982;215:1053–1060. doi: 10.1126/science.7063839. [DOI] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Vaccination against rubella and measles: quantitative investigations of different policies. J. Hyg. Camb. 1983;90:259–325. doi: 10.1017/s002217240002893x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- André J.B, Gandon S. Vaccination, within-host dynamics and virulence evolution. Evolution. 2006;60:13–23. [PubMed] [Google Scholar]
- Andreasen V. Virulence management and disease resistance in diploid hosts. In: Dieckmann U, Metz J.A.J, Sabelis M.W, Sigmund K, editors. Adaptive dynamics of infectious diseases. Pursuit of virulence management. Cambridge University Press; Cambridge, UK: 2002. pp. 222–232. [Google Scholar]
- Andreasen V, Christiansen F.B. Slow coevolution of a viral pathogen and its diploid host. Phil. Trans. R. Soc. B. 1995;348:341–354. doi: 10.1098/rstb.1995.0072. [DOI] [PubMed] [Google Scholar]
- Beck K. Coevolution: mathematical analysis of host-parasite interactions. J. Math. Biol. 1984;19:63–77. doi: 10.1007/BF00275931. [DOI] [PubMed] [Google Scholar]
- Blows M.W. A tale of two matrices: multivariate approaches in evolutionary biology. J. Evol. Biol. 2006;20:1–8. doi: 10.1111/j.1420-9101.2006.01164.x. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt W.-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J.J, Pfennig D.W, Wang I. Genetic details, optimization and phage life histories. Trends Ecol. Evol. 2004;19:76–82. doi: 10.1016/j.tree.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Cody M.L, Overton J.McC. Short-term evolution of reduced dispersal in island plan populations. J. Ecol. 1996;84:53–61. doi: 10.2307/2261699. [DOI] [Google Scholar]
- Crowell M.J. Morphological and physiological variations in the descendants of a single diphtheria Bacillus. J. Bacteriol. 1926;11:65–74. doi: 10.1128/jb.11.1.65-74.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C.M, Webster J.P, Woolhouse M.E.J. Tradeoffs in the evolution of virulence in an indirectly transmitted macroparasite. Proc. R. Soc. B. 2001;268:251–257. doi: 10.1098/rspb.2000.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T. Virulence evolution and the timing of disease life-history events. Trends Ecol. Evol. 2003;18:113–118. doi: 10.1016/S0169-5347(02)00049-6. [DOI] [Google Scholar]
- Day T, Gandon S. Insights from Price's equation into evolutionary epidemiology. In: Feng Z, Dieckmann U, Levin S, editors. Disease evolution: models, concepts, and data analyses. American Mathematical Society; Providence, RI: 2006. [Google Scholar]
- Day T, Proulx S. A general theory for the evolutionary dynamics of virulence. Am. Nat. 2004;163:E40–E63. doi: 10.1086/382548. [DOI] [PubMed] [Google Scholar]
- de Roode J.C, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushoff J. Incorporating immunological ideas in epidemiological models. J. Theor. Biol. 1996;180:181–187. doi: 10.1006/jtbi.1996.0094. [DOI] [PubMed] [Google Scholar]
- Earn D.J.D, Rohani P, Bolker B.M, Grenfell B.T. A simple model for complex dynamical transitions in epidemics. Science. 2000;287:667–670. doi: 10.1126/science.287.5453.667. [DOI] [PubMed] [Google Scholar]
- Fenner, F., Henderson, D. A., Arita, I., Jezek, Z. & Ladnyi, I. D. 1988 Smallpox and its eradication, pp. 332–335. Geneva, Switzerland: World Health Organization.
