Abstract
H-NS, a nucleoid-associated DNA-binding protein of enteric bacteria, was discovered thirty-five years ago and subsequently found to exert widespread and highly pleiotropic effects on gene regulation. H-NS binds to high-affinity sites and spreads along adjacent AT-rich DNA to silence transcription. Preferential binding to sequences with higher AT-content than the resident genome allows H-NS to repress the expression of foreign DNA in a process known as “xenogeneic silencing.” Counter-silencing by a variety of mechanisms facilitates the evolutionary acquisition of horizontally transferred genes and their integration into pre-existing regulatory networks. This review will highlight recent insights into the mechanism and biological importance of H-NS-DNA interactions.
H-NS-- A Nucleoid Protein
H-NS is an abundant DNA-binding protein implicated in the organization of the bacterial chromosome [1]. H-NS and other nucleoid-associated proteins can affect DNA topology at specific loci, thereby modulating gene transcription. Binding by these proteins is proposed to selectively direct supercoiling effects to promoters [2•]. Mutation of hns alters the responsiveness of transcription to changes in DNA superhelicity. Regulatory effects of supercoiling are linked to metabolic and environmental conditions. Thus, H-NS has been viewed as a nucleoid structuring protein with global effects on gene expression [3].
H-NS exists primarily as a dimer at low concentrations but can multimerize into higher order complexes [4, 5] that form bridges between adjacent DNA helices [6]. Optical tweezers have been used to demonstrate that H-NS dimers or multimers can simultaneously interact with separate DNA binding sites [7••]. H-NS-coated DNA is not self-interactive, suggesting that dimerization or oligomerization must precede DNA binding for bridging to occur. Force measurements suggest that transcription barriers created by H-NS binding are weak (∼7 pN) and readily overcome, so that H-NS oligomers pose only a relative barrier to translocating RNA polymerase. DNA bridging is a conserved feature of H-NS homologs found in most gram-negative bacteria [8] that appears to constrain large DNA loops [9••] and helps to account for the effects of H-NS on transcription. However, it must be noted that the relationship between bridging as a general effect and the silencing of specific promoters is not yet clear.
H-NS is more appropriately viewed as a determinant of chromosomal architecture rather than as a general structural component. The analysis of nucleoids treated with urea suggests that neither H-NS nor the nucleoid-associated proteins Fis, Dps or StpA is required for a cooperative transition between compacted and partially expanded forms of the chromosome [10]. The bacterial chromosome is organized into topological domains averaging 10kbp, resulting in approximately 400 domains per chromosome. Although H-NS bridging constrains short loop formation, a possible role of H-NS in the formation or maintenance of larger topological domain barriers has been recently highlighted [11, 12]. DNA looping has been proposed as a fundamental mechanism for action at a distance in the control of gene expression by proteins. DNA resists bending and twisting due to its inherent rigidity, and a major function of architectural proteins is to reshape DNA and/or modify its stiffness. It has been suggested that looping might require a binding protein to act not only at the end of a loop but also on intervening DNA to enhance flexibility [9]. An assay to study the effects of the nucleoid proteins H-NS, HU and IHF on the in vivo flexibility of DNA demonstrated that H-NS destabilizes rather than stabilizes small loops [9]. In contrast, HU promotes DNA looping in vivo. Changes in superhelicity do not appear to explain the effects of architectural proteins on DNA looping. Since looping can affect DNA affinity for regulatory molecules and promote cooperative interactions [13], H-NS constraints on looping may account for some of its repressive actions.
Effects of H-NS and other nucleoid-associated proteins on superhelicity are proposed to have important regulatory consequences during changes in environmental conditions [2]. H-NS has long been regarded as a global modulator of gene expression in response to pH, temperature, osmolarity and growth phase [14-16]. A large proportion of Salmonella genes induced upon temperature shift from 25°C to 37°C are dependent on H-NS [17], an effect attributed to putative conformational changes in H-NS and reduced DNA binding. However, a large number of H-NS-repressed genes remain repressed at 37°C, suggesting that a simple effect of temperature on H-NS multimerization or DNA binding cannot account for the temperature-dependent derepression of a subset of genes. Moreover, H-NS oligomerization has been reported to be higher at 37°C than 25°C [18] and uninfluenced by further temperature increases to 48°C or pH variation between 4.0 and 9.0. Similarly, while H-NS represses the transcription of a number of osmoregulated genes [14], many H-NS-silenced genes are unaffected by changes in osmolarity. These observations indicate that the relationship between H-NS and environmentally dependent alterations in gene expression is complex, and that H-NS should not be viewed as a simple temperature or osmolarity sensor.
