Abstract
When one considers the fine-scale spread of an epidemic, one usually knows the sources of biological variability and their qualitative effect on the epidemic process. The force of infection on a susceptible unit depends on the locations and the strengths of the infectious units, and on the environmental and intrinsic factors affecting infectivity and/or susceptibility. The infection probability for the susceptible unit can then be modelled as a function of these factors. Thus, one can build a conceptual model at the fine scale. However, the epidemic is generally observed at a larger scale and one has to build a model adapted to this larger scale. But how can the sources of variation identified at the fine scale be integrated into the model at the larger scale? To answer this question, we present, in the context of plant epidemiology, a multi-scale approach which consists of defining a base model built at the fine scale and upscaling it to match the scale of the sampling and the data. This approach will enable comparing experiments involving different observational processes.
Keywords: disease spread, epidemiology, multi-scale modelling, propagule dispersal, spatial
1. Introduction
In plant, animal or human epidemiology and population genetics, dispersal models can be used when a spatial component is considered. In epidemiology, dispersal models are needed to evaluate the spatial spread of a disease from already infected individuals and to improve control strategies. In population genetics, these models enable estimating gene dispersal, a typical case being the dispersal of pollen from genetically modified plants to other plants.
In such contexts, different scales appear naturally: the phenomenon scale; the sampling scale; and the modelling scale (Dungan et al. 2002). Precisely describing the phenomenon and collecting the corresponding data are generally impossible, especially if the phenomenon is not completely known. So, one has to resort to realistic data collection, i.e. changing the sampling scale, and to simplify the description of the phenomenon, i.e. changing the modelling scale. Then, the three scales do not automatically coincide and a model is generally the result of a compromise between (i) a description of the physical or biological processes and (ii) the temporal and spatial features of the observed dataset.
In studies of pollen dispersal for trees, for example, the mating model (Smouse & Sork 2004) is a popular one as soon as all the trees are known in the surroundings of the mother trees of interest. Assuming that the pollen is similarly dispersed around each father tree, the pollination probability of a seed of a mother by a given father is described as a function of a dispersal function and the locations of the father trees. But when the locations of the possible fathers are not observed, an alternative is to assume that these locations are drawn from a Poisson point process, and to integrate the stochasticity generated by this assumption (Smouse & Sork 2004).
In plant epidemiology, spore dispersal at short time scales can be described by a Brownian motion (Stockmarr 2002; Bicout & Sache 2003) if one assumes that the behaviour of the spores is a diffusion process, which implies very specific wind conditions. At large time scales, many different wind conditions may appear, and the interest is not so much in spore dispersal as in disease spread. Dispersal will then be described through empirical disease dispersal curves (Aylor 1990; McCartney & Fitt 2006; Soubeyrand et al. 2007, in press).
In animal epidemiology, when an epidemic is studied within a farm composed of pens, the spread of the disease between animals can be modelled by a system of transmission probabilities: one probability for animals located in the same pen; one probability for animals located in neighbouring pens; and a zero probability otherwise (Höhle et al. 2005). But when the epidemic is studied at the scale of a country with irregularly located farms, the individuals of interest can become farms instead of animals, and a spatial dispersal kernel accounting for the heterogeneity of inter-farm distances can be used (Keeling et al. 2001).
In all these cases, where the dispersal process is of primary interest, each model is obtained by a direct translation from a conceptual model to a mathematical model suitable for the specific framework of interest. In particular, the model is adapted to (i) the type of disease observation (e.g. the disease presence/absence measure), (ii) the scale (or support) of disease observation (e.g. the farm) and (iii) the covariates that are observed and which explain a part of data variability (e.g. the locations of the infectious units). Thus, these dispersal models are well adapted to specific situations. However, in general, their outputs cannot be compared in a quantitative way because they do not share a common construction base. This is a major problem for comparative studies involving different survey strategies.
In this paper, we propose to develop specific but coherent models: specific because each of them is tailored for a given situation and coherent because they all stem from a single base model. For this purpose, we suggest translating a conceptual model into a mathematical model at a fine scale, i.e. a scale at which describing the sources of variations is natural, inherent and intuitive. Then, models at larger scales are built based on the mathematical model, using an approach similar to the multi-scale modelling approach developed in physics where a macroscopic model is derived from a microscopic model (Weinan & Engquist 2003; Weinan et al. 2003). Explicit links between model structures at the fine scale and each specific scale are then exhibited. In particular, links between parameters at different scales are made explicit. These parameters can then be used to compare model outputs obtained in different situations.
We illustrate this proposal in the case of the spread of plant diseases, when two observation dates are available. Section 2 presents the conceptual model, its biological assumptions and its mathematical translation at the finest of the considered scales. This fine-scale model describes the probabilistic behaviour of the presence/absence of the disease on small-scale susceptible units. The model includes the effects of spatially unstructured and structured covariates (e.g. due to genotype, physiology, climate) affecting the infectiousness of the infectious units and the receptivity of the susceptible units. Then, the fine-scale model is scaled up to build larger-scale models adapted to situations where
the type of disease observation is changed (e.g. from the presence/absence to the number of symptoms),
the scale (or support) of disease observation is changed (e.g. from the plant to the agricultural plot), and
the unstructured and structured covariates are censored (i.e. unobserved or partially observed), so reducing the information content of the observed data.
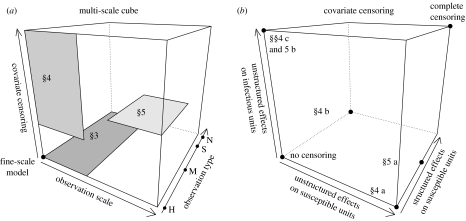
Thus, in §§3, 4 and 5 of this paper, we study different larger-scale models adapted to various sampling schemes. This study corresponds to exploring some parts of the cube drawn in figure 1a. The first axis of this cube represents the observation scale, the second axis the observation type and the third axis the covariate censoring. The fine-scale model is represented by the point at the origin. The zones of the cube which are explored in the paper are coloured in grey. In §3 we will look at different types of disease measures made at relatively small scales (dark grey zone). In §4 we will study what happens when one ignores some of the covariates associated with relatively small-scale observation units. In §5 we will consider larger-scale observation units within which the covariates are varying. Figure 1b breaks down what sorts of covariate censorings are considered and where some of the subsections of the paper are located in this space. We discuss, in §6, the interests and limits of the proposed methodology developed in a conceptual context of plant epidemiology. In §7 we will see how this context can be extended, especially to animal and human epidemiology.
