Abstract
Transcription factors from the family of Signal Transducers and Activators of Transcription (STAT) are activated by numerous cytokines. Two members of this family, STAT5A and STAT5B (collectively called STAT5), have gained prominence in that they are activated by a wide variety of cytokines such as interleukins, erythropoietin, growth hormone, and prolactin. Furthermore, constitutive STAT5 activation is observed in the majority of leukemias and many solid tumors. Inactivation studies in mice as well as human mutations have provided insight into many of STAT5’s functions. Disruption of cytokine signaling through STAT5 results in a variety of cell-specific effects, ranging from a defective immune system and impaired erythropoiesis, the complete absence of mammary development during pregnancy, to aberrant liver function. On a molecular level, STAT5 has been linked to cell specification, proliferation, differentiation, and survival. Evidence is growing that the diverse outcomes of STAT5 signaling are not only determined by the expression of specific receptors but also by the interaction of STAT5 with cofactors and the cell-specific activity of members of the SOCS family, which negatively regulate STAT function. In this review, we focus on emerging concepts and challenges in the field of Janus kinase (JAK)–STAT5 signaling. First, we discuss unique functions of STAT5 in three distinct systems: mammary epithelial cells, hepatocytes, and regulatory T cells. Second, we present an example of how STAT5 can achieve cell specificity in hepatocytes through a physical and functional interaction with the glucocorticoid receptor. Third, we focus on the relevance of STAT5 in the development and progression of leukemia. Next, we discuss lessons derived from human mutations and disease. Finally, we address an emerging issue that the interpretation of experiments from STAT5-deficient mice and cells might be compromised as these cells might reroute and reprogram cytokine signals to the “wrong” STATs and thus acquire inappropriate cues. We propose that mice with mutations in various components of the JAK–STAT signaling pathway are living laboratories, which will provide insight into the versatility of signaling hardware and the adaptability of the software.
[Keywords: Mammary epithelium, hepatocytes, immunoregulation, body growth, cytokine]
Historical perspective
In 1994, Bernd Groner and colleagues (Wakao et al. 1994), then at the Friedrich Miescher Institute in Basel, cloned a cDNA from lactating ovine mammary tissue that encoded a transcription factor promoting prolactin-induced transcription of milk protein genes in mammary epithelium. The respective protein had been coined Mammary Gland Factor (MGF), implying specificity to the physiology of mammary tissue. However, based on its similarity to the family of Signal Transducers and Activators of Transcription (STAT), it was renamed appropriately STAT5. In 1995, three groups independently cloned the mouse homolog, now named STAT5A, and a closely related protein, called STAT5B (Azam et al. 1995; Liu et al. 1995; Mui et al. 1995). It turned out that both STAT5A and STAT5B are widely expressed and activated by a plethora of cytokines, including prolactin (PRL), growth hormone (GH), erythropoietin, thrombopoietin, and several interleukins (ILs). Since the first publication in 1997 in these pages of mice that carried either an inactivated Stat5a (Liu et al. 1997) or Stat5b (Udy et al. 1997) gene, we have witnessed great advances in our understanding of how cytokines transmit their message and control physiology and pathophysiology. However, we are still far from understanding how a limited set of generic components can trigger specific events ranging from lactation to leukemia.
Background
STAT5A and STAT5B are two closely related members of the family of STAT proteins. STAT5A and STAT5B are 96% conserved at the protein level. The two proteins contain 793 and 786 amino acids, respectively, and the highest degree of divergence is found in the C-terminal transactivation domain. STAT5A and STAT5B are encoded by two genes located on chromosome 11 (in mouse) and chromosome 17 (in humans) in a locus that also contains the Stat3 gene.
STATs are the mediators of signals that emanate from cytokine receptors. Considered latent transcription factors, they are activated upon binding of a ligand to the receptor by phosphorylation of a critical tyrosine residue through Janus kinases (JAK). Activated STATs bind to specific DNA sequences, named GAS (γ-interferon-activated sequences), and initiate transcription of target genes. In the cytokine signaling pathway there are a large number of ligands and receptors that funnel into a limited number of transcription factors. STAT5 is implicated in a wide variety of signaling events foremost in the immune system, mammary epithelial cells, and hepatocytes.
