Abstract
N-glycosylated proteins were isolated from Arabidopsis thaliana mature stems using affinity chromatography on Concanavalin A Sepharose, separated by 2D-electrophoresis and identified using nanoHPLC-MS/MS and MALDI-TOF MS. 102 glycoproteins were identified. 94% of these proteins were predicted by bioinformatics to be targeted to the secretory pathway and 87% of them were predicted to be localized in the cell wall or at the plasma membrane. 30% of these proteins belong to glycoside hydrolases (GHs) families with some of them possibly involved in the hydrolysis of cell wall polysaccharides. The second major class of identified proteins comprises aspartyl and serine proteases. Other proteins are predicted to be oxido-reductases, contain interacting domains, are potentially involved in signalling or have an unknown function. This is to our knowledge the first survey of plant cell wall N-glycosylated proteins.
Keywords: Arabidopsis, enzymology, growth & development, Arabidopsis Proteins, classification, metabolism, physiology, Cell Wall, enzymology, Chromatography, Affinity, methods, Chromatography, High Pressure Liquid, Computational Biology, Concanavalin A, chemistry, Glycoside Hydrolases, metabolism, physiology, Glycosylation, Plant Stems, enzymology, growth & development, Proteome, Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization
Keywords: Arabidopsis thaliana, cell wall, concanavalin A, glycoside hydrolase, glycosylation, plant stem
Introduction
Plant stems play different roles in growth and development, transport of nutrients and water and constitute a physical support for the plant. Major components of stem cell walls are polysaccharides (Carpita and Gibeault, 1993; Roberts, 1994). Polysaccharides represent up to 90% of plant cell walls and constitute three different kinds of polymers: cellulose, hemicelluloses and pectins. Plant cell wall polysaccharide composition and structure change during plant development and are different from one plant species to another (Cosgrove, 1997; Popper and Fry, 2003). Cell wall proteins (CWPs) contribute to wall architecture or are involved in the regulation of growth and development, or defence against biotic or abiotic stresses (Lee et al., 2004; Jamet et al., 2006). Cell wall modifying proteins such as glycoside hydrolases (GHs), esterases, transglycosylases, lyases and peroxidases are involved in the construction, remodelling or turnover of cell wall components (Heredia et al., 1995; Cosgrove, 1997; Fry, 2004; Stolle-Smits et al., 1999; Obel et al., 2002; Reiter, 2006).
The enzymes of GH and transglycosylase superfamilies are particularly important for the reorganization of cell wall polysaccharides after their deposition (Fry, 2004; Minic and Jouanin, 2006). They fall into several families whose distinction is based on amino acid sequence similarities (Henrissat, 1991; 1998). Exo-glycanases attack polysaccharides progressively from the non-reducing end or substituted side groups, thus releasing monosaccharides. Endo-glycanases attack polysaccharide backbones in an endo-fashion. They have a large impact on the molecular mass of polysaccharides. A third group of hydrolases can break some substituted non-carbohydrate groups linked to wall polysaccharides such as O-acetyl, O-methyl and O-feruloyl groups (Fry, 2004). Xyloglucan transglycosylase hydrolases (XTHs) can exhibit both endo-glycanase and transglycosylase activities (Fry, 2004).
Significant progress has been made in proteomic analysis of plant cell walls (Jamet et al., 2006). Interesting results were obtained using cell cultures, culture medium of seedlings, leaves, etiolated hypocotyls, protoplasts and roots (Chivasa et al., 2002; Borderies et al., 2003; Boudart et al., 2005; Charmont et al., 2005; Kwon et al., 2005; Jamet et al., 2006; Zhu et al., 2006). One cell wall proteome of mature stems was described in Medicago sativa L. (Watson et al., 2004). All these studies were based either on elution of cell wall proteins from living cells or on extraction of proteins from purified cell walls with salt solutions. However, since all CWPs are secreted proteins, they can be N-glycosylated with sugars such as D-glucose and D-mannose during their passage through endoplasmic reticulum and Golgi (Lerouge et al., 1998). It should be possible to trap them on Concanavalin A (Con A) which is a lectin extracted from Canavalia ensiformis L. able to bind molecules containing α-D-mannopyranosyl, β-D-glucopyranosyl or sterically-related residues (Carlsson et al., 1998). Recently, the N-glycoproteomes of human urine and human bile were analysed using Con A Sepharose affinity chromatography followed by 2D-electrophoresis and mass spectrometry (Kristiansen et al., 2004; Wang et al., 2006). The majority of the proteins identified were predicted to be extracellular or membrane components. Con A affinity chromatography was also used for the characterisation of N-linked glycoproteins of Ceanorhabditis elegans (Kaji et al., 2003) and of GHs from various plant organs (Sheldon et al., 1998; Wilson and Altmann, 1998; Minic et al., 2004; Li and Kushad, 2005; Minic et al., 2006; Van Riet et al., 2006). In this work, we have developed a new proteomic approach starting from a crude protein extract and using Con A Sepharose affinity chromatography to identify soluble cell wall N-linked glycoproteins. This glycoproteome is significantly enriched for putative cell wall GHs compared to previous cell wall proteomes.
Materials and methods
Plant material
Wild-type Arabidopsis thaliana, Wassilewskija ecotype, was grown in the greenhouse at 20°C to 22°C with a 16 h-photoperiod at 150 μE.m−2.s−1. Inflorescence stems of plants at mature stage (18–22 cm) were used for analysis.
Preparation of protein extracts from stems of A. thaliana
Mature stems of A. thaliana measuring 18–22 cm in length at the late flowering stage were used for analysis. Approximately 10 g of stem tissues were suspended in 12 mL of ice-cold extraction buffer and grinded in a mortar with a pestle for 5 min. The extraction buffer consisted in 25 mM BisTris pH 7.0 (HCl), 200 mM CaCl2, 10% (v/v) glycerol, 4 μM Na-cacodylate, 1/200 (v/v) protease inhibitor cocktail (P-9599, Sigma Chemical, St Louis, MO, USA). The ground material was centrifuged twice at 4°C for 3 min at 10,000 g, and the supernatant was further centrifuged for 15 min at 17,000 g. The resulting supernatant was used for chromatographic analyses.
Con A Sepharose affinity chromatography
A 1 x 6-cm column was filled with 3 mL of Con A Sepharose (Sigma Chemical, St Louis, MO, USA) and washed with 6 mL of 20 mM Tris pH 7.4 (HCl), 1 mM CaCl2/MgCl2/MnCl2 and 0.5 M NaCl buffer. The soluble protein extract (10 mL) was added and then washed with 15 mL of this buffer at a flow rate of 5 mL.h−1. Proteins were eluted with 0.2 M methyl-α-glucopyranoside in the same buffer. The eluate was collected, concentrated by “Ultrafree-CL” (10 kDa) (Sigma Chemical, St Louis, MO, USA) to 300 μL and dialysed against 7 M urea, 5 mM K2CO3, 0.125% SDS, 0.6% Triton X-100, 1 mM DTT, 2% carrier ampholytes 3–10 (GE Healthcare Europe GmbH, Orsay, France).