Figure 1.
(a) Cube representing the space partly explored in this paper. The fine-scale model is at the origin (units with infinitesimal areas, presence/absence for the disease measure, no covariate censoring). The grey rectangles are the zones of the cube which are explored in §§3, 4 and 5. On the observation-type axis the letters H, M, S and N denote, respectively, the following disease measures: presence/absence, count of infected subunits, severity and count of lesions (notation introduced in §3).(b) Cube representing the decomposition of the censoring axis drawn in the cube of (a). At the origin of the cube, there is no censoring; at the opposite point the censoring is complete; the other points correspond to intermediate situations tackled in this paper.
2. At the fine scale
2.1 Biological assumptions
We will subsequently focus on the spread of a plant disease between two dates corresponding to the beginning and the end of an epidemic cycle. We focus on diseases for which the entity responsible for disease transmission is called a propagule. The propagule can be a specialized cell (spore), a whole organism (bacterium) or a structure embedding a pathogen (pollen grain and vector).
We assume that the variability of the disease cycle duration is negligible, and that a common starting point in time exists for transmission from all infectious plants. Then, the cycles can be distinguished and observed at the time scale we are interested in.
We assume that at the starting point the infectious plant units are detectable, and that they remain infectious during the cycle. At the end of the cycle, we assume that the newly infected plant units, thereafter called infected units, are detectable. The newly infected units are not infectious during the cycle.
We assume that the rules governing the transmission mechanisms are the same at all the spatial scales we are looking at.
2.2 Conceptual model
The conceptual model describes spatial spread by identifying the different spatial and temporal elements without actually completely specifying them.
From a spatial point of view, plants or plant units are considered as points in space, and with a specific qualitative status: either healthy, infected or infectious. We assume that no new plant units are generated during the period of interest (the generation of plant units can however be handled in given situations; e.g. Soubeyrand et al. 2006b).
From a temporal point of view, time is discrete, i.e. each time step corresponding to the beginning of a cycle.
Epidemic spread is understood as a three-step mechanism. First, propagules are dispersed from each infectious plant or plant unit. Second, the accumulation of propagules over a given susceptible unit defines a local infectious potential. Third, the susceptible unit becomes infected with a success probability depending on the local infectious potential.
2.3 Mathematical translation
We denote the location of the ith unit in the considered region by xi. For a given time t, we denote δit=0 if the health status of unit i is not observed at time t, δit=1 if it is observed.
Health status of unit i at time t is denoted by Iit and Hit with Iit=1 if unit i is infectious, Iit=0 if it is not and Hit=1 if unit i is infected, Hit=0 if it is healthy.
Propagule dispersal from a given infectious unit i is described by a dispersal function fθ(x−xj) where x is any location in the region of interest and θ a set of parameters. Different shapes for the dispersal function have been proposed (e.g. Tufto et al. 1997; Klein et al. 2003). Local infectious potential at location x at time t is then written as the following convolution (Mollison 1977):
| (2.1) |
i.e. the sum of the values at location x of the dispersal functions centred on the infectious units.
The probability of infection of a healthy unit at point xj is described by a function depending on the local infectious potential
where g is a link function from R+ to [0,1].
If all infectious units are observed and if the observations are made at the beginning and the end of a cycle, parameter estimation can then be carried out by maximizing the log-likelihood
| (2.2) |
Depending on the shape of fθ, (2.2) is the log-likelihood of a generalized linear or nonlinear model (McCullagh & Nelder 1989; Collett 1991; Harrell 2001; Huet et al. 2004).
Note that in (2.2) the sum is computed only for units j such that Hjt=0 because the other units, already infected at time t, do not bring information on the parameters in the framework of interest here (see Soubeyrand et al. (2006b) for a framework where already infected units bring information on the dispersal parameters).
2.3.1 Introduction of covariates
In fact, infection success depends on many local factors (Rapilly 1991) such as plant characteristics (e.g. genotype, individual variations within a genetically homogeneous plantation, age, size), the environment (e.g. the soil and the climate which can influence plant physiology), randomness in source infectivity (some infectious plants may be more infectious than others owing to a larger production of propagules on this plant or a larger local population of vectors for a vector-borne disease).
These factors can be introduced into the model as penalties acting on the infectious potential L whose initial mathematical expression is written in (2.1); then, as above, the link between the new L and the probability of success of an infection will be described by the link function g. Specifically, we propose to model the effect of the factors mentioned above as multiplicative factors: if j is a susceptible unit located at xj, then
| (2.3) |
where aj denotes a spatially unstructured effect associated with the susceptible unit j; b(xj) denotes the effect of spatially structured factors on unit j; and ci denotes the spatially unstructured effect associated with an infectious unit i. b(xj) may just depend on location, or may depend on explicit covariates (e.g. soil composition). Similarly, aj and ci may depend on plant characteristics. Note that we could also have added spatial penalties by taking into account possible spatially structured effects affecting the infectious units. Thereafter, we omit the word ‘spatially’ for the sake of shortness.
2.3.2 Examples of specifications
In practice, one must specify the nature of the infectious and susceptible units, the dispersal function fθ and other elements of the model. Typical specifications might be the following.
An infectious unit can be an agricultural plot, a plant, a leaf or a lesion. Each infectious unit spreads around its location a random number of propagules, for example a Poisson number of propagules with mean λ.
Propagules which are dispersed around any infectious unit are, for example, independently distributed from a two-dimensional exponential law with parameter ρ: the probability density to find a propagule deposited at location y is where x is the infectious unit location. Thus, the random field of propagules generated by an infectious unit at x is a non-stationary Poisson point process with intensity and .
Many parametric forms for fθ have been proposed (Tufto et al. 1997; Klein et al. 2003; Austerlitz et al. 2004). The shape of the dispersal function is known to influence the epidemic dynamics and the statistical estimation. This point, already discussed for example by Fitt et al. (1987) and Austerlitz et al. (2004), is not tackled in this paper.
The argument in the dispersal function fθ is very often the Euclidian distance, as in the above example. However, other types of arguments can be used depending upon the context. Indeed, fθ can be a function of the distance and the direction (Soubeyrand et al. in press) if there is a prevailing wind for example. If the disease spreads through contacts between individuals, relations between individuals can be modelled in a network, and distances on this network used as the argument of the dispersal function (Hufnagel et al. 2004; Dargatz et al. 2005; Parham & Ferguson 2006).