Recognition of DNA by H-NS
H-NS was regarded for many years as a DNA binding protein without a specific consensus sequence for binding, and the well-known ability of H-NS to affect a wide range of genes was attributed to a preference for particular DNA structures [1]. H-NS recognition sites typically display planar curvature specified by AT-rich motifs, as commonly found at promoters. However, quantitative analysis of the H-NS binding site at the osmoregulated E. coli proU promoter, which regulates an osmoprotectant uptake locus, has demonstrated that H-NS specifically recognizes a 10bp sequence with a KD of ∼15 nM [19••]. This motif is also present at two locations downstream of the proU promoter. Synergy between these sites and lower affinity sites in the region favors the formation of a specific nucleoprotein complex that efficiently represses transcription. The 10bp high-affinity binding motif has been proposed as a nucleation site for H-NS binding. Upon binding to high affinity sites, H-NS spreads along DNA using sites of lower affinity to occupy the promoter region, allowing the formation of higher order structures. Moreover, insertion of a high-affinity site into a GC-rich sequence enhances H-NS binding, excluding the possibility that high affinity binding requires a more extensive structure for recognition. It has therefore been suggested that an H-NS binding region contains several sites with variable affinity, and that the organization of binding sites determines the formation of a repressive nucleoprotein complex.
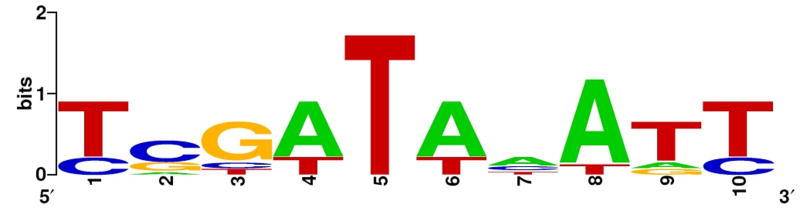
Comparison of the 10bp high-affinity binding motif with a sequence inferred by footprinting experiments using the isolated DNA-binding domain of H-NS [20•] and with predicted and confirmed H-NS sites at the fis promoter allowed the identification of a putative consensus recognition sequence for H-NS. This experimentally deduced motif is presented in figure 1. The presence of this motif has been correlated with H-NS binding in vivo in chromatin immunoprecipitation on microarray (ChIP-on-chip) experiments [21•]. A statistically significant correlation was observed between the proposed consensus sequence and observed H-NS binding. Some promoters such as bgl display more than 10 potential binding sites for H-NS, while others like nir contain fewer binding motifs. This suggests that the regulation of gene expression by H-NS can be modulated by variations in the number and organization of binding sites. In summary, although DNA recognition by H-NS was long thought to be based on structure rather than sequence, the discovery of a specific high affinity H-NS-binding motif suggests that H-NS can bind to one or more high affinity sites and subsequently polymerize and bind adjacent sites to spread regionally along AT-rich DNA [20•]. Once bound, the predominant effect of H-NS on gene expression is repressive [22••].
Figure 1. Logo representation of the high-affinity H-NS binding motif.
Mechanism of Transcriptional Silencing by H-NS
Although some investigators did not find a correlation between H-NS and RNAP binding sites [23•], others have observed a significant correlation [21•, 24] consistent with a model in which H-NS nucleoprotein complexes can trap RNAP to prevent transcription. Under conditions favoring repression, H-NS prevents open complex formation by RNAP at the proU promoter, and appears to exert similar actions at the bgl promoter [25••]. In other instances, such as the hdeAB promoter, H-NS appears to trap the open complex once it has formed [26•]. Thus, H-NS appears to be capable of silencing transcription by a variety of mechanisms.