Glycoside hydrolase activities
The reaction mixture contained 2 mM pNP-glycosides (Sigma Chemical, St Louis, MO, USA), 0.1 M acetate buffer (pH 5.0), 2 mM sodium azide, and 50 μL of protein extract in a total volume of 0.5 mL. The reaction was carried out at 37°C for 5 to 60 min (depending on activity) and stopped by the addition of 0.5 mL 0.4 M sodium chloride. Controls were stopped at time 0. Concentration of the resulting pNP was determined spectrophotometrically at 405 nm by comparison to a calibration curve. Standard deviations values for 3 replicate assays were less than 5%.
2D-electrophoresis
Isoelectric focusing (IEF) was performed using 24 cm-immobilized pH gradient (IPG) strips (GE Healthcare Europe GmbH, Orsay, France) with a linear pH gradient from 4 to 7 and 250 μg of protein were applied on an IPG strip for in-gel rehydration in 7 M urea, 2 M thiourea, 2% CHAPS, 10 mM DTT, 2% IPG buffer pH 4–7 (Méchin et al., 2004). Focusing was achieved using a Protean IEF Cell (Bio-Rad, Hercules, CA, USA). An active rehydration was performed at 22°C during 12 h at 50 V prior to focusing. To improve sample entry, the voltage was increased step by step from 50 to 10,000 V (0.5 h at 200 V, 0.5 h at 500 V, 1 h at 1000 V then 10,000 V for a total of 94,000 V h). After IEF, IPG strips were successively incubated in 50 mM Tris pH 8.8 (HCl), 6 M urea, 30% glycerol, 2 % SDS, 1% DTT for 15 min, and in 50 mM Tris pH 8.8 (HCl), 6 M urea, 30% glycerol, 2 % SDS, 2.5% iodoacetamide for 15 min (Görg et al., 1987). Strips were further sealed on top of the 1 mm-thick second dimensional gel (24 x 24 cm) with the help of 1% low-melting agarose in SDS–electrophoresis buffer (25 mM Tris, 0.2 M glycine, 0.1% SDS). Continuous gels (11% T, 2.67% C gels with PDA as cross-linking agent) were used. Separation was carried out at 20 V for 1 h and subsequently at a maximum of 30 mA/gel, 120 V overnight, until the bromophenol blue front had reached the end of the gel.
Protein staining
Following 2D-electrophoresis, gels were stained with colloidal Coomassie blue G250 according to Mechin et al. (2004).
Identification of proteins by mass spectrometry
The individual protein spots obtained after 2D-electrophoresis were excised and in-gel digested with trypsin according to a standard protocol (Santoni et al., 2003). Tryptic peptides from each protein were analyzed by nanoHPLC-MS/MS or MALDI-TOF MS as previously described (Minic et al., 2004; Mechin et al., 2004). Proteins analysed by MALDI-TOF MS were identified via automated NCBI non redundant protein database (http://www.ncbi.nlm.nih.gov/) searching using the MASCOT programme (http://www.matrixscience.com/search_form_select.html). Only mowse scores exceeding threshold (p<0.5) were considered as positive results. Identification of proteins with nanoHPLC-MS/MS (ion trap) was performed with Biowoks™ (Thermo scientific, San Jose, USA). The main search parameters were methionine oxidation as differential modification and trypsin as enzyme. One miss cleavage was allowed. The A. thaliana protein database was downloaded from the mips website (http://mips.gsf.de/projects/plants). Identification was considered significant when the proteins were identified with at least 2 different tryptic peptides as first candidate, Xcorr > 1.7, 2.2 and 3.3 for respectively mono-, di- and tri-charged peptides and delta Cn >0.1.
Bioinformatics analyses
Sub-cellular localization and length of signal peptides were predicted using PSORT (http://psort.nibb.ac.jp/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Nielsen et al., 1997; Emanuelsson et al., 2000). Prediction of transmembrane domains was done with Aramemnon (http://aramemnon.botanik.uni-koeln.de/) (Schwacke et al, 2003). Molecular masses and pI values were calculated using the aBi program (http://www.up.univ-mrs.fr/~wabim/d_abim/compo-p.html). Homologies to other proteins were searched for using BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al., 1990). Identification of protein families and functional domains was performed using MyHits (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and InterProScan (http://www.ebi.ac.uk/InterProScan/) (Quevillon et al., 2005). GHs and CEs were classified according to the CAZy database (http://www.cazy.org/CAZY/) (Coutinho et al., 1999). Peroxidases were named as in the PeroxiBase (http://peroxidase.isb-sib.ch/index.php) (Bakalovic et al., 2006).
Protein measurements
Protein concentration was determined by the method of Bradford (1996) using bovine serum albumin dissolved in extraction buffer as the standard.
Results and discussion
Extraction of glycoside hydrolases from stem tissues of A. thaliana
In a first attempt to study GHs from stem tissues of A. thaliana by using a proteomic approach it was necessary to establish a protocol for the extraction of these enzymes. Based on published experimental data on the purification of GHs from various plant organs (Sheldon et al., 1998; Wilson and Altmann, 1998; Minic et al., 2004; Li and Kushad, 2005; Van Riet et al., 2006), we developed a 2-step extraction procedure. Stems were ground in a buffer containing 200 mM CaCl2, followed by Con A Sepharose affinity chromatography. CaCl2 was chosen as the most efficient salt for CWP extraction (Boudart et al., 2005). This protocol is different from those used in previous cell wall proteomic studies (Feiz et al., 2006): (i) the initial step is a grinding in a buffer containing 200 mM CaCl2 to release CWPs instead of a low ionic strength buffer usually used to prevent CWP elution; (ii) there is no step of cell wall isolation to avoid loosing CWPs weakly-bound to cell walls during the centrifugation steps required for cell wall isolation; (iii) the last step is an affinity chromatography to trap N-glycosylated proteins. Results show that the affinity chromatography step resulted in a significant increase in specific activities of several exo-GHs using artificial substrates compared to what was measured in the dialysed crude protein extract. These increases varied from 2.0 for β-D-xylosidase to 6.1 for β-D glucosidase (Table 1). On the basis of this observation, this protocol was adapted to analyze the N-glycoproteome of mature stems.
Table 1. Specific activities of several glycoside hydrolases after Con A Sepharose affinity chromatography.