The random field of propagules generated by all infectious units is an inhomogeneous Poisson random field whose intensity at point y is the local infectious potential , where xi is the location of the infectious unit i. Note that with such a specification, the unstructured and structured effects aj, b(xj) and ci are constant.
The susceptible unit, at the fine scale, can be an infinitesimal susceptible zone with area dx. The health status Hj,t+1 is defined, in this case, by the presence or the absence of the disease at time t+1 on the susceptible unit j with area dx and location xj. The area dx captures a Poisson number of propagules with intensity . Assuming that propagules land independently and that the failure probability is , then , where the exponential shape for the link function g comes from the Poisson assumption. A Taylor expansion of the previous expression (justified because dx is infinitesimal) yields .
3. Deriving the fine-scale model to build models adapted to various disease-observation scales
The model proposed above (§2.3.2) describes the presence/absence of a disease on infinitesimal units. In practice, various sorts of disease measures corresponding to various observation scales are encountered. A review on relationships between disease intensity measurements was proposed for example in plant epidemiology by McRoberts et al. (2003). Using the same type of derivations, we study how the fine-scale model, where the base susceptible units are infinitesimal parts of healthy plants, can be derived to obtain models adapted to various disease measures acquired from larger susceptible units, such as a leaf. These larger susceptible units are assumed to be small enough that the local infectious potential is constant within any unit (§5 will present a situation where the infectious potential varies within the units).
3.1 Counting the lesions on susceptible units
Consider a larger susceptible unit with area sj at point xj. It captures a Poisson number of propagules with intensity , and the number Nj,t+1 of lesions generated at time t+1 from the propagules is then Poisson distributed with mean , i.e. .
So, if lesions can be identified then the disease measure can be lesion counts, and the log-likelihood used to estimate the parameters becomes
where the summation is performed on units observed at times t and t+1 (i.e. ) and healthy at time t (i.e. Hjt=0).
Remark: the sum in this log-likelihood is computed only for healthy units at time t. However, already infected units at time t could also be considered in the log-likelihood. Indeed, they can be affected by propagules dispersed from the infectious units and, consequently, contain information on the parameters. But, in order to account for this information, the autoinfection must be modelled as well as its interaction with the alloinfection (i.e. the process of infection from other units). This point will not be tackled in this paper.
3.2 Measuring the infected areas of susceptible units
When lesions are hardly distinguishable, counting them is impossible and one relies on severity measures, the most classical one being the infected area on the susceptible unit, say Sjt for unit j at time t. Suppose that the area is a random variable depending on Nj,t+1 and sj: . The function F is a random function which can be selected empirically and/or based on mechanistic assumptions about the disease. For example, Sj,t+1 can be derived from a spatial Boolean process (Stoyan et al. 1995; Molchanov 1996) on the unit if lesions are assumed to be independent surface areas. The density probability function of Sj,t+1 is
where is the conditional density probability function of F(N, s) given N and s. The log-likelihood is then
The remark written in §3.1 is also valid here.
Remark: susceptibility of units is already incorporated in the local infectious potential (2.3) through the unstructured effects aj. We could also account for susceptibility in the density function f because it can affect not only the number of lesions but also their sizes.
3.3 Observing the presence/absence of the disease on susceptible units
The easiest way to measure the disease on a given susceptible unit is to observe whether it is present or not on the unit. To avoid cumbersome notation, we denote the presence/absence of the disease on the susceptible unit j by Hjt, the same notation as for the infinitesimal units. The disease is not on unit j if no propagule succeeds in infecting the unit, which occurs with probability because Nj,t+1 follows a Poisson distribution with mean (§3.1). Thus, Hj,t+1 is Bernoulli distributed with probability .
In this case, we obtain the log-likelihood
| (3.1) |
This formula is similar to the log-likelihood (2.2), with depending on the unit characteristics sj and q.
3.4 Counting the infected subunits of susceptible units
Sometimes, the observation unit (e.g. a plant) is split into mj subunits (e.g. the leaves) and the disease measure is the number of infected subunits Mjt. The interest of such a measure is to obtain a variable which can be mapped because it is less noisy than the presence/absence variable. Let Hjkt denote the health status of subunit k of unit j. Following §3.3, Hjk,t+1 is Bernoulli distributed with probability , where sjk is the area of subunit k. In this section all subunits of unit j are submitted to the same infectious potential L(xj). In addition, the Hjk,t+1 are independent because under the Poisson assumption, the propagules land independently on the subunits. This yields the following.
- In the case where the subunit areas are the same (i.e. ), Mj,t+1 follows a binomial distribution with size mj and success probability . Thus, the log-likelihood becomes
where .(3.2) In the case where the subunit areas are different and cannot be measured individually, one can, for example, consider the areas as independently and identically distributed with probability density function fs. Then, Hjkt is Bernoulli distributed with probability , Mj,t+1 follows a binomial distribution with size mj and success probability pj, and the log-likelihood can be written as in (3.2) by replacing pj by its new expression.
3.5 Conclusion: estimating relevant biological parameters
As mentioned before, the interest of the derivation from a basic model is in allowing (i) the estimation of biologically relevant parameters, those defined at the fine-scale model and (ii) the comparison of experiments performed at different scales. Indeed, for each constructed model, we have written a log-likelihood on which the inference on the parameters can be based. In particular, inference on the parameters included in the infectious potential L is possible in each case since L appears in each expression of the log-likelihood. Moreover, each context offers the possibility to infer other parameters which are specific to the context; for example, the parameters which could enter in the random function F linking the lesion count to the infected area (§3.2), or those which could enter in the probability density function fs of the subunit areas (§3.4).
Applying this derivation-based approach requires interactions between biologists and statisticians, in particular, to define the crucial fine-scale model, to incorporate the context-specific parameters and to decide which model components can be neglected.
4. Deriving the fine-scale model to study the consequences of ignoring covariates
In §3, we built models adapted to different disease-observation scales, i.e. when the information content of the observations is changed. In the present section, we study the consequences of reducing the information content on the covariates.
Suppose that we observe the presence/absence of a disease on susceptible units; so, we consider the model of §2.3
| (4.1) |
| (4.2) |
This model is quite simple, but estimating its parameters using the log-likelihood (3.1) requires the computation of the potential (4.2) for each susceptible unit, i.e. requires the knowledge of all the infectious units and the precise status of the sampled units (location, health status and covariates). In practice, collecting all these data can be a very cumbersome task, and the covariates denoted by aj, b(xj) and ci in (4.2) are usually not observed, in particular because their identify is often unknown. A common approach consists of ignoring the covariate which is not observed and adopting a simplified model where the ignored covariate is replaced by a constant value. In this section we study the consequences of this approach and how these consequences can be used in a residual analysis to build a relevant model from the simplified model.