Molecular structure of STAT molecules
All members of the STAT family share a highly conserved molecular structure. Structural studies of STAT1, the best understood member of the family, have revealed an N-terminal domain, followed by an α-helical coiled-coil and DNA-binding domain and a linker that connects to the C terminus. The C terminus contains the src homology 2 (SH2) domain, followed by a short region containing a tyrosine residue, which is critical for the activation by phosphorylation through JAKs and a transactivation domain, which is the most divergent part within the STAT family. The molecule contains two flexible loops, one between the N terminus and the core fragment and a second loop connecting the C terminus to the core. These loops are thought to allow conformational changes in the transition from the inactive to the activated state. The three-dimensional structure of an unphosphorylated STAT5A core fragment (residues 129–712) lacking 129 amino acids from the N terminus and the C-terminal transactivation domain revealed overall similarity to other STAT molecules (Neculai et al. 2005). The N-terminal coil-coil domain forms a four-helix bundle followed by a β-barrel domain that connects to the α-helical linker and SH2 domain. The unphosphorylated STAT5A forms anti-parallel dimers in the cytoplasm through hydrophobic interactions of the four-helix bundle and β-barrel domains. Activation of STAT5A by tyrosine phosphorylation induces major rearrangement, a dissociation of the rather weak unphosphorylated dimer, and generation of phosphorylated dimers, which are formed through interaction of the SH2 domains. This conformation allows accumulation of STAT in the nucleus and binding to STAT response elements in DNA defined by a TTCN3GAA consensus sequence.
Mutating the Stat5a/b locus in mice
The genes encoding STAT5A and STAT5B are juxtaposed, and the transcriptional start sites are within 10 kb of each other. Their close relative Stat3 is located immediately downstream from the Stat5a gene. Although the Stat5a and Stat5b gene promoters might share certain regulatory elements, cell-preferential transcriptional patterns have emerged (Liu et al. 1995). While STAT5A is the prevalent STAT5 protein in mammary tissue, STAT5B is more abundant in muscle and liver. Other tissues, such as heart and salivary gland, have an equivalent distribution of STAT5A and STAT5B. In 1997, mice with disabling mutations in the genes encoding STAT5A (Liu et al. 1997) and STAT5B (Udy et al. 1997) were reported. Mice lacking the individual genes were viable and displayed distinct defects. Loss of STAT5A resulted in impaired mammary development and differentiation during pregnancy. The absence of STAT5B led to stunted body growth. These gene-specific defects either reflect the tissue distribution of STAT5A and STAT5B or tissue-specific functions of the two proteins. In 1998, Teglund and colleagues (Teglund et al. 1998) introduced mutations in both genes in mice revealing redundant and nonredundant roles of both isoforms. However, immunological defects were not as severe as had been anticipated from mutations in receptors that signal through the JAK–STAT5 pathway. As it turned out, the gene targeting mutations resulted in translation of hypomorphic N-terminally truncated STAT5A and STAT5B proteins (referred to as STAT5ΔN) that were able to form dimers but not tetramers. To overcome this potential problem, mice were generated that carried the 110-kb Stat5a/b locus flanked by loxP sites (Cui et al. 2004). This permitted Cre-recombinase-based complete deletion of both genes. These mice have been used extensively to explore cytokine–STAT5 signaling in specific cell types.
Mammary epithelium
The importance of STAT5A in the mammary gland is reflected in its original assignment as MGF, a transcription factor that stimulates prolactin-induced expression of mammary-specific milk protein genes (Fig. 1). However, gene deletion revealed a much more profound role in mammary epithelial cells. Deletion of STAT5A (Liu et al. 1997) caused an attenuation of mammary alveolar development and milk secretion, while absence of STAT5B (Udy et al. 1997) did not affect mammary development. The introduction of mutations into both STAT5 genes had a much stronger effect, demonstrating some functional redundancy in this system (Teglund et al. 1998; Miyoshi et al. 2001; Cui et al. 2004).
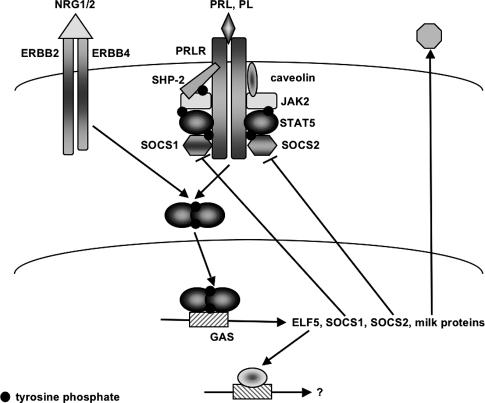
Figure 1.
Regulation of STAT signaling in mammary epithelial cells. Binding of prolactin (PRL) and placental lactogen (PL) to the PRLR or neuregulin1/neuregulin2 (NRG1/2) to ERBB4 activates the receptor-associated JAK kinase and induces the phosphorylation of STAT5A and STAT5B. Upon phosphorylation, STAT5 dimers translocate to the nucleus, where they bind to GAS elements and induce transcription of the genes encoding ELF5, SOCS1, SOCS2, and milk proteins. SOCS1/2 are recruited to the receptor and attenuate STAT5 signaling. Membrane-associated caveolin-1 modulates STAT5 activation by regulating JAK2 accessibility. The phosphatase SHP-2 regulates STAT5 phosphorylation by binding to the receptor and STAT5. ETS5 is a transcription factor in its own right that activates additional genes required for normal mammary function.