All enzyme activities were measured in vitro at 37 °C, using 50 μL of protein and pNP-glycosides as substrates.
| Enzyme | Specific activities (nmol/min/mg protein)
|
Recovery (%)
|
Ratio of specific activities
|
|
|---|---|---|---|---|
| Crude protein extract | After Con A Sepharose | Con A Sepharose/Crude protein extract | ||
| β-D-xylosidase | 32 | 89 | 43 | 2.8 |
| α-L-arabinofuranosidase | 103 | 442 | 66 | 4.3 |
| β-D-glucuronidase | 10 | 22 | 34 | 2.0 |
| β-D-mannosidase | 26 | 108 | 64 | 4.2 |
| α-D-mannosidase | 47 | 172 | 56 | 3.6 |
| β-D-glucosidase | 82 | 497 | 93 | 6.1 |
| α-D-galactosidase | 85 | 468 | 84 | 5.5 |
| β-D-galactosidase | 393 | 825 | 32 | 2.1 |
| α-D-glucosidase | 8 | 31 | 59 | 3.9 |
Proteomic analysis after enrichment of the soluble protein extract in glycoside hydrolases by lectin affinity chromatography
The proteomic analysis was performed using prot ein extracts from 18–22 cm mature stems at the late flowering stage. About 10 g of stem tissues were used for the extraction of proteins. After grinding and centrifugation, the crude protein extract contained 2.5 mg of protein as determined by the Bradford method (1996). A fraction of this protein extract was subjected to Con A Sepharose affinity chromatography. Eluted proteins were concentrated and dialysed, resulting in a fraction of 400 μL containing about 300 μg of protein. A sample containing 250 μg of protein was subjected to 2D-electrophoresis. Proteins were detected by colloidal Coomassie blue staining (Fig. 1). The number of resolved spots was about 200. Fifty-seven spots resolved by 2D-electrophoresis were analyzed using MALDI-TOF. Spots corresponding to proteins having molecular mass smaller than 20 kDa were not analyzed since they are not expected to contain GHs on the basis of calculations made from genes predicted to encode such proteins (Minic and Jouanin, 2006). The other proteins visible on the gel were also analyzed, but due to small quantity or mixture with other proteins their scores were not significant. Fifteen spots localized at the basic side of the gel were subjected to tryptic digestion and proteins were identified using nanoHPLC-MS/MS. Each of them was expected to contain more than one protein since previous studies showed that most CWP are basic (Jamet et al., 2006). A total number of 102 different proteins was identified (Table 2; Tables S1 and S2 at JXB online). Many of these proteins were present in several of the spots resolved by 2D-electrophoresis suggesting post-translational modifications such as glycosylations. On the basis on these results, those spots were collected into thirty-five groups as shown in Fig 1. Conversely, as expected, most of the 15 spots at the basic side of the gel contained more than one protein.
Fig 1.
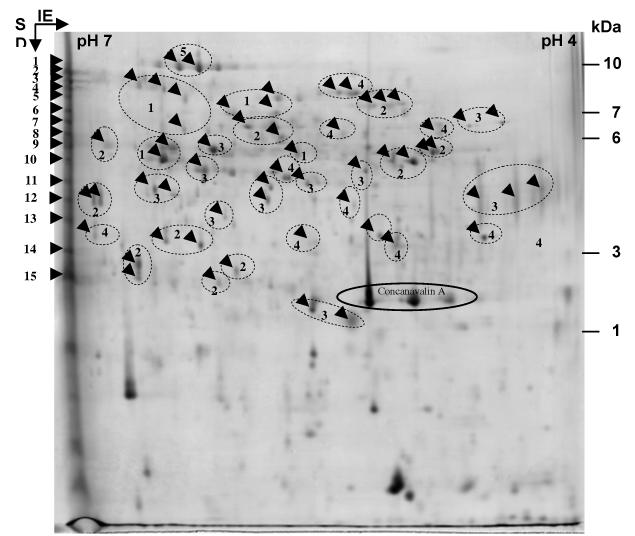
Analysis of A. thaliana proteins by 2D-electrophoresis. The 2D-gel was loaded with 250 μg of the fraction obtained after Con A Sepharose affinity chromatography from stem tissues of A. thaliana. The gel was stained with colloidal Coomassie blue. Fifteen spots were picked for nanoHPLC-MS/MS (1 to 15) and fifty-seven for MALDI-TOF analyses. In the latter case, numbering refers to groups of spots containing the same protein. Arrows in the circles represent same identified proteins. Molecular mass markers are indicated on the right.
Table 2.
Proteins identified through proteomic analysis of A. thaliana mature stems
| Predicted or known functionsa | Accession | Spot numberb |
|---|---|---|
| Proteins acting on polysaccharides | ||
| GH family 1 | At2g44450 | 7 |
| GH family 1 | At3g18080 | 13, 14, 15 |
| GH family 1 | At5g54570 | 47 |
| GH family 1 (thioglucoside hydrolase 1) (TGG1) | At5g26000 | 41 |
| GH family 3 (BXL1) (beta-xylosidase 1) | At5g49360 | 5, 9, 11, 13 |
| GH family 3 (BXL2) (beta-xylosidase 2) | At1g02640 | 13, 15 |
| GH family 3 | At5g20950 | 5, 6, 8 |
| GH family 16 (xyloglucan endotransferase) (EXGT-A1) (At-XTH4) | At2g06850 | 13,14, 15 |
| GH family 16 (xyloglucan endotransferase) (Meri5) (At-XTH24) | At4g30270 | 15 |
| GH family 17 | At3g07320 | 6, 7 |
| GH family 17 | At5g56590 | 11, 13 |
| GH family 17 | At4g31140 | 33 |
| GH family 19 (chitinase), carbohydrate-binding module family 18 (chitin binding function) | At2g43570 | 31 |
| GH family 20 (N-acetyl-beta-glucosaminidase) | At1g65590 | 7, 10 |
| GH family 20 (N-acetyl-beta-glucosaminidase) | At3g55260 | 22 |
| GH family 28 (polygalacturonase) | At1g80170 | 8, 9, 10 |
| GH family 28 (polygalacturonase) | At3g07830 | 24 |
| GH family 28 (polygalacturonase) | At3g16850 | 29 |
| GH family 31 (alpha-glucosidase) (AtGLU1) | At5g11720 | 9, 13 |
| GH family 31 (alpha-xylosidase) (XYL1) | At1g68560 | 17 |
| GH family 32 (beta-fructofuranosidase) (ATBETAFRUCT4) | At1g12240 | 28 |
| GH family 35 (beta-galactosidase) (BGAL1) | At3g13750 | 8 |
| GH family 35 (beta-galactosidase) (BGAL9) | At2g32810 | 10 |
| GH family 35 (beta-galactosidase) (BGAL10) | At5g63810 | 7, 13 |
| GH family 38 (alpha-mannosidase) | At5g13980 | 13 |
| GH family 38 (alpha-mannosidase) | At3g26720 | 42 |
| GH family 51 (alpha-L-arabinofuranosidase) | At3g10740 | 27 |
| GH family 79 (endo-beta-glucuronidase/heparanase) | At5g07830 | 10 |
| GH family 79 (endo-beta-glucuronidase/heparanase ) | At5g34940 | 7 |
| CE family 8 (pectin methylesterase) | At3g43270 | 10 |
| CE family 13 (pectin acylesterase) | At5g23870 | 9 |
| Oxido-reductases | ||
| homolog to SKU5 (SKS4) (multicopper oxidase domain) | At4g22010 | 6 |
| homolog to SKU5 (SKS5) (multicopper oxidase domain) | At1g76160 | 7, 8 |