- Frank S.A. Models of parasite virulence. Q. Rev. Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Frank S.A. Princeton University Press; Princeton, NJ: 2002. Immunology and evolution of infectious diseases. [PubMed] [Google Scholar]
- Gandon S. Evolution of multihost parasites. Evolution. 2004;58:455–469. doi: 10.1554/03-390. [DOI] [PubMed] [Google Scholar]
- Gandon S, Mackinnon M.J, Nee S, Read A.F. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- Gandon S, Mackinnon M.J, Nee S, Read A.F. Imperfect vaccination: some epidemiological and evolutionary consequences. Proc. R. Soc. B. 2003;270:1129–1136. doi: 10.1098/rspb.2003.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov V.V, Antia R. Imperfect vaccines and the evolution of pathogens causing acute infections in invertebrates. Evolution. 2006;60:957–969. doi: 10.1554/05-504.1. [DOI] [PubMed] [Google Scholar]
- Geritz S.A.H, Kisdi E, Meszena G, Metz J.A.J. Evolutionary singular strategies and the adaptive growth and branching of the evolutionary tree. Evol. Ecol. 1998;12:35–57. doi: 10.1023/A:1006554906681. [DOI] [Google Scholar]
- Goulder P.J.R, Watkins D.I. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- Gupta S. Pathogen evolution: the case of malaria. In: Dieckmann U, Metz J.A.J, Sabelis M.W, Sigmund K, editors. Adaptive dynamics of infectious diseases. Pursuit of virulence management. Cambridge University Press; Cambridge, UK: 2002. pp. 347–361. [Google Scholar]
- Hanski I, Saastamoinen M, Ovaskainen O. Dispersal-related life-history trade-offs in a butterfly metapopulation. J. Anim. Ecol. 2006;75:91–100. doi: 10.1111/j.1365-2656.2005.01024.x. [DOI] [PubMed] [Google Scholar]
- Hsu H, Chang M, Liaw S, Ni Y, Chen H. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–1317. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- Hughes C.L, Hill J.K, Dytham C. Evolutionary trade-offs between reproduction and dispersal in populations at expanding range boundaries. Biol. Lett. 2003;270:S147–S150. doi: 10.1098/rsbl.2003.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joklik W.K, Willet H.P, Amos D.B, editors. Zinsser microbiology. 19th edn. Appleton and Lange; Norwalk, CA: 1988. [Google Scholar]
- Lande R. Natural selection and random genetic drift in phenotypic evolution. Evolution. 1976;30:314–334. doi: 10.2307/2407703. [DOI] [PubMed] [Google Scholar]
- Lande R. A quantitative genetic theory of life history evolution. Ecology. 1982;63:607–615. doi: 10.2307/1936778. [DOI] [Google Scholar]
- Lenski R.E, May R.M. The evolution of virulence in parasites and pathogens: reconciliation between two competing hypotheses. J. Theor. Biol. 1994;169:253–265. doi: 10.1006/jtbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- Mackinnon M.J, Read A.F. The effects of host immunity on virulence–transmissibility relationships in the rodent malaria parasite. Plasmodium chabaudi. Parasitology. 2003;126:103–112. doi: 10.1017/S003118200200272X. [DOI] [PubMed] [Google Scholar]
- McLean A.R. Vaccination, evolution and changes in the efficacy of vaccines: a theoretical framework. Proc. R. Soc. B. 1995a;261:389–393. doi: 10.1098/rspb.1995.0164. [DOI] [PubMed] [Google Scholar]
- McLean A.R. After the honeymoon in measles control. The Lancet. 1995b;345:272. doi: 10.1016/S0140-6736(95)90272-4. [DOI] [PubMed] [Google Scholar]
- McLean A.R. Vaccines and their impact on the control of disease. Br. Med. Bull. 1999;54:545–556. doi: 10.1093/oxfordjournals.bmb.a011709. [DOI] [PubMed] [Google Scholar]
- McLean A.R, Anderson R.M. Measles in developing countries. Part II: the predicted impact of mass vaccination. Epidemiol. Infect. 1988;100:419–442. doi: 10.1017/s0950268800067170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A.R, Blower S.M. Modelling HIV vaccination. Trends Microbiol. 1995;3:458–463. doi: 10.1016/S0966-842X(00)89010-1. [DOI] [PubMed] [Google Scholar]