Complete null, hypomorphic (STAT5ΔN), and conditional alleles were used to investigate the requirement for STAT5 signaling. Since the complete absence of STAT5 caused perinatal lethality (Cui et al. 2004), mammary epithelial transplants into wild-type hosts were used to evaluate epithelial development in adult stages. STAT5-deficient epithelial cells were able to form simple ducts that grew at a normal rate. However, cell proliferation and alveolar development during pregnancy were severely impaired. By a conditional gene deletion approach, STAT5 was deleted before the onset of pregnancy with Cre recombinase driven by the MMTV-Cre transgene (Cui et al. 2004). This resulted in a phenotype similar to the one observed in STAT5-null epithelia. Females developed a ductal system in puberty, but alveolar development in pregnancy was inhibited, and dams were unable to rear litters. When STAT5 was deleted with a Cre transgene that is active only in epithelial cells that are differentiated and start to express milk protein genes, the loss of STAT5 caused rapid cell death. These results demonstrate stage-specific requirements for STAT5 for proliferation, differentiation, and maintenance of secretory mammary epithelium.
Signaling through the ERBB4 receptor also induces STAT5 phosphorylation, and deletion of a conditional Erbb4 allele in differentiated mammary epithelial cells inhibited cell proliferation and functional differentiation (Long et al. 2003). Interestingly, ERBB4 signaling also induced phosphorylation of serine residues in STAT5A (Clark et al. 2005). Phosphorylation of Ser 779 stabilized the interaction of STAT5A and ERBB4 and stimulated gene expression. Presence of the N-terminal Ser 127/128 residues was necessary for phosphorylation of the critical tyrosine at position 694 and for DNA binding of STAT5A. This raises the possibility that STAT5A activated by the ERBB4 pathway executes alternate functions by exhibiting different target gene selection.
Activation of STAT5A increases strongly in the course of pregnancy when levels of PRL and placental lactogens (PLs) rise to induce development of alveoli and transcription of milk protein genes. Both hormones activate the prolactin receptor (PRLR) and JAK2, and deletion of these genes resulted in the same inhibition of alveolar development (Ormandy et al. 1997; Miyoshi et al. 2001; Shillingford et al. 2002; Wagner et al. 2004; Sakamoto et al. 2007). The levels of PRLR seem to be the rate-limiting step since mice that only contain one Prlr gene displayed haploinsufficiency and were unable to nurse their litters (Ormandy et al. 1997). This system was used to study the distinct roles of Suppressor of Cytokine Signaling (SOCS) molecules in the negative feedback regulation of PRLR signaling. SOCS are a family of proteins that are induced by cytokines. They negatively regulate JAK–STAT signals by binding to cytokine receptors and the activation loop of JAK through their central SH2 domain. A short kinase inhibitory domain in SOCS1 and SOCS3 also interacts with JAK. All SOCS proteins contain a SOCS-box domain, which targets proteins for ubiquitination and degradation. It has not been resolved whether STATs or JAKs or both are regulated by this mechanism and to what extent it contributes to the regulation of STAT signaling in vivo.
By genetic means, SOCS1 and SOCS2 were shown to balance STAT5 activation through the PRLR. Deletion of one Socs1 allele restored normal levels of STAT5A activity and corrected lactation failure in Prlr+/− mice (Lindeman et al. 2001). Complete absence of SOCS1 led to precocious activation of STAT5A and accelerated alveolar proliferation and differentiation. Socs2 was identified as a direct target of STAT5A transcription in the mammary gland (Harris et al. 2006). It also attenuated PRLR signaling, but deletion of both alleles was required for a rescue of development of Prlr+/− epithelium. Furthermore, absence of both Socs2 alleles did not affect mammary development (Harris et al. 2006). The mechanisms underlying the inhibition of signaling could explain this difference. In contrast to SOCS1, SOCS2 does not contain the kinase inhibitor domain, which is thought to be involved in JAK inhibition. SOCS2 has also been shown to regulate signaling through the insulin-like growth factor 1 (IGF-1) receptor. Alternatively, the different behavior could also reflect partial redundancy or overlapping expression patterns of SOCS1 and SOCS2 during pregnancy.
Additional ways of regulating STAT5A activity in the mammary gland have been described. Access of JAK molecules to the receptors is affected by caveolins. These membrane-bound proteins interact with JAK through a caveolin scaffolding domain and attenuate kinase activity (Fig. 1). STAT5A activity in mammary epithelium of caveolin-1-deficient mice was elevated and led to accelerated development and premature lactation in pregnancy (Park et al. 2002). This indicates that prolactin signaling is enhanced. In addition, caveolin-1-deficient mice also displayed defects in innate immunity and inflammatory responses (Medina et al. 2006). Secretion of inflammatory agents by macrophages was increased in these mice in response to bacterial pathogens, but the mechanism of this enhanced immune response is unclear.