4.1 Ignoring the unstructured effects of the susceptible units
Measuring individual characteristics of the plant units is a difficult task, in particular if the relevant characteristics are not known in advance, so that many of them have to be measured. In practice, individual characteristics are simply not observed and ignored in the modelling. In this subsection we study the consequence of ignoring the unstructured effects affecting the susceptible units.
Let be the conditional event ; aj does not appear in since it is not observed. The unstructured effects, supposed to be independently distributed, can be written as where a is the mean value of the effects and ϵj is a centred random unstructured variable with variance . The infectious potential (4.2) affecting unit j can be written
where . The random variables ϵjAj are independent, centred and with variances . Then, conditionally on the events j, the infected status at time t+1 of units susceptible at time t remain independent (as they were conditional on the events ). In addition, supposing that , then a Taylor expansion yields the following approximation:
where g(2) is the second derivative of g.
If the ϵj are ignored, the infectious potential is and the infection probability of a unit susceptible at time t is . Hence, the absence of the correction factor , which should compensate the non-observation of the unstructured effect aj. For example, with the link function obtained in §3, and for the true set of parameters, the probability , used when the unstructured effects are ignored, is higher than it should be. So, ignoring the unstructured covariate will lead to biased parameter estimators.
To avoid the bias in the estimator when the unstructured covariate cannot be measured, a possible approach consists of specifying a parametric distribution for the unstructured effects aj viewed as random effects, as in the frailty model of Soubeyrand et al. (2007). The distribution of the random effects can sometimes be difficult to specify and results can be sensitive to its form. To overcome this difficulty, methods like the ones developed by Ritz (2004), Soubeyrand et al. (2006a, 2007) and Waagepetersen (2006) can be used to specify the unobserved random effects.
4.2 Ignoring the structured effects on the susceptible units
For large-scale studies, spatially structured factors due to physical environment (soil, climate) or genetics are often neglected in a first step, the observation effort being focused on disease detection. These structured effects, taken into account through b(x) in (2.1) (or (4.2)), are often considered as constant. We go on to study the consequences of ignoring the variations of , following the asymptotic approach applied in §4.1.
Suppose that the b(xj) are not observed, the conditional events we are working with are then . Set and suppose that the ϵx are small. The random values ϵx cannot be considered as independent, but are assumed to form a stationary random field with variance and spatial autocorrelation function r(d). Calculations similar to those carried out above yield
where . Furthermore, conditionally on the events , a spatial dependence appears among the health status at time t+1 of susceptible units at time t
where djk is the distance between xj and xk.
If the variations of the structured covariate are ignored, is simply g(bBj), the covariance is zero; consequently, the parameter estimators will be biased, as in §4.1.
To avoid the bias in the estimator, if the spatial distribution of b(x) can be specified up to a given set of unknown parameters, we obtain a hierarchical model including spatially correlated random effects, and a likelihood-based or Bayesian estimation procedure using Monte Carlo sampling can be performed (Diggle et al. 1998; Zhang 2002; Desassis et al. 2005).
4.3 Ignoring the unstructured effects of the infectious units
Very often, locations are the only available data on infectious units. Time and level of infection, or individual unit characteristics are not known although they can influence greatly the infectiousness of a given unit. So, suppose that the unstructured effects ci associated with the infectious units are not observed; the conditional events we are working with are then . Set , the infectious potential can be written
where . Suppose that the independent random variables ϵj are centred and with variance . Set . Then using Taylor's expansions,
and a spatial dependence appears between health status,
because susceptible units near more infectious units have a higher chance of being infected.
As before, if the distribution of the ci are known up to a given number of parameters, the model is a hierarchical one and a procedure based on Monte Carlo sampling can be used to estimate the parameters.
4.4 Detection of the main departure from the simplest model
If a departure from the simplest model, i.e. the model where covariates are replaced by constants, is known to be due to one specific reason, then the statistical treatment will depend on the situation: (i) if the absent covariate can be measured, the model including the covariate will be fitted using an estimation procedure for generalized linear or nonlinear models and (ii) if the covariate cannot be measured, then hierarchical models and the associated estimation procedures as those mentioned at the ends of §§4.1–4.3 can be applied.
Very often, however, one does not know if the simplest model is suitable or if any departure must be taken into account. In such a case, a common strategy consists of (i) estimating the simplest model in a first step and (ii) checking on residuals to examine whether this model is acceptable or whether it must be modified. By doing so, one generally assumes that the dispersal as described by the simplest model captures the main features of the observed dispersal, and that departures are not due to many reasons but that one reason is more important than the others.
A residual analysis based on the results presented above can then be used to point out the main departure. Consider the conditional event . Under the simplest model, the conditional probability for the unit j to be infected is , where is the local infectious potential affecting j. Under the model with unstructured effects for the susceptible units (§4.1), . Under the model with structured effects for the susceptible units (§4.2), . Under the model with unstructured effects for the infectious units (§4.3), , where .
Plotting against (respectively, ) or (respectively ) can help in deciding whether there is a departure from the simplest model, and if the most important departure is due to effects on the susceptible units or the infectious units. Indeed, the expected value of Wj is zero under the simplest model, positive and constant (either or ) under the model with either the unstructured effects or the structured effects for the susceptible units, and a space-varying function () under the model with the unstructured effects on the infectious units.
To distinguish between departures due to unstructured or structured effects on the susceptible units, the conditional covariance between health status at time t+1 given can be used. Indeed, this covariance is zero under the simplest model and the model with the unstructured effects for the susceptible units, whereas it is under the model with structured effects for the susceptible units. Hence, if the plot above shows that there are (unstructured or structured) effects for the susceptible units, plotting against the distance djk between xj and xk can help in deciding between the unstructured or structured effects. We present in appendix A how these statistics could be used on a simulated example.
5. Sampling through large spatial units
When dealing with datasets collected on a large spatial scale, looking at individual plants or leaves becomes impossible, and the observation unit is the agricultural plot for example. At this scale, the unstructured and structured covariates, which are not observed or partially observed, are varying within the observed spatial units. In the following, we study the consequences of these variations when the susceptible plants or the infectious plants are grouped in larger spatial units.