| homolog to SKU5 (SKS6) (multicopper oxidase domain) | At1g41830 | 7, 8, 12 |
| homolog to SKU5 (SKS7) (multicopper oxidase domain) | At1g21860 | 7, 12, 13 |
| homolog to SKU5 (SKS17) (multicopper oxidase domain) | At5g66920 | 10 |
| SKU5 (multicopper oxidase domain) | At4g12420 | 3, 6, 7, 8, 9 |
| homolog to peroxidase (AtPrx22) | At2g38380 | 11 |
| homolog to peroxidase (AtPrx28) | At3g03670 | 49 |
| homolog to peroxidase (AtPrx33) | At3g49110 | 11, 13 |
| homolog to peroxidase (AtPrx34) | At3g49120 | 23 |
| homolog to C. pepo ascorbate oxidase (P37064) | At5g21100 | 11 |
| homolog to berberine bridge enzyme (S)-reticulin:oxygen oxido-reductase | At4g20830 | 13 |
| homolog to germin (AtGER3) | At5g20630 | 32 |
| Proteins with interacting domains | ||
| homolog to carrot EDGP and tomato XEGIP | At1g03230 | 9, 10 |
| homolog to carrot EDGP and tomato XEGIP | At1g03220 | 39 |
| expressed protein (LRR repeats) | At1g33590 | 26 |
| expressed protein (LRR repeats) | At1g33600 | 8, 9 |
| expressed protein (LRR repeats) | At2g42800 | 14 |
| polygalacturonase inhibiting protein (PGIP2) | At5g06870 | 13, 15 |
| homolog to lectin (legume lectin beta domain) | At1g53070 | 13 |
| homolog to lectin (legume lectin beta domain) | At1g53080 | 13, 14, 15 |
| homolog to lectin (legume lectin beta domain) | At3g16530 | 1, 13, 14, 15 |
| homolog to lectin (curculin-like) | At1g78850 | 7, 8 |
| homolog to lectin (curculin-like) | At1g78860 | 7, 8 |
| homolog to lectin (curculin-like) | At5g18470 | 7, 8, 10 |
| Signalling | ||
| fasciclin-like arabinogalactan protein (AtFLA8) | At2g45470 | 1, 13, 14, 15 |
| fasciclin-like arabinogalactan protein (AtFLA13) | At5g44130 | 30 |
| Proteases | ||
| homolog to aspartyl protease | At1g09750 | 11, 12, 13 |
| homolog to aspartyl protease | At3g52500 | 8, 9 |
| homolog to aspartyl protease | At5g07030 | 10, 11, 12, 13 |
| homolog to aspartyl protease | At3g54400 | 21 |
| homolog to serine protease | At1g30600 | 7 |
| homolog to serine protease | At2g39850 | 50 |
| homolog to serine protease | At4g21630 | 12, 13, 14, 15 |
| homolog to serine protease | At4g21650 | 10, 13, 14, 15 |
| homolog to serine protease | At5g67360 | 16 |
| homolog to serine protease | At3g14067 | 18 |
| xylem serine peptidase 1 (XSP1) | At4g00230 | 9 |
| homolog to serine carboxypeptidase (SCPL11) | At2g22970 | 14, 15 |
| homolog to serine carboxypeptidase (SCPL12) | At2g22920 | 15 |
| homolog to serine carboxypeptidase (SCPL34) | At5g23210 | 12, 13 |
| Miscellaneous | ||
| homolog to anther specific proline-rich protein APG (lipase acylhydrolase domain) | At1g54000 | 13 |
| homolog to anther specific proline-rich protein APG (lipase acylhydrolase domain) | At1g67830 | 12 |
| homolog to purple acid phosphatase | At2g16430 | 7 |
| homolog to purple acid phosphatase | At2g27190 | 13 |
| homolog to purple acid phosphatase | At5g34850 | 19 |
| homolog to Chlamydomonas reinhardtii apospory associated protein (aldose-1-epimerase domain) | At4g25900 | 13 |
| homolog to glycerophosphoryl diester phosphodiesterase | At4g26690 | 35 |
| Unknown function | ||
| expressed protein | At1g21680 | 5, 6 |
| expressed protein | At1g21670 | 2, 3 |
| expressed protein | At3g14920 | 14, 15 |
| expressed protein (DUF642) | At4g32460 | 44, 13 |
| expressed protein (DUF642) | At5g11420 | 10, 11, 12 |
| expressed protein (DUF642) | At5g25460 | 12 |
| expressed protein (DUF1005) | At4g29310 | 13 |
| expressed protein (DUF1184) | At4g18080 | 46 |
| expressed protein (DUF1680) | At5g12950 | 5 |
| expressed protein (cupin domains) | At3g22640 | 34 |
| Intracellular proteins | ||
| homolog to thioredoxin | At1g21750 | 45 |
| GH family 1 | At2g25630 | 7 |
| homolog to serine protease | At2g05920 | 3, 9 |
| homolog to serine carboxypeptidase | At4g36195 | 43 |
| homolog to phosphorylase | At4g24350 | 12 |
| homolog to gamma-glutamyltranspeptidase | At4g39640 | 12 |
| homolog to aldo-keto reductase | At2g27680 | 37 |
| homolog to peroxiredoxin | At3g52960 | 25 |
| homolog to copper amine oxidase | At4g12990 | 21 |
| expressed protein (PH domain) | At2g30880 | 36 |
| expressed protein (RNA recognition motif) | At3g52980 | 40 |
| expressed protein | At2g41950 | 48 |
| expressed protein (CBS domains) | At3g48530 | 38 |
Functions are predicted as described in Experimental
Spot number refers to Fig. 1.
Bioinformatic prediction of sub-cellular localization and N-glycosylation of identified proteins
PSORT, TargetP and Aramemnon programmes were used to predict the sub-cellular localization of the identified proteins. Seventy-seven out of the 102 identified proteins (77%) were predicted to be localized in the cell wall matrix, 13 at the plasma membrane, 6 into the endoplasmic reticulum, 2 in the cytoplasm and 3 in the chloroplast. Six proteins were predicted to be either targeted to vacuoles or to the cell wall. However, vacuolar targeting is not well-established in plants and the predictions are not yet very reliable (Hadlington and Denecke, 2000). Altogether, about 90% of the proteins have a predicted N-terminal signal peptide, which means that all of these proteins are targeted to the secretory pathway. Two proteins could not be assigned to any sub-cellular compartment due to discrepancies between predictions with PSORT and TargetP. Seven proteins were predicted to harbour a glycosyl phosphatidyl inositol (GPI) anchor. As expected, all of the identified proteins contained N-glycosylation sites as predicted by the MyHits programme (see Supplementary Table S1 at JXB).