STAT5 activity is also regulated by SHP-2, a SH2 domain-containing phosphatase that binds to activated STAT5. The outcome of this interaction seems to be dependent on the cell context. SHP-2 deficiency in mouse embryonic fibroblasts has been shown to result in prolonged STAT5A activity due to delayed dephosphorylation in the absence of SHP-2 activity (Chen et al. 2003). A positive role for SHP-2 in the regulation of STAT5A activity in the mammary gland was reported recently. The viability and growth of pups nursed by SHP-2-deficient dams were impaired due to reduced development of the alveolar epithelial cell compartment of the mammary gland (Ke et al. 2006). The levels of activated STAT5A in the gland during lactation were reduced. Association of JAK2 with the PRLR was also attenuated, suggesting that SHP-2 may regulate STAT5 activation on multiple levels.
In an attempt to identify genes that are targets of STAT5, Harris et al. (2006) compared the expression profile of transplanted wild-type and Prlr−/− mammary epithelial cells at early pregnancy stages. Among the genes that were differentially expressed, they identified the ETS family transcription factor ELF5 (Harris et al. 2006). This protein is induced in epithelial cells during pregnancy. Its function in the mammary gland has only been studied in heterozygous mice as Elf5−/− embryos die at early embryonic stages. In Elf5+/− animals, a similar reduction of alveolar development was observed as in Prlr+/− epithelial cells, implying an important role in development during pregnancy. Expression of ELF5 in Prlr−/− epithelial cells rescued development and milk gene expression, demonstrating that this transcription factor mediates a crucial function of STAT5A in mammary epithelium.
Hepatocytes
GH controls to a large extent the physiology of hepatocytes (for signaling diagram, see Fig. 2). Based on mice that carry various GH receptor (GHR) mutations (Zhou et al. 1997; Rowland et al. 2005), Stat5b-null mice (Udy et al. 1997; Teglund et al. 1998) and liver-specific Stat5-mutant mice (Cui et al. 2007; Engblom et al. 2007), it can be concluded that the majority of the GH signaling is conveyed by the transcription factor STAT5B. Similarly, the glucocorticoid receptor (GR) controls numerous physiological parameters of hepatocytes (Tronche et al. 2004), which overlap with those controlled by GH to a large extent. Recent studies (Engblom et al. 2007) demonstrated that much of the liver physiology controlled by GR and STAT5B depends on their physical interaction, which in itself controls the activation of very specific gene patterns. While STAT5 controls the PRL-induced proliferation, differentiation, and function of mammary secretory epithelium, its role in hepatocytes is confined to modulating cellular metabolism and the production of IGF-1, a key regulator of postnatal body growth.
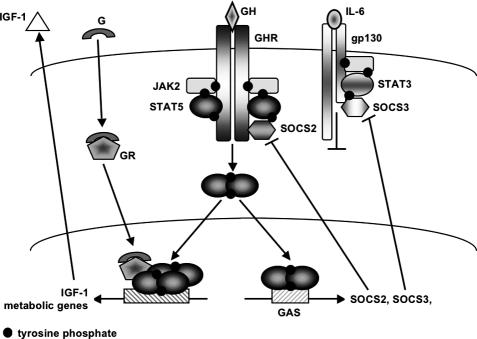
Figure 2.
Interaction of GHR and GR signaling through STAT5 in hepatocytes. STAT5B is activated upon binding of GH to its receptor by the associated JAK2. Tyrosine-phosphorylated STAT5B dimers bind to GAS and induce transcription of the negative regulators SOCS2 and SOCS3. SOCS2 and SOCS3 modulate signaling from the GHR and gp-130 receptors, respectively. Transcription of IFG-1 and metabolic genes is induced through additional signals from the GR and depends on the direct interaction of GR and STAT5B tetramers. (G) Glucocorticoid; (gp) glycoprotein.
Body growth
Severe growth retardation in humans as a consequence of GH insensitivity (GHI) was first reported by Laron et al. (1966) more than 40 years ago. Underlying mutations have been identified in genes that are at the core of GH signaling, namely, those encoding GHR, STAT5B, and IGF-1 (Rosenfeld et al. 2007). No mutations in the JAK2 gene have been associated with GHI, possibly because loss of JAK2 would lead to fetal death due to impaired erythropoiesis, as has been shown in mice (Neubauer et al. 1998; Parganas et al. 1998).
Lessons from mice
Evidence from mice and man has established that GH-induced postnatal body growth is mediated mainly by STAT5B, which controls IGF-1 gene expression in hepatocytes and muscle (Fig. 2). While loss of STAT5A/B in hepatocytes resulted in reduced circulating IGF-1 levels and concomitant stunted growth (Engblom et al. 2007), loss of STAT5A/B from muscle abrogated local IGF-1 levels with only minor effects on circulating IGF-1 levels (Klover and Hennighausen 2007). However, these mice had a reduced body size, thus confirming a critical role for paracrine IGF-1 in the muscle.