5.1 Grouping susceptible subunits in spatial units
Suppose that susceptible subunits are regularly spaced in a spatial unit. They correspond to trees in an orchard for instance, the orchard being the spatial unit under consideration. Suppose there are mj susceptible subunits in the spatial unit j, their locations being xjk for . The disease measure at the spatial unit level can be the number of infected subunits, or the presence/absence of at least one infected subunit ( if , zero otherwise).
If, conditionally on the set of parameters to be estimated, the local infectious potential can be computed for each subunit, the are mutually independent with probability distributions satisfying
and the are mutually independent with probability distributions satisfying
| (5.1) |
Based on these expressions, a likelihood can be built as in §3 for estimating the model parameters. This assumes that either the simplest model (without varying covariates, see §4.4) accurately describes the epidemic spread or that the covariates have been measured at the subunit level.
In practice, the covariates will not be measured at the subunit level: for instance, the unstructured variations described by the coefficients ajk will not be observed, and the structured variations b(xjk) will be measured only at a given location of the spatial unit, say its centre zj. In this case, one has to tackle two problems: (i) the non-observation of the unstructured covariate as in §4 and (ii) the so-called change-of-support problem (Chilès & Delfiner 1999) since the disease measure is an areal datum, in the sense that the disease notation is common for all the unit area, whereas the structured covariate is a point datum.
By an asymptotic development, as in §4, one can investigate what the distributions of and become when problems (i) and (ii) occur. Consider, for example, the case of the presence/absence variable . Assume that the ajk have variance , that is a stationary random field independent of the ajk, with variance and autocorrelation function . Set . It can be shown that, conditionally on the event , the probability that is asymptotically the sum of and a function depending on , and . So, the probability that is the sum of a term analogous to the right-hand side of (5.1) where the covariates associated with the susceptible units would be assumed to be constant, and a correction factor depending on the characteristics of these covariates. Moreover, the covariance between the measures and made at two spatial units j and j′ is not zero but equal to a function of , and .
5.2 Grouping infectious subunits in spatial units
Infectious unit recording is not always done at the individual level, but can be done at larger spatial units. For example, one will not observe the location of individual infectious trees, but only the central locations of infectious orchards and the number of infectious trees in each orchard. Then, the infectious potential
affecting the susceptible unit j cannot be computed since the exact locations xik as well as the unstructured effects cik of the infectious trees of any orchard i are not observed. Therefore, the probability that which is equal to cannot be computed either.
However, an asymptotic development can also be used here for approximating the probability that given, for each infectious spatial unit i, the location zi of its centre and the number of infectious subunits Ni,t at time t. Asymptotically, this probability is the sum of where
and a correction factor depending on the variance of the unstructured effects cik associated with the infectious subunits. is a hypothetical infectious potential where the subunits are supposed to be clustered at point zi and the unstructured effects are supposed to be constant. Moreover, the covariance between the measures and made at two susceptible units j and j′ at time t is not zero but equal to a function of .
5.3 Small-scale versus large-scale spatial units
Pooling subunits in small-scale spatial units (§3.4) or in large-scale spatial units (§§5.1 and 5.2) have different consequences for the probability distributions of the health status of susceptible units at time t. Indeed, when the spatial units are large, the infection probabilities are changed and, furthermore, there is a non-zero covariance between the health status of different susceptible units. These changes occur because the covariates associated with the subunits are not observed at this level.
The fact that the infection probabilities and the covariance between health status depend on the characteristics of the covariates shows that the data contain information on the unstructured and structured effects even if these effects are not observed or partially observed. Consequently, these characteristics can be inferred from the collected data, at least in principle. Nevertheless, regarding inference, the likelihood obtained when the spatial units are large is not tractable in practice. To overcome this problem, an estimating equation can be built based on (i) a pseudo-likelihood which will ignore the spatial dependence between the health status and (ii) a least square criterion between the empirical and the theoretical covariance of the health status. A hierarchical model and an appropriate estimation procedure can also be applied; the random effects included in the hierarchical model would then be the unobserved locations of the subunits in the observed spatial units and the values of the covariates for these subunits.
6. Overview of the proposed multi-scale approach
6.1 Summary
In this paper, we have presented a multi-scale modelling approach to building on epidemic models which takes into account the sources of variation of the epidemic and which matches the scale of the sampling and the data. The multi-scale approach consists of (i) defining a base model describing an epidemic at a fine scale and (ii) upscaling it in order to build models at larger scales. In this paper, we have studied various larger-scale models adapted to various sampling schemes. The considered sampling schemes were characterized by the type of disease observation (e.g. presence/absence of the disease), the scale (or support) of disease observation (e.g. the plant) and the censoring level of the covariates (e.g. censored structured effects but observed unstructured effects). This study has allowed us to explore a part of the cube drawn in figure 1a.
6.2 What is the interest of a multi-scale approach?
In epidemiological studies where the spatial component is considered, the dispersal process is often of primary interest and a model including a description of the dispersal process is generally used to analyse the data. The model is in fact based on the mathematical translation of a conceptual model from which several derivations are possible; the derivations consist, for instance, in adding covariates, changing the disease measure and/or changing the sampling scheme. Using a base model, namely the fine-scale model, from which others can be derived is useful from several viewpoints.
If measurements are done at various scales for different experiments (§§3 and 5), the multi-scale approach helps in exhibiting the link between (1) the characteristics of the models built for the different experiments and (2) the parameters and functions defining the fine-scale model. Then, experiments can be compared by going back to the parameters and functions of the fine-scale model.
If covariates are known to influence the dispersal process, then the multi-scale modelling approach offers a framework where the covariates can be included into the model in a biologically relevant way instead of adding them empirically, as covariates are added into a statistical linear model.
The model validation step (Cook & Weisberg 1982; McCullagh & Nelder 1989), based on residual analysis, can be guided by the expected deviations from the fine-scale model instead of just looking at empirical links between residuals and covariates as is usually the case. Thus, the multi-scale approach enables one to check the hypotheses made in the conceptual model.
6.3 Using asymptotic developments
Many developments in this paper have been performed in an asymptotic framework, by assuming that disturbances are of secondary importance with respect to the dispersal effect. The advantage of this assumption is in generating explicit formulae which can be easily interpreted. If this assumption is not suitable, bias and covariances (similar to those exhibited in this paper) remain and can be assessed by simulation. However, in cases where validation statistics are needed, the statistics proposed in the asymptotic framework can be used to check the goodness-of-fit of the model (see §4.4). In other words, the asymptotic context helps to propose validation statistics which can then be used in more general contexts. The asymptotic formulae can also be used to modify and improve the model as in Soubeyrand et al. (2006a) and Soubeyrand & Chadœuf (in press).