These results show that the proposed protocol allowed the isolation of a protein fraction essentially composed of N-linked glycoproteins targeted to either the cell wall or to the plasma membrane. Despite the absence of a cell wall purification step, it should be noted that the proportion of proteins with a predicted intracellular localization was very low (12%). This protocol thus appears as an efficient alternative to previously described protocols used for A. thaliana cell wall proteomic analyses. Previous protocols include: (i) non-destructive methods such as analysis of culture media (Borderies et al., 2003; Charmont et al. 2005), washing of cells cultured in liquid medium with salt solutions (Borderies et al., 2003; Kwon et al. 2005) and vacuum infiltration of leaves (Boudart et al., 2005); (ii) destructive methods, i.e. cell wall purification, prior to CWP extraction with various buffers (Chivasa et al. 2002; Bayer et al., 2006; Feiz et al., 2006). The choice of a particular protocol will depend on the aim of the study and on the plant organ of interest.
Identification and functional classification of proteins
Identification of protein families and functional domains were performed using several bioinformatic programmes. Proteins were classified according to the predicted functional classes of CWPs proposed by Jamet et al. (2006) (Tables 2, 3). Proteins belonging to seven functional classes were found according to the presence of functional domains predicted as described in the Material and methods: (i) proteins acting on polysaccharides include GH and esterases; (ii) oxido-reductases mainly include peroxidases and multicopper oxidases; (iii) proteins with interacting domains include proteins with lectin or LRR (leucine rich repeat) domains as well as enzyme inhibitors; (iv) proteins involved in signaling processes include fasciclin AGPs (arabinogalactan proteins); (v) proteases; (vi) proteins of yet unknown function; (vii) miscellaneous. However, this classification is provisional since the biological role of many of these proteins remains to be determined (Tatosov et al., 1997). For example, an enzyme of the GH 3 family (XYL3) shows amino acid homology with β-D-xylosidase. However, it was identified as an enzyme that efficiently hydrolyzed arabinosyl residues from arabinans, suggesting that it works as an α-L-arabinofuranosidase (Minic et al., 2006).
Table 3. Predicted functional classes of proteins in the A. thaliana stem sub-proteome.
Functional classes have been defined according to Jamet et al. (2006). A list of all proteins identified in this study is provided in Table 2. The detailed bioinformatics functional analysis is given in Supplementary Table S1 at JXB online.
| Functional classes | Number of proteins | |
|---|---|---|
| Proteins acting on polysaccharides | 31 | |
| Glycoside hydrolases | 29 | |
| Esterases | 2 | |
| Oxido-reductases | 13 | |
| Peroxidases | 4 | |
| Multicopper oxidases | 6 | |
| Others | 3 | |
| Proteins with interacting domains | 12 | |
| Lectin domains | 6 | |
| LRR domains | 3 | |
| Others | 3 | |
| Signalling | 2 | |
| Proteases | 14 | |
| Serine proteases | 10 | |
| Aspartyl proteases | 4 | |
| Miscellaneous | 7 | |
| Homologs to phosphatase | 3 | |
| Homologs to proline-rich protein (lipase acid hydrolase domain) | 2 | |
| Others | 2 | |
| Unknown function | 10 | |
| Intracellular proteins | 13 | |
| Total | 102 | |
Thirty-three proteins were expected to act on polysaccharides (Table 2). Furthermore, 30 proteins (29%) belong to the superfamily of GHs, 29 of which were predicted to be extracellular or plasma membrane-associated. The second largest group comprises 16 proteases, 14 of which were predicted to be localized in the extracellular matrix. Together GHs and proteases represent 47% of identified proteins. Among other proteins, oxido-reductases, proteins with interacting domains, miscellaneous proteins, proteins of unknown function, signalling and intracellular proteins were identified.
Proteins from the same functional classes as in previous cell wall proteomic studies were found, but 37 proteins were not identified before (Chivasa et al. 2002; Borderies et al. 2003; Borner et al., 2003; Schultz et al., 2004; Boudart et al., 2005; Charmont et al. 2005; Kwon et al. 2005; Bayer et al., 2006; Feiz et al. 2006). This stem N-glycoproteome thus appears to be very specific. Among the 90 proteins predicted to be at the plasma membrane or in the cell wall, only 5 also have been found in previously described proteomes: cell suspension cultures, rosette leaves and etiolated hypocotyls (Jamet et al., 2006). They encode a β-xylosidase that belongs to the GH 31 family (At1g68560), a multicopper oxidase (At1g76160), two lectins (At1g78850, At1g78860) and a protein homologous to the carrot extracellular dermal glycoprotein (EDGP) and to the tomato xyloglucan specific endoglucanase inhibitor protein (XEGIP) (At1g03220) (Qin et al., 2003). However, some protein families are missing. Since proteins with molecular masses lower than 20 kDa were not analyzed, it was not possible to identify homologs to protease or pectin methylesterase inhibitors, non-specific lipid transfer proteins, and blue copper binding proteins. Only one protein homolog to germins was identified. Although several expansins, which molecular masses are between 25 and 30 kDa, were previously identified in cell wall proteomes (Jamet et al., 2006), none was found in this study. This might be explained either by their low abundance or their low level of N-glycosylation. Finally, no structural protein could be identified either because of their strong binding to the extracellular matrix, or the absence of N-glycans.
Possible roles of proteins identified in stem tissues of A. thaliana
Proteins acting on polysaccharides constitute the major functional class. According to Coutinho et al. (1999), they belong to 12 GH families and to 2 carbohydrate esterase (CE) families (Fig. 2, Table 2). These enzyme families have diverse biological functions in defence, signalling, hydrolysis of starch, and cell wall modifications (Minic and Jouanin, 2006). A total of 18 GHs belonging to 7 different GH families were found. Three of them, αL-arabinofuranosidase (At3g10740), α-L-arabinofuranosidase/β-D-xylosidase (At5g49360) and β-glucosidase AtGLU1 (At5g11720), were recently purified and characterized (Minic et al., 2004; Monroe et al., 1999). The XYL1 β-xylosidase and two XTHs (Meri5/At4g30270, EXGT-A1/At2g06850) have been studied previously using both biochemical and genetic approaches (Akamatsu et al., 1999; Sampedro et al., 2001; Rose et al., 2002). A pectin methyl- and a pectin acyl-esterase were also identified. Previous studies have shown that pectin methylesterase activity is inversely correlated to the growth rate of expanding tissues, suggesting its possible involvement in wall rigidification (McQueen-Mason and Cosgrove, 1995).
Fig. 2.