Evidence that STAT5B is the main mediator of body growth originally came from mice carrying either a nonfunctional Stat5a or Stat5b gene. Stat5b-null males but not females were reduced in size, demonstrating a sexual dimorphism of the mutation (Udy et al. 1997). In contrast, Stat5a-null mice exhibited normal body growth (Liu et al. 1997). Disruption of both Stat5 genes resulted in a more pronounced reduction of body growth in females than in males (Teglund et al. 1998), suggesting an additional role of STAT5A that was not seen in the Stat5a-null mice. The reduction of body growth in the Stat5-deficient mice was similar to that identified in GHR-null mice (Zhou et al. 1997) and mice that expressed a GHR mutant lacking STAT5 docking sites, but still retaining JAK2 activity and ERK signaling (Rowland et al. 2005). Identification of functional GAS sites in the second intron of the IGF-1 gene (Woelfle et al. 2003) completed the link between GH and IGF-1 signaling in body growth. Additional support that GH–STAT5 signaling is critical for body growth came from Socs2-null mice. The elimination of this negative regulator of STAT5 caused gigantism (Greenhalgh et al. 2005).
To clarify whether the GH–STAT5 axis controls postnatal body growth through liver-derived systemic IGF-1 or locally produced and acting IGF-1, the two Stat5 genes were specifically deleted either in hepatocytes (Cui et al. 2007; Engblom et al. 2007) or in muscle tissue (Klover and Hennighausen 2007) using Cre-loxP-mediated recombination. For deletion of the Stat5 genes in hepatocytes, Engblom et al. (2007) used a Cre transgene that was activated by a combination of the albumin gene promoter and the α-fetoprotein gene enhancer, while Cui et al. (2007) used a Cre transgene under control of the albumin gene promoter (Yakar et al. 1999). Engblom et al. (2007) observed growth retardation in both males and females as early as 2–3 wk after birth, which correlated with reduced circulating IGF-1 levels. Despite comparably reduced levels of circulating IGF-1, the study by Cui et al. (2007) did not observe reduced body growth. Since the two studies used Cre transgenes under control of different regulatory elements with different temporal activity, and possibly cell specificity within the liver, the exact contribution of liver-derived system IGF-1 in body growth is still not completely clear. Moreover, there may be differences in strain background. Deletion of the Stat5 genes from muscle tissue resulted in ∼20% reduction of body growth despite almost normal levels of circulating IGF-1 (Klover and Hennighausen 2007). IGF-1 levels were greatly reduced in muscle only, and an additional paracrine function in muscle rather than an exclusive liver-derived regulation in body growth was suggested.
Cui et al. (2007) found that the deletion of liver STAT5 resulted in impaired cell proliferation, development of fatty livers, and increased GH levels. This could be attributed to a reduced expression of Socs2 and aberrant activation of STAT1 and STAT3, leading to inappropriate transcription of STAT1 and STAT3 target genes as discussed in more depth below (Cui et al. 2007).
Lessons from humans
The identification of disabling mutations in the STAT5B gene in six patients with severe growth retardation (Kofoed et al. 2003; Rosenfeld et al. 2007) conclusively established a central role of STAT5B in GH-mediated postnatal growth. Loss of functional STAT5B is associated with severe IGF-1 deficiency, further demonstrating that this pathway is responsible for most of the GH-induced IGF-1 production. Growth retardation in individuals carrying either STAT5B or GHR mutations was indistinguishable, demonstrating that in this case STAT5A could not compensate for the loss of STAT5B. In contrast to mice, where loss of STAT5B resulted in reduced size of males only, five of the six patients were females, suggesting that in humans, STAT5B does not display sexual dimorphism of body growth.
Unlike patients with the classical Laron syndrome (mutations in GHR or IGF-1), individuals homozygous for STAT5B mutations also displayed symptoms of immune dysfunction, such as severe eczema and herpes keratitis as discussed below (Kofoed et al. 2003; Hwa et al. 2005). This is in agreement with the obligate role of STAT5 in the activation of regulatory T cells (Tregs) and the response to interleukins. Notably, Stat5b-null mice had profound defects in NK cells (Imada et al. 1998). However, loss of STAT5B function in humans does not necessarily associate with immunological problems as shown in one patient (Vidarsdottir et al. 2006).
STAT5 and regulatory T cells
Lessons from mice
STAT5A and STAT5B play essential redundant and nonredundant roles in orchestrating immunoregulation and the development of immune cells. Notably, in the complete absence of STAT5, mice failed to develop T, B, and natural killer (NK) cells (Hoelbl et al. 2006; Yao et al. 2006). Mice that express N-terminally truncated STAT5 displayed less severe immunological defects (Teglund et al. 1998; Moriggl et al. 1999; Sexl et al. 2000). Recent studies have demonstrated that STAT5 is the critical link between the IL-2/15 and FOXP3 (Burchill et al. 2007; Yao et al. 2007), the master regulator of regulatory T (Treg) cells (for signaling diagram, see Fig. 3).