6.4 Dealing with reduced information content in the dataset
Taking into account a covariate effect is similarly difficult regardless of whether the covariates act on the infectious or the susceptible plant units. When dealing with censoring on the locations, the situation is much more tricky when the censoring affects the infectious units rather than the susceptible ones. The main reason is that the pattern of the infection of susceptible units is the result of a dispersal process for which the main driving factors, namely the locations of the infectious units, are supposed to be known. So, dealing with reduced information on the locations of the susceptible units remains basically a statistical power problem: there is less data than one could actually have. In addition, when the locations of the infectious units are not known, one needs to restore them. This can lead to hierarchical models in which the infectious units will be randomly distributed in a given space, as long as no information is available on the processes explaining the spatial repartition of these units; note however that the influence of such a choice is difficult to evaluate in practice.
6.5 Tackling other deviations from the fine-scale model
Deviations from the fine-scale model can appear in many ways and not only those considered in this paper and summed up in figure 1. We have chosen, for example, to consider a structured effect only on susceptible plant units; but a similar development could have been made by considering a structured effect on infectious plant units. Another interesting situation which has not been tackled in this paper is the situation where some of the infectious units are not recorded. This situation is particularly expected to occur when the spatial scale of interest is large owing to the cost of an exhaustive mapping of infectious units. This problem could be approached by restoring the unobserved infectious units. This sort of problem (detection of unknown sources) was handled by Martin et al. (2006) when the number of unknown sources is small.
7. Beyond an approach developed in a conceptual context of plant epidemiology
We now discuss in which other directions the proposed approach could be developed.
7.1 Application to data
The approach we presented has been developed in a conceptual context. Even if some components of this approach have been applied to real datasets (see some of the references cited before), its multi-scale feature has not been fully exploited to compare datasets collected at different scales. More precisely, we think that the approach we presented could be especially useful in performing a multi-scale meta-analysis enabling a better understanding of multi-scale phenomena like epidemics.
7.2 Changing the time scale
We have chosen to consider the simplest situation where only one epidemic cycle happens. This leads to a considerable simplification as, under this assumption, all infection events are independent and the fine-scale model can be derived relatively easily. When the time scale is changed such that several cycles may arise between two observation dates, the infection events are not independent anymore. Indeed, if a susceptible unit is infected, it can then be infectious and the infection events due to this new source of propagules are conditional on the initial infection event. Dealing with such a situation is not easy, even with a base model without covariates. Gibson (1997), Fewster (2003) and Jamieson et al. (2005) proposed an estimation based on the modelling of the successive infection (or colonization) events, whereas Keeling et al. (2004) proposed an empirical procedure to estimate the dispersal function by minimizing the difference between an observed spatial pattern and the one obtained under individual pattern changes guided by the dispersal function. Chadœuf et al. (1992) proposed to model the spatial dependence between infectious events. Applying an approach similar to the one developed in this paper could help in analysing which part of the observed dependence is due to dispersal, and which one is due to covariates or measurement pooling.
7.3 Extending the approach to animal and human epidemiology
Accounting for potential mismatches between the scale at which biological processes operate and that at which data are acquired is important in plant, animal and human epidemiology if models are to accurately explore the mechanisms that give rise to biological variability. We looked at this challenge in the context of plant diseases, where the individuals are sedentary, by developing a multi-scale framework. This simple case could be expanded to the analysis of epidemiological data collected at nested scales (e.g. individuals within families (or other social units) within settlements within counties within countries; or animals within fields within farms within parishes). The multi-scale framework developed in this paper is not directly applicable to animal and human epidemiology because, for instance, animals and humans can move, and disease measurements and transmission processes will probably be different. In such cases, the main difficulty is the fact that transmission does not necessarily originate from the point at which an infectious individual is observed, but from every point of its path, which is generally unknown. Nevertheless, the ideas presented in this paper (upscaling a fine-scale model, studying the consequences of ignoring covariates and of sampling across larger spatial units) can be applied to these disciplines. The application would be particularly facilitated in situations where an infectious potential can be defined as in equation (2.3). Precisely this concept of infectious potential has been developed in a number of different contexts: in plant (e.g. Gibson 1997; Jamieson et al. 2005); livestock (Gerbier et al. 2002; Keeling et al. 2004; Diggle 2005; Höhle et al. 2005; Höhle & Feldmann in press); and human epidemiology (Neal & Roberts 2004), as well as in other disciplines (Lescouret et al. 1998; Medlock & Kot 2003; Parham & Ferguson 2006), and is causing epidemiologists to shift their perspective from that of a single transmission process to a multi-scale transmission system.
Acknowledgments
We would like to thank Dan Haydon and the reviewers for their comments on this article.
Appendix A.
Here we show in a simulated example how the residuals proposed in §4.4 can be used to look at departures from the simplest model, where the structured and unstructured effects are assumed to be constant. We first simulated an initial pattern of susceptible and infectious units in the following way.
A first set of points was simulated using a Strauss point process (Stoyan et al. 1995) with intensity 100, interaction distance 0.02 and complete inhibition distance on a square of side 2.7. This leads to a regular set of points which could represent centres of orchards. Then, for each orchard, 10 points (representing trees) were independently and uniformly spread in a 0.02×0.02 square centred on the previous points.
Each orchard was labelled as either infectious or susceptible with probability 0.1 and 0.9, respectively. All the trees of an orchard have the same status.
Then, the final states of initially healthy trees were simulated under the model described by equations (4.1) and (4.2) specified as follows.
The dispersal function fθ was chosen to be a centred bivariate isotropic Gaussian density with standard deviation 0.05.
The unstructured effects of the infectious units were all equal to one.
The unstructured effects of the susceptible units satisfied , where Ui were independently drawn from the LogNormal(0,5) distribution.
The structured effects on the susceptible units were drawn using a Boolean disc model (Stoyan et al. 1995; Molchanov 1996) with intensity 15 and radius 0.075, the effect value being 0.1 outside the discs and 3.96 inside the discs.
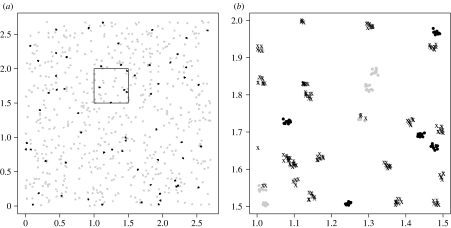
Figure 2a shows the locations of orchards (dots) and their initial health status (grey, susceptible; black, infectious). The detail of the zone delimited by a square is shown in figure 2b. Here we can see how the trees are scattered within the orchards. We can also see the final health status of the trees (grey dots, susceptible trees; black dots, infectious trees; crosses, newly infected trees).