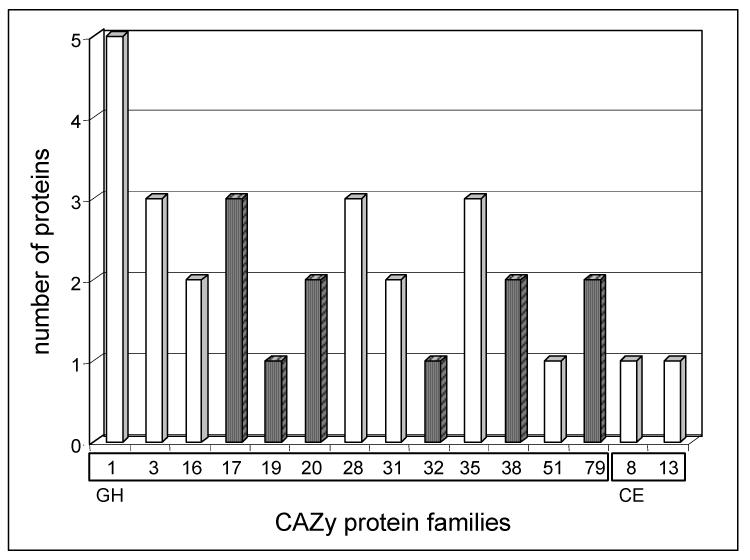
Distribution of families of glycoside hydrolases (GH) and carbohydrate esterases (CE) in the A. thaliana stem sub-proteome. Proteins are listed in Table 2. Proteins have been classified according to the CAZy nomenclature (Henrissat et al., 1998; http://www.cazy.org/). Glycoside hydrolase (GH) families from 1 to 79 are on the left whereas carbohydrate esterase (CE) families 8 and 13 are on the right. White bars correspond to families that might participate in cell wall modification and reorganization.
Possible substrates in muro of the majority of these enzymes are xyloglucans and pectins. Most of them act as exo-enzymes whereas only enzymes belonging to GH families 16 and 28 act as endo-GHs on xyloglucans and homogalacturonans, respectively. Other cell wall GHs can hydrolyse β1,4 glucan, arabinoxylan and xylan. These results suggest that xyloglucan and pectins, that are composed of homogalacturonans (HG), arabinans and galactans (RG-I), undergo structural changes in mature stem. However, some GH families can act on several natural polysaccharides showing broad substrate specificity. This low specificity has been reported in the case of several purified cell wall GHs (Leach et al, 1995; Kim and al., 2000; Sampedro et al., 2001; Steele et al., 2001; Rose et al., 2002; Lee et al., 2003; Minic et al, 2004; Minic et al., 2006). It has been hypothesized that it allows efficient modification of complex cell wall polysaccharides without requiring an extremely high number of enzymes (Minic et al, 2004; 2006).
Many GH families described here could have other functions than cell wall modifications. Predicted extracellular GHs such as β-D-glucuronidase (GH 79), α-D-mannosidase (GH 38) and acetyl-N-hexasaminase (chitinase-like enzymes, GH 19) could be involved in post-translational modifications of glycoproteins. Recently, an A. thaliana β-D-glucuronidase (AtGUS) was shown to hydrolyze glucuronic acids from carbohydrate chains of AGPs (Eudes, personal communication). Kinetic and structural analyses of Ginkgo α-D-mannosidase acting on a pyridylamino derivative of oligo mannosides strongly suggested its involvement in the catabolism and turnover of N-linked glycoproteins (Woo et al., 2004). The pumpkin endo-β-N-acetylglucosaminidase, partially purified from cotyledons, was highly active towards high-mannose type glycans (Kimura et al., 2002).
Among other GHs, one thioglucosyl hydrolase (GH 1), 3 β-1,3-D-glucanase (GH 17) and 3 chitinase-like enzymes (GH 19 and 20) were identified. Thioglucosidases, also known as myrosinases, play diverse roles in cruciferous plant during growth, development and defence against microorganisms and insects (Rodman, 1991). Chitinase-like enzymes are able to degrade chitin in cell walls of fungal pathogens. However, the substrates and functions of most chitinase-like enzymes are not completely known. For example, a mutation in the chitinase-like gene classified in GH 19 family (AtCTL1/At1g05850) caused a cellulose deficiency as well as aberrant patterns of lignification with incomplete cell walls in the stem pith (Zhong et al., 2002; Rogers et al., 2005).
The second largest class of identified proteins comprises proteases. Seventeen putative proteinases including aspartyl and serine type proteases were found (Table 2). Fifteen of them were predicted to be secreted (Supplementary Table S1 at JXB online). The abundance of proteases in mature stem suggests that these enzymes may be actively involved in secondary wall formation. Proteases may play various roles in plant development and during plant pathogen interactions through maturation of CWPs and generation of active peptides. It has been shown that the extracellular subtilisin-like serine protease SDD1 (STOMATAL DENSITY AND DISTRIBUTION 1) is involved in the regulation of stomatal density and distribution in A. thaliana (Berger and Altmann, 2000). ALE1 (ABNORMAL LEAF EPIDERMIS1) is also predicted to encode a subtilisin-like serine protease and is assumed to produce a peptide required for proper differentiation of epidermis (Tanaka et al., 2001). CDR1 (CONSTITUTIVE DISEASE RESISTANT 1) encodes a putative aspartic protease (Xia et al., 2004). Overexpression of CDR1 causes dwarfing and resistance to virulent Pseudomonas syringae. It was shown that CDR1 generates a small mobile signal (3–10 kDa) sensitive to heating and to protease. The substrates of these three proteases are yet unknown. On the contrary, it was shown that the CLE (CLV3/ESR-related) basic secreted proteins are processed at their C-terminus to generate 14 amino-acid peptides that carry a biological activity (Ito et al., 2006; Kondo et al. 2006). In those cases, the proteases have not yet been identified. Finally, several plant cell wall proteomic analyses show a large discrepancy between the observed and the expected molecular masses of proteins (Boudart et al., 2005; Kwon et al., 2005; Zhu et al., 2006). Proteases could be involved in processing and/or turnover of cell wall proteins.
Several oxido-reductases such as multicopper oxidase-like (6 proteins), peroxidases (4 proteins), germin-like protein (1 protein) and a homolog to berberine bridge enzyme were identified. In contrast to previously characterized cell wall proteomes, this study allowed the identification of numerous multicopper oxidase-like proteins. They catalyse full, four-electron reduction of dioxygen (O2) to water (H2O) using a variety of substrates (Solomon et al., 1997). They belong to a large gene family of 19 members in A. thaliana (Jacob and Roe, 2005). Only two members of the family have been previously studied, SKU5 (At4g12420) and SKS6 (At1g41830). It was shown that SKU5 is involved in the control of root growth (Sedbrook et al., 2002) and that SKS6 contributes to cotyledon vascular patterning during development (Jacob and Roe, 2005). Peroxidases are involved in many physiological and developmental processes that have been reviewed recently (Passardi et al., 2004). They can be involved in both cell elongation processes and in their arrest. In the latter case, they catalyze the formation of bridges across phenolic residues of lignins and between lignins and adjacent cell wall proteins or polysaccharides.