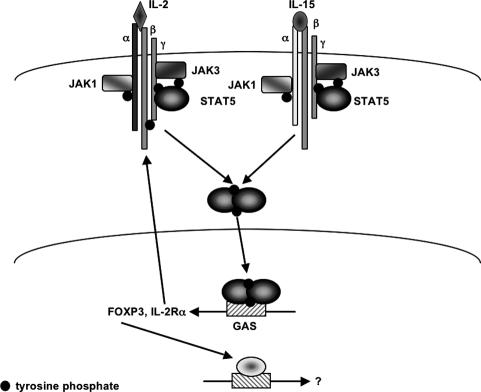
Figure 3.
STAT5 signaling in Treg cells. In regulatory T cells, STAT5 is phosphorylated by JAK3 upon binding of IL-2 and IL-15 to their respective receptors. STAT5 induces the expression of the IR-2Rα chain and Foxp3 genes.
CD4+CD25+ Tregs suppress T-cell proliferation and function and thus restrict immune responses against self- and nonself-antigens. These cells express the transcription factor FOXP3, which has been recognized as an essential component for both Treg development and function (Fontenot et al. 2003, 2005b; Hori et al. 2003). Notably, FOXP3 is sufficient to convert CD25− into CD25+ cells. While mice lacking IL-2Rα (CD25) have slightly reduced numbers of Tregs (Malek et al. 2000), the absence of the IL-2Rγc (Fontenot et al. 2005a) and Il-2Rβ (Suzuki et al. 1995) chains resulted in greatly reduced numbers of CD4+CD25+ Tregs and autoimmunity. The IL-2 and IL-15 receptors share the IL-2Rγc and IL-2Rβ subunits, and it is now accepted that both cytokines are redundant in the development of Tregs. Only the loss of both cytokines results in a loss of Tregs (Burchill et al. 2007). In complete and T-cell-specific Stat5-null mice, the reduction of CD25- and FOXP3-expressing cells was similar to that observed in Jak3-null and Il2rγ-null mice. Moreover, binding of STAT5 was detected to GAS sites in the promoter of the Foxp3 gene (Zorn et al. 2006; Burchill et al. 2007; Yao et al. 2007), confirming a direct transcriptional regulation by STAT5.
Lessons from humans
The previously mentioned six cases with disabling STAT5B defects were not only characterized by stunted body growth, they also had a history of immunological dysfunction (Kofoed et al. 2003; Hwa et al. 2005; Rosenfeld et al. 2007). Case 1 displayed low numbers of CD4+CD25high Tregs (Cohen et al. 2006). Notably, these cells had low levels of FOXP3 and a diminished ability to suppress the proliferation of CD4+CD25− T cells. In contrast, there was only a modest impact on circulating CD4 and CD8 T cells, NK cells and B cells, suggesting that in these compartments, STAT5A and STAT5B are redundant. A second case, carrying a different disabling STAT5B mutation, presented itself with a moderate T-cell lymphopenia, diminished CD4+CD25high Tregs and very low numbers of NK cells and γδ T cells (Bernasconi et al. 2006), which is in agreement with lower NK-cell numbers in Stat5b-null mice (Imada et al. 1998). Both the number and function of B cells were normal in this patient, suggesting a T-cell-restricted role for STAT5B. Two patients had decreased numbers of CD4+CD25high Tregs, supporting a role of STAT5B in IL-2-mediated accumulation of functional Tregs (Bernasconi et al. 2006; Cohen et al. 2006). These mutations may provide an inroad into a better understanding of the complex roles of STAT5 in regulating the development of the immune system.