The existence of specific binding sites for H-NS only partially explains the silencing effect of H-NS at various promoters. This question has been nicely approached by both experimental and computational work on the bgl promoter, and to a lesser extent, the proU promoter. The effect of H-NS binding on regions upstream (URE) and downstream (DRE) from the transcription start site on the activity of these promoters [27] and models derived from these results lead to the conclusion that repression by H-NS is inversely correlated to the transcription elongation rate across the H-NS binding region, suggesting that RNA polymerase engaged in elongation can disrupt the repressing nucleoprotein complex formed by H-NS. In the case of the bgl promoter, silencing requires synergy between H-NS bound at the URE and DRE sequences. H-NS bound to the DRE alone would not be predicted to impede RNA polymerase translocation. Thus, bgl transcriptional silencing by H-NS is intimately linked to protein-protein interactions between H-NS bound upstream and downstream of the promoter. The feedback loops created by H-NS repression of transcription initiation and downstream anti-termination mediated by the BglG protein in this model help to explain how small changes in bgl promoter activity can have large effects on overall operon expression [28•]. At the proU promoter, the mechanism by which a similar regulatory loop might be formed is not yet clear [25••, 28•].
Interactions between H-NS and other Nucleoid Proteins
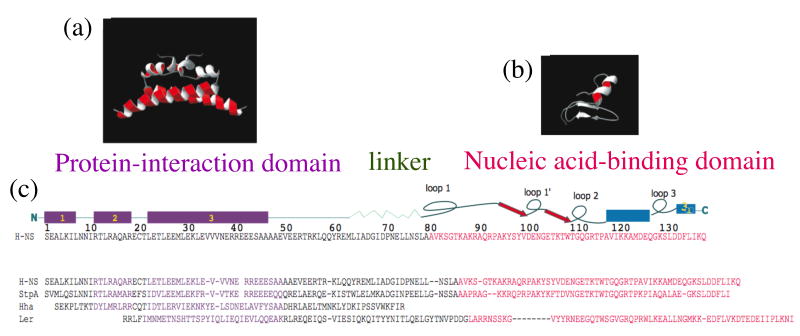
In addition to forming homodimers and homooligomers, H-NS can form heteromeric complexes by interacting with other nucleoid-associated proteins such as Hha and the H-NS paralog StpA (figure 2). Phenotypes associated with Hha include altered plasmid supercoiling and insertion sequence transposition [29]. Hha is a member of the Hha/YmoA family of proteins, which by themselves do not bind DNA but structurally resemble the N-terminus of H-NS (containing the dimerization domain) and are capable of associating with H-NS to enhance its repressive action at some but not all loci [30, 31]. Hha/YmoA proteins are less abundant than H-NS, so heterodimers are the major form of Hha/YmoA in the cell [30].
Figure 2. Sequence alignment of the four E.coli H-NS like proteins cited in the text.
Structural modularity is evident in this family of gene-regulating proteins. For clarity, sequences have been aligned with the known structures of E. coli H-NS amino- and carboxyl-terminal domains. Models are based on RMN studies of the N- terminal dimer (a) coiled coil interaction and the C-terminal (b) domains of H-NS [3]. A schematic representation of the domain organization of H-NS (c) is aligned with the primary sequence of H-NS, StpA, Hha and Ler. The N-terminal domain contains three alpha helices (α1, α2 and α3). A linker region separating the N and C termini consists of two β strands intercalated between loops 1 and 1′ and loops 1′ and 2 terminating in an α helix and a 310 α helix that form a nucleic acid binding domain.
Levels of StpA in wild-type cells are also low, because stpA expression is inhibited by H-NS. Furthermore, in its homodimeric form StpA is degraded by the Lon protease. Consequently the heterodimer form of StpA is predominant. A mutation in stpA has no discernable phenotype on its own in E. coli but derepresses a subset of genes in the absence of H-NS [32]. StpA can repress its own expression but cannot repress the bgl operon unless at least the N-terminus of H-NS is present to allow formation of heterodimers [33]. H-NS and StpA repress the expression of genes encoding the type II secretory system required for secretion of E. coli heat-labile enterotoxin (LT) [34] by inhibiting open complex formation. H-NS also represses eltAB encoding the enterotoxin itself [35]. H-NS can act synergistically as co-repressor with LRP, the leucine-responsive regulatory protein, to inhibit expression of the ribosomal RNA promoter [36], although no direct interaction between LRP and H-NS has been reported. Much remains to be learned about the physiological significance of H-NS interactions with other nucleoid proteins.