Figure 2.
Simulated data set. (a) Locations and initial health status of orchards (grey dots, susceptible orchards; black dots, infectious orchards). (b) Close-up of the zone delimited by the square on (a). The symbols are now located at tree locations and give the final health status of the trees (grey dots, susceptible trees; black dots, infectious trees; crosses, newly infected trees).
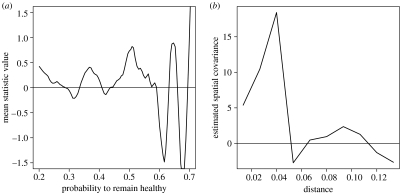
We then estimated the parameters of the model with constant structured and unstructured effects. Finally, we computed the normalized residuals . The evolution of its mean value with respect to the probability to remain healthy is given in figure 3a. It mostly remains above 0 for probabilities less than 0.55, then oscillates with a large amplitude for larger probabilities. The estimated autocovariance of which estimates the expectation of is given in figure 3b. It remains positive from 0 to 0.11, except at distance 0.05 where it is equal to −1, whereas it begins at 5 approximately 0 and peaks at 15 at distance 0.05. Note that, for this Boolean model, the value of two points are independent as soon as their separating distance is greater than 0.15.
Figure 3.
Residual plots. (a) Mean value of the normalized residuals and (b) autocovariance of the normalized residuals.
Following the procedure proposed above, one decides first that a structured or unstructured effect exists, due to the positive value of the local mean of the normalized residuals, second that it is a structured effect due to the presence of an autocovariance. Note that this statistical test needs to be formalized in order to take into account the random variations of the statistics, as the oscillation observed on the first curve. In this simulation, zones with probability larger than 0.55 are scarce. This fact, together with the presence of a spatial structure can lead to large structured variations as the one in figure 3a.
Footnotes
One contribution of 20 to a Theme Issue ‘Cross-scale influences on epidemiological dynamics: from genes to ecosystems’.
References
- Austerlitz F, Dick C.W, Dutech C, Klein E.K, Oddou-Muratorio S, Smouse P.E, Sork V.L. Using genetic markers to estimate the pollen dispersal curve. Mol. Ecol. 2004;13:937–954. doi: 10.1111/j.1365-294X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- Aylor D.E. The role of intermittent wind in the dispersal of fungal pathogens. Annu. Rev. Phytopathol. 1990;28:73–92. doi: 10.1146/annurev.py.28.090190.000445. [DOI] [Google Scholar]
- Bicout D.J, Sache I. Dispersal of spores following a persistent random walk. Phys. Rev. E. 2003;67:031913.1–031913.7. doi: 10.1103/PhysRevE.67.031913. [DOI] [PubMed] [Google Scholar]
- Chadœuf J, Nandris D, Geiger J.P, Nicole M, Pierrat J.C. Modélisation spatio-temporelle d'une épidémie par un processus de Gibbs: estimation et tests. Biometrics. 1992;48:1165–1175. doi: 10.2307/2532707. [DOI] [Google Scholar]
- Chilès J.-P, Delfiner P. Modeling spatial uncertainty. Wiley; New York, NY: 1999. Geostatistics. [Google Scholar]
- Collett D. Chapman & Hall; London, UK: 1991. Modelling binary data. [Google Scholar]
- Cook R.D, Weisberg S. Chapman & Hall; New York, NY: 1982. Residuals and influence analysis. [Google Scholar]
- Dargatz C, Georgescu V, Held L. Technical report. Ludwig-Maximilians Universität; Munich, Germany: 2005. Stochastic modelling of the spatial spread of influenza in Germany. [Google Scholar]
- Desassis N, Monestiez P, Bacro J.N, Lagacherie P, Robbez-Masson J.M. Mapping unobserved factors on vine plant mortality. In: Renard P, Demougeot-Renard H, Froidevaux R, editors. Geostatistics for environmental applications. Springer; Berlin, Germany: 2005. pp. 125–136. [Google Scholar]
- Diggle P.J, Tawn J.A, Moyeed R.A. Model-based geostatistics. J. R. Stat. Soc. C. 1998;47:299–350. doi: 10.1111/1467-9876.00113. [DOI] [Google Scholar]
- Diggle, P. J. 2005 A partial likelihood for spatio-temporal point processes. Johns Hopkins University, Department of Biostatistics working paper 75.
- Dungan J.L, Perry J.N, Dale M.R.T, Legendre P, Citron-Pousty S, Fortin M.-J, Jakomulska A, Miriti M, Rosenberg M.S. A balanced view of scale in spatial statistical analysis. Ecography. 2002;25:626–640. doi: 10.1034/j.1600-0587.2002.250510.x. [DOI] [Google Scholar]
- Fewster R.M. A spatiotemporal stochastic process model for species spread. Biometrics. 2003;59:640–649. doi: 10.1111/1541-0420.00074. [DOI] [PubMed] [Google Scholar]
- Fitt B.D.L, Gregory P.H, Todd A.D, McCartney H.A, Macdonald O.C. Spore dispersal and plant disease gradients: a comparison between two empirical models. J. Phytopathol. 1987;118:227–242. [Google Scholar]
- Gerbier G, Bacro J.-N, Pouillot R, Durand B, Moutou F, Chadœuf J. A point pattern model of the spread of foot-and-mouth disease. Prevent. Vet. Med. 2002;56:33–49. doi: 10.1016/S0167-5877(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Gibson G.J. Markov chain Monte Carlo methods for fitting spatiotemporal stochastic models in plant epidemiology. J. R. Stat. Soc. C. 1997;46:215–233. doi: 10.1111/1467-9876.00061. [DOI] [Google Scholar]
- Harrell F. Springer; Berlin, Germany: 2001. Regression modeling strategies with applications to linear models, logistic regression and survival analysis. [Google Scholar]
- Höhle, M. & Feldmann, U. In press. RLadyBug. An R package for stochastic epidemic models. Comput. Stat. Data Anal. [DOI] [PMC free article] [PubMed]
- Höhle M, Jørgensen E, O'Neill P.D. Inference in disease transmission experiments by using stochastic epidemic models. J. R. Stat. Soc. C. 2005;54:349–356. doi: 10.1111/j.1467-9876.2005.00488.x. [DOI] [Google Scholar]
- Huet S, Bouvier A, Poursat M.A, Jolivet E. 2nd edn. Springer; Paris, France: 2004. Statistical tools for nonlinear regression. [Google Scholar]
- Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc. Natl Acad. Sci. USA. 2004;101:15 124–15 129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson, L., Brooks, S. P. & Gilligan, C. A. 2005 Joint estimation of spatial and temporal parameters for epidemic dynamics from successive snapshots. Technical report.