STAT5, glucocorticoids, and metabolism
In a recent study in this journal, Günther Schütz and colleagues (Engblom et al. 2007) demonstrated that the physical interaction of STAT5 and the GR is required for many of the functions exerted by either transcription factor (Fig. 2). Glucocorticoids execute the hypothalamus–pituitary–adrenal (HPA) axis and control homeostasis during physiological challenges. They bind to the widely expressed GR, which targets glucocorticoid response elements (GREs) and activates or represses transcription of target genes. In addition, GR can bind to other transcription factors and modulate their activities without interacting with GREs (Reichardt et al. 1998, 2001). In vitro studies had demonstrated the binding of GR to STAT5A and pointed to a physiological significance in the expression of milk protein genes (Stocklin et al. 1996). However, the deletion of GR specifically in mammary epithelium did not interfere with lactation (Wintermantel et al. 2005), questioning an essential role of the GR in mammary function. Yet, deletion of GR from hepatocytes resulted in stunted body growth congruent to that observed in liver-specific Stat5b-null mice (Tronche et al. 2004), suggesting a functional interaction between the two transcription factors. More than 40% of the genes whose expression was reduced in the absence of GR were also down-regulated in Stat5-null liver. Mice from which both GR and STAT5 were deleted from liver tissue further established that the transcriptional- activating effect on a large set of genes was dependent on the presence of STAT5. Gene sets, which require GR–STAT5 interaction, included those controlling body growth and maturation. Gene sets repressed by the GR–STAT5 complex included those controlling fatty acid metabolism. GR was found to bind to the STAT5 N terminus, and the biological relevance was further established (Engblom et al. 2007) in Stat5ΔN mice (Teglund et al. 1998). The GR–STAT5 interaction is a selective requirement for the activation of specific gene sets in hepatocytes, and it remains to be seen whether other cell types rely on this or similar interactions.
Cytokine–STAT5 signaling in pathophysiology
A variety of solid tumors, leukemias, and myeloproliferative disorders (MPDs) are characterized by the presence of constitutively active STAT5, which has been suggested to provide survival and proliferation stimuli and by that token may represent an attractive therapeutic target. Evidence is accumulating that the presence of STAT5 is required for the development of, at least, some tumors. The general topic of cytokine signaling in leukemia has recently been reviewed extensively (Van Etten 2007).
Already a decade ago, activated STAT5 had been detected in BCR-ABL-induced chronic myeloid leukemia (CML) (Ilaria and Van Etten 1996) and was seen more recently in blast cells from patients with acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL) (Van Etten 2007). Notably, deregulated kinases, such as internal tandem duplication of FLT3 (FLT3/ITD), are responsible for the activation of STAT5. A subset of patients with chronic myelomonocytic leukemia (CMML) carries chromosomal translocations that result in TEL/PDGFβR fusion proteins with constitutive kinase activity. Recruitment of SH2-containing proteins, including STATs, results in their activation. Approximately 40% of patients with severe congenital neutropenia (SCN) carry mutations in the gene encoding the G-CSF receptor (CSF3R) that truncate part of the cytoplasmic domain of the receptor. Acquisition of these mutations is associated with constitutively active STAT5 and the development of AML and myelodysplastic syndrome (MDS) (Germeshausen et al. 2007). Lastly, the somatic activating mutation (V617F) in the JAK2 tyrosine kinase identified in the majority of patients with the MPD polycythemia vera (PV) has been associated with constitutive STAT5 activation (Wernig et al. 2006).
Mice with targeted mutations in the Stat5a and Stat5b genes were instrumental in addressing the contribution of STAT5 in leukemia and solid tumors. Originally, the results from mice that still express N-terminally truncated STAT5 (Teglund et al. 1998) provided a mixed picture. While the development of the myelo- and lymphoproliferative disease induced by TEL/KAK2 was dependent on the presence of the hypomorphic STAT5ΔN (Schwaller et al. 2000), another study suggested that STAT5 was not absolutely required for the induction of BCR-ABL-induced CML-like leukemia (Sexl et al. 2000). More recent experiments with fetal liver cells from mice in which the entire Stat5 locus had been deleted strongly argued that the presence of STAT5 is also required for the development of leukemia upon introduction of BCR-ABL (Hoelbl et al. 2006). Even in the absence of only STAT5A, a reduced incidence of CML was observed (Ye et al. 2006), suggesting a dose requirement for tumor formation. Similarly, STAT5-null fetal liver cells were refractory to transformation by TEL-PDGFβR in methylcellulose colony assays, and, again, a dose dependency was observed (Cain et al. 2007). Studies with primary cells have demonstrated that the thrombopoietin–STAT5 signaling cascade is a critical regulator of normal hematopoietic stem cells (HSCs) (Kato et al. 2005; Seita et al. 2007) and possibly leukemic stem/progenitor cells (Schepers et al. 2007). In the mouse, expression of mutated Csf3R found in SCN led to a clonal HSC advantage, which again was dependent on the presence of STAT5 (Liu et al. 2008). In contrast to its role as growth stimulator, in some instances STAT5A can be considered a tumor suppressor as proposed by a recent study on the tyrosine kinase NPM1-ALK in a subset of T-cell lymphomas (Zhang et al. 2007). It was found that NPM1-ALK epigenetically silences the STAT5A but not the STAT5B gene and that STAT5A itself can inhibit the expression of NPM1-ALK. As in these hematopoietic disorders, the presence of STAT5A, at least in mice, was also required for the development of oncogene-induced mammary cancers (Humphreys and Hennighausen 1999; Ren et al. 2002).