Counter-silencing of H-NS and its Functional Consequences
A striking correlation has been observed between AT content and H-NS binding [22••, 23•]. High AT-content relative to the resident genome has long been recognized as a hallmark of horizontally transferred DNA [37]. An hns mutation in Salmonella is detrimental to cell viability unless accompanied by mutations in positive regulators of virulence gene expression (rpoS, phoP) or pathogenicity islands (SPI2), suggesting that overexpression of H-NS-repressed loci is harmful to the cell.
The ability of H-NS to recognize foreign DNA with higher AT content than the resident genome [22••, 23•] has been referred to as “xenogeneic silencing” [38•]. The paradigm of xenogeneic silencing is providing a new framework for understanding the genomic evolution of pathogenic bacteria and specifically how alien genes can be integrated into existing regulatory networks [38•]. The recognition and transcriptional silencing of AT-rich DNA by H-NS simultaneously protects bacteria from adventitious effects of foreign gene expression [16, 22••] and facilitates the incorporation of such genes. Moreover, xenogeneic silencing provides a rationale for the conservation of GC content by bacterial species, as a means of differentiating self and non-self DNA and an explanation for the strong tendency of foreign sequences to have higher AT content than the resident genome (allowing such sequences to be more readily harnessed). The inability of horizontally-transferred GC-rich DNA to be recognized and silenced by H-NS would make such sequences less likely to be tolerated by the recipient. Given the prominent role of horizontal gene transfer in the evolution of pathogenic bacteria [39], xenogeneic silencing is of particular importance in understanding the regulation of virulence gene expression [22••].
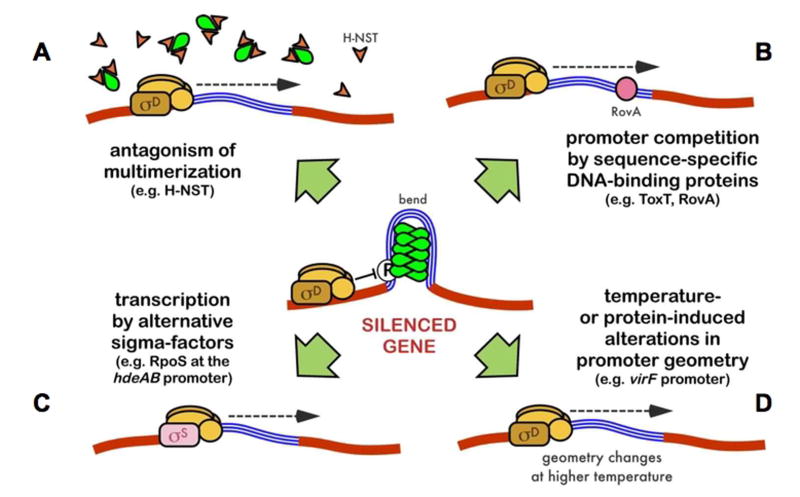
Regulated expression of H-NS-silenced genes requires specific mechanisms of counter-silencing, such as activation by an alternative sigma factor or competition from sequence-specific DNA-binding proteins and H-NS homologs (figure 3). Direction of transcription by the alternative sigma factor σS appears to be an important mechanism of counter-silencing. The specific ability of H-NS to assemble nucleoprotein complexes with Eσ70 but not Eσs RNAP holoenzyme may account in part for the selectivity of σs promoters [26•, 40•]. In the absence of H-NS, it appears that many σS-activated promoters can be transcribed by either form of RNAP. For example, the csgBA and hdeAB loci of E. coli require Eσs for expression in the presence of H-NS, but can be transcribed by Eσ70 in an hns rpoS mutant background [41]. Hsp31, encoded by hchA, is regulated by both Eσ70 and Eσs. H-NS silences hchA expression during exponential growth, when expression is driven by Eσ70, but does not effectively repress hchA expression in stationary phase when Eσs becomes dominant [42]. Interestingly, H-NS also appears to be required for the normal proteolytic turnover of σs [43]. Circumvention of H-NS silencing provides an explanation for σs involvement in the regulation of horizontally-acquired virulence genes [44, 45], and complex interactions between σs and H-NS are responsible for their co-involvement in the expression of certain osmoregulated genes [41, 46, 47].