- Moxon E.R, Jansen V.A. Phage variation: understanding the behaviour of an accidental pathogen. Trends Microbiol. 2005;13:563–565. doi: 10.1016/j.tim.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Olivieri I, Gouyon P.H. Seed dimorphism for dispersal: theory and implications. In: Haeck J, Woldendorp J.W, editors. Structure and functioning of plant populations. North-Holland Publishers; Amsterdam, The Netherlands: 1985. pp. 77–90. [Google Scholar]
- Pappenheimer A.M, Murphy J.R. Studies on the molecular epidemiology of diphtheria. The Lancet. 1983;2:923–926. doi: 10.1016/S0140-6736(83)90449-X. [DOI] [PubMed] [Google Scholar]
- Peroni P.A. Seed size and dispersal potential of Acer rubrum (Aceraceae) samaras produced by populations in early and late successional environments. Am. J. Bot. 1994;81:1428–1434. doi: 10.2307/2445316. [DOI] [Google Scholar]
- Porco T.C, Blower S.M. HIV designing vaccination policies: subtypes and cross-immunity. Interfaces. 1998;28:167–190. [Google Scholar]
- Porco T.C, Blower S.M. HIV vaccines: the effect of the modes of action on the coexistence of HIV subtypes. Math. Popul. Studies. 2000;8:205–229. [Google Scholar]
- Price G. Selection and covariance. Nature. 1970;227:520–521. doi: 10.1038/227520a0. [DOI] [PubMed] [Google Scholar]
- Restif O, Grenfell B. Integrating life-history and cross-immunity into the evolutionary dynamics of pathogens. Proc. R. Soc. B. 2006;273:409–416. doi: 10.1098/rspb.2005.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani P, Earn D.J.D, Grenfell B.T. Impact of immunisation on pertussis transmission in England and Wales. The Lancet. 2000;355:285–286. doi: 10.1016/S0140-6736(99)04482-7. [DOI] [PubMed] [Google Scholar]
- Rouderfer V, Becker N.G, Hetcote H.W. Waning immunity and its effects on vaccine schedules. Math. Biosci. 1994;124:59–82. doi: 10.1016/0025-5564(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Scherer A, McLean A. Mathematical models of vaccination. Br. Med. Bull. 2002;62:187–199. doi: 10.1093/bmb/62.1.187. [DOI] [PubMed] [Google Scholar]
- Shimizu Y.K, Hijikata M, Iwamoto A, Alter H.J, Purcell R.H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A.D, Thomas C.D. Changes in dispersal during species' range expansions. Am. Nat. 2004;164:378–395. doi: 10.1086/423430. [DOI] [PubMed] [Google Scholar]
- Smith T, Lehmann D, Montgomery J, Gratten M, Riley I.D, Alpers M.P. Acquisition and invasiveness of different serotypes of Streptococcus pneumoniae in young children. Epidemiol. Infect. 1993;111:27–39. doi: 10.1017/s0950268800056648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.D, Bodsworth E.J, Wilson R.J, Simmons A.D, Davies Z.G, Musche M, Conradt L. Ecological and evolutionary processes at expanding range margins. Nature. 2001;411:577–581. doi: 10.1038/35079066. [DOI] [PubMed] [Google Scholar]
- Tildesley M.J, Savill N.J, Shaw D.J, Deardon R, Brooks S.P, Woolhouse M.E.J, Grenfell B.T, Keeling M.J. Optimal reactive vaccination strategies for an outbreak of foot-and-mouth disease in Great Britain. Nature. 2006;440:83–86. doi: 10.1038/nature04324. [DOI] [PubMed] [Google Scholar]
- van Boven M.F.R, Mooi J.F.P, Schellekens H.E, de Melker, Kretzschmar M. Pathogen adaptation under imperfect vaccination: implications for pertussis. Proc. R. Soc. B. 2005;272:1617–1624. doi: 10.1098/rspb.2005.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner P.L, Waldor M.K. Bacteriophage control of bacterial virulence. Infect. Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D, Gavrilets S. 20 questions on adaptive dynamics. J. Evol. Biol. 2005;18:1139–1154. doi: 10.1111/j.1420-9101.2005.00948.x. [DOI] [PubMed] [Google Scholar]
- Wilson J.N, Nokes D.J, Carman W.F. Current status of HBV vaccine escape variants—a mathematical model of their epidemiology. J. Viral Hepatitis. 1998;s2:25–39. doi: 10.1046/j.1365-2893.1998.0050s2025.x. [DOI] [PubMed] [Google Scholar]
- Wilson J.N, Nokes D.J, Carman W.F. The predicted pattern of emergence of vaccine-resistant hepatitis B: a cause for concern? Vaccine. 1999;17:973–978. doi: 10.1016/S0264-410X(98)00313-2. [DOI] [PubMed] [Google Scholar]