Three extracellular acid phosphatases were identified in this work. The presence of phosphorylated proteins and phosphatases in plant cell wall has been reported in several proteomes (Chivasa et al, 2002; Kwon et al., 2005; Jamet et al., 2006). However, no extracellular kinase has yet been found (Chivasa et al., 2005). Acid phosphatases may participate in extracellular signalling events or in regulation of cell wall proteins.
Some proteins contained interacting domains, such as LRRs. The polygalacturonase-inhibiting protein (PGIP2/At5g06870) plays a role in plant defence (Di Matteo et al., 2006). Two proteins identified as fasciclin-like AGPs (AtFLA8/At2g45470, AtFLA13/At5g45130) can participate in cell-to-cell adhesion in plant (Johnson et al., 2003; Groover and Robischon, 2006). Finally, 10 proteins of unknown function were found.
Concluding remarks
This study demonstrates the effectiveness of the purification procedure to isolate cell wall glycoproteins. This includes novel GHs, multicopper oxidases and proteases. In contrast to these analyses, we did not identify homologs to protease or pectin methylesterase inhibitors, non-specific lipid transfer proteins, blue copper binding proteins, expansins and structural proteins. The abundance of GHs suggests a great plasticity of polysaccharides in cell walls, even in well-differentiated tissues such as mature stems. Finally, the presence of phosphatases, proteases and GHs suggests a complex regulation of cell wall proteins involving various types of post-translational modifications such as de-phosphorylation and hydrolytic processing by proteases or glycosidases.
Supplementary data
Supplementary data are available at JEB online. Table S1: Proteins identified in Arabidopsis thaliana mature stems. Table S2: LC-MS/MS data allowing identification of proteins in spots 1 to 15.
Acknowledgments
This work was partly funded by the Génoplante program Af2001-009. We thank Bruno Letarnec for growing A. thaliana plants, Dr Christain Malosse for mass spectrometry analyses, Drs Jorun Johansen and Herman Höfte for improving this manuscript.
References
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda Y. Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiology. 1999;121:715–722. doi: 10.1104/pp.121.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bakalovic N, Passardi F, Ioannidis V, Cosio C, Penel C, Falquet L, Dunand C. PeroxiBase: A class III plant peroxidase database. Phytochemistry. 2006;67:534–539. doi: 10.1016/j.phytochem.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics. 2006;6:301–311. doi: 10.1002/pmic.200500046. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes and Development. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya L, Brewer CF. Formation of homogeneous carbohydrate-lectin cross-linked precipitates from mixtures of D-galactose/N-acetyl-D-galactosamine-specific lectins and multiantennary galactosyl carbohydrates. European Journal of Biochemistry. 1992;208:179–185. doi: 10.1111/j.1432-1033.1992.tb17172.x. [DOI] [PubMed] [Google Scholar]
- Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerre-Tugaye MT, Boudet A, Pont-Lezica R. Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis. 2003;24:3421–3432. doi: 10.1002/elps.200305608. [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiology. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerré-Tugayé MT, Pont-Lezica R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics. 2005;5:212–221. doi: 10.1002/pmic.200400882. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Janson J-C, Sparrman M. Affinity chromatography. In: Janson J-C, Ryden L, editors. Protein Purification, Principles, High Resolution and Applications. VCH Publishers Inc; New York: 1998. pp. 275–329. [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Charmont S, Jamet E, Pont-Lezica R, Canut H. Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings: improved recovery following removal of phenolic compounds. Phytochemistry. 2005;66:453–461. doi: 10.1016/j.phytochem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Lindsey K, Sladas AR. Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. The Plant Cell. 2005;17:3019–3034. doi: 10.1105/tpc.105.036806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR. Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis. 2002;23:1754–1765. doi: 10.1002/1522-2683(200206)23:11<1754::AID-ELPS1754>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annual Review of Cell Developmental Biology. 1997;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. The Royal Society of Chemistry; Cambridge: 1999. pp. 3–12. [Google Scholar]
- Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006;67:528–533. doi: 10.1016/j.phytochem.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Feiz L, Irshad M, Pont-Lezica RF, Canut H, Jamet E. Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods. 2006;2:10. doi: 10.1186/1746-4811-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytologist. 2004;161:641–675. doi: 10.1111/j.1469-8137.2004.00980.x. [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash S, Somerlot L. Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis. 1987;8:122–124. [Google Scholar]
- Hadlington JL, Denecke J. Sorting of soluble proteins in the secretory pathway of plants. Current Opinion in Plant Biology. 2000;3:461–468. doi: 10.1016/s1369-5266(00)00114-x. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemistry Journal. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. Glycosidase families. Biochemical Society Transactions. 1998;26:153–156. doi: 10.1042/bst0260153. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jacobs J, Roe JL. SKS6, a multicopper oxidase-like gene, participates in cotyledon vascular patterning during Arabidopsis thaliana development. Planta. 2005;222:652–666. doi: 10.1007/s00425-005-0012-3. [DOI] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF. Cell wall proteins: a new insight through proteomics. Trends in Plant Sciences. 2006;11:33–39. doi: 10.1016/j.tplants.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nature Biotechnology. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kim JB, Olek AT, Carpita NC. Cell wall and membrane-associated exo-beta-D-glucanases from developing maize seedlings. Plant Physiology. 2000;123:471–486. doi: 10.1104/pp.123.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Matsuo S, Tsurusaki S, Kimura M, Hara-Nishimura I, Nishimura M. Subcellular localization of endo-β-N-acetylglucosaminidase and high-mannose type free N-glycans in plant cell. Biochimica and Biophysica Acta. 2002;1570:38–46. doi: 10.1016/s0304-4165(02)00149-6. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Kristiansen TZ, Bunkenborg J, Gronborg M, Molina H, Thuluvath PJ, Argani P, Goggins MG, Maitra A, Pandey A. A proteomic analysis of human bile. Molecular Cell Proteomics. 2004;3:715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- Kwon HK, Yokoyama R, Nishitani K. A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiology. 2005;46:843–857. doi: 10.1093/pcp/pci089. [DOI] [PubMed] [Google Scholar]