Figure 3. Mechanisms of Counter-silencing.
Potential mechanisms of counter-silencing include: (A) disruption of H-NS complexes by multimerization antagonists such as H-NST; (B) competition for DNA binding by a high-affinity sequence specific DNA binding protein such as ToxT or RovA; (C) activation of certain promoters such as the E. coli hdeAB promoter by the alternative sigma factor σs; (D) changes in promoter geometry due to protein binding or environmental changes that disrupt H-NS complex formation such as may occur at the virF promoter in Shigella. These mechanisms are not mutually exclusive, and it is possible that more than one mechanism may be operating at any given promoter. Reproduced with permission from (38).
The ability of other DNA-binding proteins to act as counter-silencers by antagonizing the effects of H-NS on transcription and the ability of H-NS to polymerize along AT-rich sequence tracts can rationalize the location of transcription factor binding sites downstream of translational start sites, which is difficult to reconcile with classical activation mechanisms but consistent with an ability of H-NS to mediate interactions between upstream and downstream binding sites [25••]. Many examples of counter-silencing by DNA-binding proteins have been recently described. The AraC-like activators GadX and GadW promote gadA (glutamate decarboxylase) expression, important for acid stress resistance, by antagonizing H-NS [48]. Interestingly, the RNAP-associated protein SspA has been proposed to enhance cell survival at acid pH by reducing hns expression during stationary phase [49]. The MarR family regulator SlyA counters H-NS silencing at sites both upstream and downstream of the hemolysin gene hlyE transcriptional start site in E. coli [50] by competing with H-NS for binding. Interestingly, SlyA appears to counter H-NS silencing at the E. coli K5 capsule biosynthetic gene cluster by remodeling the nucleoprotein complex rather than by inhibiting H-NS binding [51]. Thus, a single protein may counter silencing by more than one mechanism. The LysR family regulator LeuO counters H-NS silencing of the putative virulence gene yjjQ in avian pathogenic E. coli [52] and H-NS/StpA silencing of the ompS1 porin gene in Salmonella [53]. The importance of H-NS in regulating horizontal gene transfer is not limited to pathogenesis. The regulatory protein TraJ counter-silences H-NS repression of the tra genes in the F plasmid of E. coli but is dispensable for conjugal transfer in an hns mutant [54], demonstrating that H-NS can limit plasmid mobilization. Some plasmids such as the Salmonella plasmid R27 carry their own H-NS-like protein (Sfh, in the case of R27) that appears to prevent detrimental consequences of H-NS titration by AT-rich sequences [55•] and thereby allow the recipient cell to tolerate the plasmid.
H-NS homologs might act as counter-silencers by competing for binding sites or interfering with multimerization. Ler is an H-NS homolog (figure 2) encoded by the LEE (Locus of Enterocyte Effacement) pathogenicity island found in EHEC (enterohemorrhagic E. coli), EPEC (enteropathogenic E. coli) and Citrobacter rodentium. Ler can antagonize H-NS repression of LEE genes but also antagonizes the expression of Lpf long polar fimbriae [56]. EPEC and UPEC (uropathogenic E. coli) genomic islands carry additional truncated H-NS homologs called H-NST that can antagonize H-NS silencing, but with less specificity than Ler [57].