- Keeling M.J, et al. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813–817. doi: 10.1126/science.1065973. [DOI] [PubMed] [Google Scholar]
- Keeling M.J, Brooks S.P, Gilligan C.A. Using conservation of pattern to estimate spatial parameters from a single snapshot. Proc. Natl Acad. Sci. USA. 2004;101:9155–9160. doi: 10.1073/pnas.0400335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E.K, Lavigne C, Foueillassar X, Gouyon P.-H, Larédo C. Corn pollen dispersal: quasi-mechanistic models and field experiments. Ecol. Monogr. 2003;73:131–150. doi: 10.1890/0012-9615(2003)073%5B0131:CPDQMM%5D2.0.CO;2. [DOI] [Google Scholar]
- Lescouret F, Habib R, Genard M, Agostini D, Chadœuf J. Pollination and fruit growth models for studying the management of kiwifruit orchards. I. Models description. Agric. Syst. 1998;56:67–89. doi: 10.1016/S0308-521X(97)00042-5. [DOI] [Google Scholar]
- Martin R.J, Di Battista T, Ippoliti L, Nissi E. A model for estimating point sources in spatial data. Stat. Methodol. 2006;3:431–443. doi: 10.1016/j.stamet.2005.12.003. [DOI] [Google Scholar]
- McCartney H.A, Fitt B.D.L. Dispersal of foliar fungal plant pathogens: mechanisms, gradients and spatial patterns. In: Cooke B.M, Jones D.G, Kaye B, editors. The epidemiology of plant diseases. 2nd edn. Springer; Dordrecht, The Netherlands: 2006. pp. 159–192. [Google Scholar]
- McCullagh P, Nelder J.A. 2nd edn. Chapman & Hall; London, UK: 1989. Generalized linear models. [Google Scholar]
- McRoberts N, Hughes G, Madden L.V. The theoretical basis and practical application of relationships between different disease intensity measurements in plants. Ann. Appl. Biol. 2003;142:191–211. doi: 10.1111/j.1744-7348.2003.tb00242.x. [DOI] [Google Scholar]
- Medlock J, Kot M. Spreading disease: integro-differential equations old and new. Math. Biosci. 2003;184:201–222. doi: 10.1016/S0025-5564(03)00041-5. [DOI] [PubMed] [Google Scholar]
- Molchanov I.S. Wiley; Chichester, UK: 1996. Statistics of the Boolean models for practitioners and mathematicians. [Google Scholar]
- Mollison D. Spatial contact models for ecological and epidemic spread. J. R. Stat. Soc. B. 1977;39:283–326. [Google Scholar]
- Neal P.J, Roberts G.O. Statistical inference and model selection for the 1861 Hegelloch measles epidemic. Biostatistics. 2004;5:249–261. doi: 10.1093/biostatistics/5.2.249. [DOI] [PubMed] [Google Scholar]
- Parham P.E, Ferguson N.M. Space and contact networks: capturing the locality of disease transmission. J. R. Soc. Interface. 2006;3:483–493. doi: 10.1098/rsif.2005.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapilly F. INRA Editions; Paris, France: 1991. L'Epidémiologie en Pathologie Végétale. [Google Scholar]
- Ritz C. Goodness-of-fit tests for mixed models. Scand. J. Stat. 2004;31:443–458. doi: 10.1111/j.1467-9469.2004.02_101.x. [DOI] [Google Scholar]
- Smouse P.E, Sork V.L. Measuring pollen flow in forest trees: an exposition of alternative approaches. Forest Ecol. Manage. 2004;197:21–38. doi: 10.1016/j.foreco.2004.05.049. [DOI] [Google Scholar]
- Soubeyrand, S. & Chadœuf, J. In press. Residual-based specification of a hidden random field included in a hierarchical model. Comput. Stat. Data Anal.
- Soubeyrand S, Chadœuf J, Sache I, Lannou C. Residual-based specification of the random-effects distribution for cluster data. Stat. Methodol. 2006a;3:464–482. doi: 10.1016/j.stamet.2005.12.005. [DOI] [Google Scholar]
- Soubeyrand, S., Sache, I., Höhle, M. & Held, L. 2006b Modelling the spread in space and time of an airborne plant disease. Research report 24, UR546 Biostatistics and spatial process. Avignon, France: INRA.
- Soubeyrand S, Sache I, Lannou C, Chadœuf J. A frailty model to assess plant disease spread from individual count data. J. Data Sci. 2007;5:67–83. [Google Scholar]
- Soubeyrand, S., Enjalbert, J., Sanchez, A. & Sache, I. In press. Anisotropy, in direction and in distance, of the dispersal of yellow rust of wheat: experiments in large field plots and estimation. Phytopathology. [DOI] [PubMed]
- Stoyan D, Kendall W.S, Mecke J. 2nd edn. Wiley; Chichester, UK: 1995. Stochastic geometry and its applications. [Google Scholar]
- Stockmarr A. The distribution of particles in the plane dispersed by a simple 3-dimensional diffusion process. J. Math. Biol. 2002;45:461–469. doi: 10.1007/s002850200157. [DOI] [PubMed] [Google Scholar]
- Tufto J, Engen S, Hindar K. Stochastic dispersal processes in plant populations. Theor. Popul. Biol. 1997;52:16–26. doi: 10.1006/tpbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Waagepetersen R. A simulation-based goodness-of-fit test for random effects in generalized linear mixed models. Scand. J. Stat. 2006;33:721–731. doi: 10.1111/j.1467-9469.2006.00504.x. [DOI] [Google Scholar]
- Weinan E, Engquist B. Multiscale modeling and computation. Notices Am. Math. Soc. 2003;50:1062–1070. [Google Scholar]
- Weinan E, Engquist B, Huang Z. Heterogeneous multiscale method: a general methodology for multiscale modeling. Phys. Rev. B. 2003;67:09210-1. [Google Scholar]
- Zhang H. On estimation and prediction for spatial generalized linear mixed models. Biometrics. 2002;58:129–136. doi: 10.1111/j.0006-341X.2002.00129.x. [DOI] [PubMed] [Google Scholar]