- Leah R, Kigel J, Svendsen I, Mundy J. Biochemical and molecular characterization of a barley seed beta-glucosidase. Journal of Biological Chemistry. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hrmova M, Burton RA, Lahnstein J, Fincher GB. Bifunctional family 3 glycoside hydrolases from barley with α-L-arabinofuranosidase and β-D-xylosidase activity. Journal of Biological Chemistry. 2003;278:5377–5387. doi: 10.1074/jbc.M210627200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Saravanan RS, Damasceno CM, Yamane H, Kim BD, Rose JK. Digging deeper into the plant cell wall proteome. Plant Physiology and Biochemistry. 2004;42:979–988. doi: 10.1016/j.plaphy.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé AC, Gomord V, Faye L. N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Molecular Biology. 1998;38:31–48. [PubMed] [Google Scholar]
- Li X, Kushad MM. Purification and characterization of myrosinase from horseradish (Armoracia rusticana) roots. Plant Physiolology and Biochemistry. 2005;43:503–511. doi: 10.1016/j.plaphy.2005.03.015. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiology. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin V, Balliau T, Chateau-Joubert S, Davanture M, Langella O, Negroni L, Prioul JL, Thevenot C, Zivy M, Damerval C. A two-dimensional proteome map of maize endosperm. Phytochemistry. 2004;65:1609–1618. doi: 10.1016/j.phytochem.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Minic Z, Jouanin L. Plant glycosyl hydrolases involved in cell wall polysaccharide degradation. Plant Physiology and Biochemistry. 2006;44:435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Minic Z, Do C-T, Rihouey C, Morin H, Lerouge P, Jouanin L. Purification, functional characterization, cloning and identification of mutants of a seed specific arabinan hydrolase in Arabidopsis. Journal of Experimental Botany. 2006;57:2339–2351. doi: 10.1093/jxb/erj205. [DOI] [PubMed] [Google Scholar]
- Minic Z, Rihouey C, Do CT, Lerouge P, Jouanin L. Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis. Plant Physiology. 2004;135:867–878. doi: 10.1104/pp.104.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Gough CM, Chandler LE, Loch CM, Ferrante JE, Wright PW. Structure, properties, and tissue localization of apoplastic alpha-glucosidase in crucifers. Plant Physiology. 1999;119:385–397. doi: 10.1104/pp.119.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Obel N, Porchia AC, Scheller HV. Dynamic changes in cell wall polysaccharides during wheat seedling development. Phytochemistry. 2002;60:603–610. doi: 10.1016/s0031-9422(02)00148-6. [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. ; Performing the paradoxica: how plant peroxidases modify the cell wall. Trends in Plant Sciences. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Qin O, Bergmann CW, Rose JK, Saladie M, Kolli VS, Albersheim P, Darvill AG, York WS. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. The Plant Journal. 2003;34:327–38. doi: 10.1046/j.1365-313x.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Research. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD. Biosynthesis and properties of the plant cell wall. Current Opinion in Plant Biology. 2002;5:536–542. doi: 10.1016/s1369-5266(02)00306-0. [DOI] [PubMed] [Google Scholar]
- Roberts K. The plant extracellular matrix: in a new expansive mood. Current Opinion in Cell Biology. 1994;6:688–694. doi: 10.1016/0955-0674(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Rodman JE. A taxonomic analysis of glucosinolate- producing plants. Systematic Botany. 1991;16:598–618. [Google Scholar]
- Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM. Comparison of lignin deposition in three ectopic lignification mutants. New Phytologist. 2005;168:123–140. doi: 10.1111/j.1469-8137.2005.01496.x. [DOI] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Sieiro C, Revilla G, Gonzalez-Villa T, Zarra I. Cloning and expression pattern of a gene encoding an alpha-xylosidase active against xyloglucan oligosaccharides from Arabidopsis. Plant Physiology. 2001;126:910–920. doi: 10.1104/pp.126.2.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Vinh J, Pflieger D, Sommerer N, Maurel C. A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochemistry Journal. 2003;373:289–296. doi: 10.1042/BJ20030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. Journal of Biological Chemistry. 2004;279:45503–45511. doi: 10.1074/jbc.M407594200. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, Van Der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R. ARAMEMNON, a Novel Database for Arabidopsis Integral Membrane Proteins. Plant Physiology. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. The Plant Cell. 2002;14:1635–1648. doi: 10.1105/tpc.002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon PS, Keen JN, Bowles DJ. Purification and characterization of N-glycanase, a concanavalin A binding protein from jackbean (Canavalia ensiformis) Biochemical Journal. 1998;330:13–20. doi: 10.1042/bj3300013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon EI, Machonkin TE, Sundaram UM. Spectroscopy of multi-copper oxidases. In: Messerschmidt A, editor. Multicopper oxidases. World Scientific Publishing Co; Singapore: 1997. pp. 103–127. [Google Scholar]
- Steele NM, Sulova Z, Campbell P, Braam J, Farkas V, Fry SC. Ten isoenzymes of xyloglucan endotransglycosylase from plant cell walls select and cleave the donor substrate stochastically. Biochemical Journal. 2001;355:671–679. doi: 10.1042/bj3550671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle-Smits T, Beekhuizen JG, Kok MT, Pijnenburg M, Recourt K, Derksen J, Voragen AG. Changes in cell wall polysaccharides of green bean pods during development. Plant Physiology. 1999;121:363–372. doi: 10.1104/pp.121.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, Machida Y. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development. 2001;128:4681–4689. doi: 10.1242/dev.128.23.4681. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Van Riet L, Nagaraj V, Van den Ende W, Clerens S, Wiemken A, Van Laere A. Purification, cloning and functional characterization of a fructan 6-exohydrolase from wheat (Triticum aestivum L.) Journal of Experimental Botany. 2006;57:213–223. doi: 10.1093/jxb/erj031. [DOI] [PubMed] [Google Scholar]
- Wang L, Li F, Sun W, Wu S, Wang X, Zhang L, Zheng D, Wang J, Gao Y. Concanavalin A captured glycoproteins in healthy human urine. Molecular and Cell Proteomics. 2006;5:560–562. doi: 10.1074/mcp.D500013-MCP200. [DOI] [PubMed] [Google Scholar]
- Watson BS, Lei Z, Dixon RA, Sumner LW. Proteomics of Medicago sativa cell walls. Phytochemistry. 2004;65:1709–1720. doi: 10.1016/j.phytochem.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Wilson IB, Altmann F. Concanavalin A binding and endoglycosidase D resistance of beta1,2-xylosylated and alpha1,3-fucosylated plant and insect oligosaccharides. Glycoconjugate Journal. 1998;15:203–206. doi: 10.1023/a:1006932725821. [DOI] [PubMed] [Google Scholar]
- Woo KK, Miyazaki M, Hara S, Kimura M, Kimura Y. Purification and characterization of a co(II)-sensitive alpha-mannosidase from Ginkgo biloba seeds. Bioscience Biotechnology and Biochemistry. 2004;68:2547–2556. doi: 10.1271/bbb.68.2547. [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO Journal. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. The Plant Cell. 2002;14:165–179. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen S, Alvarez S, Asirvatham VS, Schachtman DP, Wu Y, Sharp R. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiology. 2006;140:311–325. doi: 10.1104/pp.105.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