H-NS silencing and counter-silencing of virulence genes is important in other pathogenic γ-proteobacteria as well. ToxT, a transcriptional activator belonging to the AraC family, promotes the production of cholera toxin and other virulence factors of Vibrio cholerae by both activating transcription and countering H-NS silencing [58, 59]. The VirB protein activates virulence gene expression in Shigella but resembles a plasmid partitioning protein rather than a classical transcriptional activator. VirB was found not to promote icsB transcription in an in vitro assay but rather to antagonize H-NS [60]. Binding of VirB upstream of the icsB promoter to a heptameric motif related to plasmid partitioning cis elements appears to result in derepression. Nearly all RovA (SlyA)-activated genes in Yersinia spp. are repressed by H-NS, and most RovA-bound promoters are also bound by H-NS [61]. The Y. enterocolitica inv gene encoding invasin is repressed by H-NS in conjunction with Hha (called YmoA in Y. enterocolitica) [62] and derepressed by RovA. Interestingly, removal of the last four amino acid residues of RovA abolishes its ability to displace H-NS even though dimerization and DNA-binding in vitro are retained, leading to the suggestion that the C-terminus of RovA might interact with H-NS directly (or stabilize interactions with RNAP or other RovA dimers) [63].
All five Salmonella enterica serovar Typhimurium pathogenicity islands (SPI1-5) are bound by H-NS. Activation of the SPI1 rtsA gene only requires the AraC family activators HilC and HilD if both H-NS and Hha are present [64, 65]. The SPI1-encoded transcriptional activator HilA not only promotes expression of SPI1 genes but also appears to counter H-NS silencing of the SPI4 pathogenicity island [66], which acts in concert with SPI1 during Salmonella interactions with the host intestinal mucosa [67]. A requirement for the transcriptional activator SsrB for SPI2 expression is reduced in the absence of H-NS, suggesting that SsrB both activates transcription and relieves H-NS-mediated repression [68]. This rationalizes the presence of SsrB binding sites both within the promoter region and downstream of the translational start site. An AT-rich gene from H. pylori introduced into S. Typhimurium was silenced by H-NS [22••], suggesting that H-NS inactivation may facilitate the overexpression of foreign genes for biotechnological applications.
Conclusions
A great deal has been learned about H-NS since its original description as a DNA-binding protein able to stimulate bacteriophage gene transcription [69]. Biophysical studies have revealed how H-NS dimers bridge DNA. Separate studies have demonstrated how cooperative binding and polymerization can impede transcription initiation. Global analyses of H-NS binding and effects on gene expression demonstrate that H-NS recognizes and preferentially binds to sequences with AT-content higher than the resident genome. A high-affinity H-NS binding site has been identified which can nucleate H-NS at a promoter region and allow subsequent spreading along adjacent lower affinity sites to silence transcription. Transcriptional silencing of sequences bound by H-NS can in turn be countered by a variety of mechanisms including activation by an alternative sigma factor, antagonism by other DNA-binding proteins, and interactions with H-NS paralogs. This provides a richer paradigm by which to understand the regulation of bacterial gene expression, in which silencing and counter-silencing complement transcriptional activation and repression. The ability of H-NS to recognize and silence the transcription of AT-rich DNA has important implications for the evolution of bacteria by horizontal gene transfer. Thus, the view of H-NS as a modulator of gene expression in response to environmental conditions is now being augmented by the appreciation that xenogeneic silencing by H-NS facilitates the integration of foreign DNA into regulatory networks.
Acknowledgments
This work was supported by the National Institutes of Health (AI39557, AI44486 and AI48622) and the Agence Nationale de la Recherche (NT05-2-42117 and PCV07-184204). The authors are grateful to William Navarre for his helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- • 2.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]; - These authors provide evidence for a link between superhelical density, nucleoid structure and the metabolome.
- 3.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr Opin Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Ceschini S, Lupidi G, Coletta M, Pon CL, Fioretti E, Angeletti M. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J Biol Chem. 2000;275:729–734. doi: 10.1074/jbc.275.2.729. [DOI] [PubMed] [Google Scholar]
- 5.Smyth CP, Lundback T, Renzoni D, Siligardi G, Beavil R, Layton M, Sidebotham JM, Hinton JC, Driscoll PC, Higgins CF, et al. Oligomerization of the chromatin-structuring protein H-NS. Mol Microbiol. 2000;36:962–972. doi: 10.1046/j.1365-2958.2000.01917.x. [DOI] [PubMed] [Google Scholar]
- 6.Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- •• 7.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]; - Optical tweezers were used to analyze the biophysical aspects of H-NS-mediated DNA bridging.
- 8.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 9.Becker NA, Kahn JD, Maher LJ., 3rd Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35:3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]; - This elegant study demonstrates the effects of nucleoid proteins on DNA stiffness and the consequences for loop formation.
- 10.Zimmerman SB. Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J Struct Biol. 2006;153:160–175. doi: 10.1016/j.jsb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol. 2007;17:R913–4. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 13.Vilar JM, Saiz L. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr Opin Genet Dev. 2005;15:136–144. doi: 10.1016/j.gde.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 15.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 17.Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J. 2005;391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J Mol Biol. 2006;355:169–174. doi: 10.1016/j.jmb.2005.10.034. [DOI] [PubMed] [Google Scholar]
- •• 19.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]; - A specific high-affinity H-NS binding site is discovered and characterized.
- • 20.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]; - Genome-wide analysis of a high-affinity H-NS binding motif shows clustering of sites in AT-rich regions.
- • 21.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]; - Approximately half of H-NS binding sites in non-coding regions found in this study were associated with RNA polymerase.
- •• 22.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]; - A striking correlation was observed between H-NS binding and sequences with increased AT-content, including virulence loci acquired by horizontal transfer.
- • 23.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]; - These observations provide strong corroboration of ref. 22.
- 24.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- •• 25.Nagarajavel V, Madhusudan S, Dole S, Rahmouni AR, Schnetz K. Repression by binding of H-NS within the transcription unit. J Biol Chem. 2007;282:23622–23630. doi: 10.1074/jbc.M702753200. [DOI] [PubMed] [Google Scholar]; - An attractive regulatory feedback loop model explains how synergy between H-NS bound to different sites can account for silencing of bgl expression.
- • 26.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]; - Eσ70 appears to facilitate H-NS-mediated DNA looping and trapping of RNA polymerase.
- 27.Madhusudan S, Paukner A, Klingen Y, Schnetz K. Independent regulation of H-NS-mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology. 2005;151:3349–3359. doi: 10.1099/mic.0.28080-0. [DOI] [PubMed] [Google Scholar]
- • 28.Radde N, Gebert J, Faigle U, Schrader R, Schnetz K. Modeling feedback loops in the H-NS-mediated regulation of the Escherichia coli bgl operon. J Theor Biol. 2008;250:298–306. doi: 10.1016/j.jtbi.2007.09.033. [DOI] [PubMed] [Google Scholar]; - See also ref. 25.
- 29.Madrid C, Balsalobre C, Garcia J, Juarez A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol. 2007;63:7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 30.Madrid C, Garcia J, Pons M, Juarez A. Molecular evolution of the H-NS protein: interaction with Hha-like proteins is restricted to enterobacteriaceae. J Bacteriol. 2007;189:265–268. doi: 10.1128/JB.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia J, Madrid C, Juarez A, Pons M. New roles for key residues in helices H1 and H2 of the Escherichia coli H-NS N-terminal domain: H-NS dimer stabilization and Hha binding. J Mol Biol. 2006;359:679–689. doi: 10.1016/j.jmb.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 32.Muller CM, Dobrindt U, Nagy G, Emody L, Uhlin BE, Hacker J. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol. 2006;188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Free A, Porter ME, Deighan P, Dorman CJ. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol Microbiol. 2001;42:903–917. doi: 10.1046/j.1365-2958.2001.02678.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Baldi DL, Tauschek M, Strugnell RA, Robins-Browne RM. Transcriptional regulation of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster, encoding the type II secretion pathway in enterotoxigenic Escherichia coli. J Bacteriol. 2007;189:142–150. doi: 10.1128/JB.01115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Tauschek M, Strugnell R, Robins-Browne RM. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology. 2005;151:1199–1208. doi: 10.1099/mic.0.27734-0. [DOI] [PubMed] [Google Scholar]
- 36.Pul U, Wurm R, Lux B, Meltzer M, Menzel A, Wagner R. LRP and H-NS--cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol Microbiol. 2005;58:864–876. doi: 10.1111/j.1365-2958.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- 37.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- • 38.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]; - These authors review evidence for the paradigm of xenogeneic silencing.
- 39.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- • 40.Typas A, Becker G, Hengge R. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]; - The ability of σS to escape H-NS trapping may account for the selectivity of σS-dependent promoters.
- 41.Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 42.Mujacic M, Baneyx F. Regulation of Escherichia coli hchA, a stress-inducible gene encoding molecular chaperone Hsp31. Mol Microbiol. 2006;60:1576–1589. doi: 10.1111/j.1365-2958.2006.05207.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Gottesman S. Modes of regulation of RpoS by H-NS. J Bacteriol. 2006;188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci U S A. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltrametti F, Kresse AU, Guzman CA. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J Bacteriol. 1999;181:3409–3418. doi: 10.1128/jb.181.11.3409-3418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 47.Weber A, Kogl SA, Jung K. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J Bacteriol. 2006;188:7165–7175. doi: 10.1128/JB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tramonti A, De Canio M, Delany I, Scarlato V, De Biase D. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J Bacteriol. 2006;188:8118–8127. doi: 10.1128/JB.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen AM, Qiu Y, Yeh N, Blattner FR, Durfee T, Jin DJ. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol Microbiol. 2005;56:719–734. doi: 10.1111/j.1365-2958.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- 50.Lithgow JK, Haider F, Roberts IS, Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol. 2007;66:685–698. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbett D, Bennett HJ, Askar H, Green J, Roberts IS. SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS. J Biol Chem. 2007;282:33326–33335. doi: 10.1074/jbc.M703465200. [DOI] [PubMed] [Google Scholar]
- 52.Stratmann T, Madhusudan S, Schnetz K. Regulation of the yjjQ-bglJ operon encoding LuxR-type transcription factors and the divergent yjjP gene by H-NS and LeuO. J Bacteriol. 2007 doi: 10.1128/JB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De la Cruz MA, Fernandez-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vazquez A, Calva E. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol. 2007;66:727–743. doi: 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- 54.Will WR, Frost LS. Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J Bacteriol. 2006;188:507–514. doi: 10.1128/JB.188.2.507-514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 55.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]; - An AT-rich plasmid carries an H-NS paralog to prevent detrimental effects on the host cell.
- 56.Torres AG, Lopez-Sanchez GN, Milflores-Flores L, Patel SD, Rojas-Lopez M, Martinez de la Pena CF, Arenas-Hernandez MM, Martinez-Laguna Y. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157:H7. J Bacteriol. 2007;189:5916–5928. doi: 10.1128/JB.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson HS, Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 58.Nye MB, Pfau JD, Skorupski K, Taylor RK. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol. 2000;182:4295–4303. doi: 10.1128/jb.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol Microbiol. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
- 60.Turner EC, Dorman CJ. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J Bacteriol. 2007;189:3403–3413. doi: 10.1128/JB.01813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cathelyn JS, Ellison DW, Hinchliffe SJ, Wren BW, Miller VL. The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol Microbiol. 2007;66:189–205. doi: 10.1111/j.1365-2958.2007.05907.x. [DOI] [PubMed] [Google Scholar]
- 62.Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol. 2006;188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran HJ, Heroven AK, Winkler L, Spreter T, Beatrix B, Dersch P. Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J Biol Chem. 2005;280:42423–42432. doi: 10.1074/jbc.M504464200. [DOI] [PubMed] [Google Scholar]
- 64.Olekhnovich IN, Kadner RJ. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol. 2006;357:373–386. doi: 10.1016/j.jmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 65.Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol. 2007;189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Main-Hester KL, Colpitts KM, Thomas GA, Fang FC, Libby SJ. Coordinate Regulation of Salmonella Pathogenicity Islands 1 (SPI1) and 4 (SPI4) in Salmonella enterica sv. Typhimurium. Infect Immun. doi: 10.1128/IAI.01224-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerlach RG, Jackel D, Geymeier N, Hensel M. Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect Immun. 2007;75:4697–4709. doi: 10.1128/IAI.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 69.Cukier-Kahn R, Jacquet M, Gros F. Two heat-resistant, low molecular weight proteins from Escherichia coli that stimulate DNA-directed RNA synthesis. Proc Natl Acad Sci U S A. 1972;69:3643–3647. doi: 10.1073/pnas.69.12.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]