There are some caveats to studies based on STAT5-null HSCs or progenitor cells as well as epithelial cells. Notably, HSCs lacking STAT5 are defective and severely impaired in their ability to repopulate bone marrow in a competitive setting (Li et al. 2007). It also cannot be excluded that secondary defects within HSCs or progenitor cells make them more refractive to in vitro transformation by certain oncogenes. Moreover, the jury is still out on whether the inactivation of STAT5 in an established tumor will result in its remission. Both of these issues can be addressed using mouse models in which STAT5 can be inactivated and reactivated at any given time during tumor development. At this point it is also not clear whether experimental loss of STAT5 will result in the activation of STAT3 (see below), which has been shown to contain potent tumor-inducing properties.
Loss of STAT5: filling the void
A central lesson of physics is that a vacuum is unstable and gets filled quickly. Similarly, deleting genes from the mouse perturbs the cellular balance, which has dual consequences. First, the absence of a protein will leave a void, and cells have to adapt to the presence of an altered balance of transcription factors. In addition, the absence of a transcription factor will potentially also disrupt downstream transcription pathways and, as in the examples reviewed here, the feedback of the signaling circuit (e.g., by SOCS proteins) is impaired (Fig. 4). In many cases, the deletion of a gene will impair the viability of a cell and lead to cell death. STAT5, however, are mainly modulators of cellular functions and upon their loss many cells appear to adapt instead of undergoing apoptosis. It has become apparent that in the absence of one STAT protein, other members of the family can be recruited to their receptor-binding sites (Fig. 4). Under physiological conditions, each cytokine receptor activates its preferred member of the STAT family, but it also has the ability to activate other STATs to some extent. While the GHR and IL-6R preferentially recruit STAT5 and STAT3, respectively, the interferon (IFN)-γR activates STAT1. Deletion of STAT5 from hepatocytes resulted in an aberrant activation of both STAT1 and STAT3, which was accompanied by the expression of the corresponding target genes (Cui et al. 2007). Similarly, interferon treatment of STAT1-null cells resulted in the aberrant activation of STAT3 and its respective target genes, including Socs3 (Gil et al. 2001; Ramana et al. 2001), and loss of STAT3 led to an IL-6-induced activation of STAT1 and transcription of its target genes (Costa-Pereira et al. 2002). A picture is emerging that in the absence of a given STAT member, receptors will recruit other STAT members instead. This will not necessarily result in compensation for the “lost” STAT, but might very well lead to a gain of function imposed by these “inappropriate” STATs with all its adverse consequences (Murray 2007).
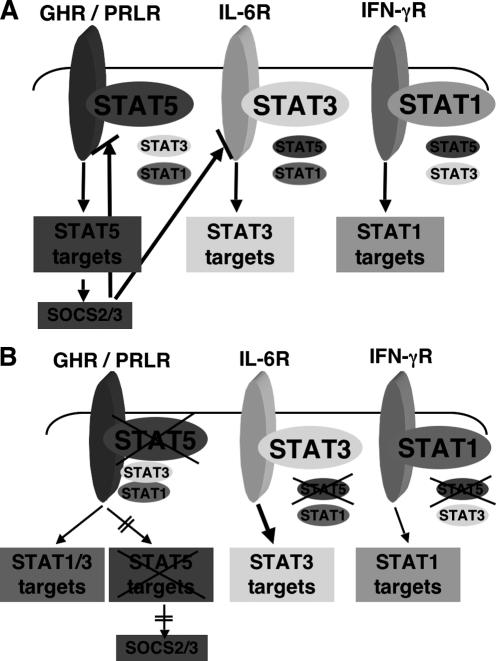
Figure 4.
(A) Specificity of cytokine–STAT signaling. Cell-specific cytokine signals result, at least in part, from the receptors that are expressed on a cell and the ligands it encounters. Each ligand binds to its cognate receptor and activates a specific STAT molecule, which then induces a particular set of target genes. Some of the target genes encode SOCS proteins, which exert a negative feedback regulation on the signals from the receptors. (B) Altered STAT signaling networks upon deletion of one family member. If one of the STAT family members is missing, inappropriate activation through binding of other family members to the receptor sites previously occupied by the original member may occur. This will induce a different set of target genes. In addition, reduced expression of SOCS proteins will lead to enhanced activation of signals from other receptors and contribute to a change in the cell’s response. In the case of a loss of STAT5, both GH and PRL are able to activate STAT1 and STAT3, which results in the induction of respective target genes. Loss of STAT5 also results in reduced expression of its prominent target genes Socs2 and Socs3 and, as a consequence, the impaired negative feedback loops will further enhance STAT3 activation.
Loss of STAT5 in hepatocytes also resulted in reduced expression of its target genes Socs2 and Socs3 (Cui et al. 2007), which led to exacerbated STAT3 signaling through gp130-based receptors (Fig. 2; Cui et al. 2007). Along the same theme, deletion of Socs3 from the mouse genome resulted in increased IL-6-mediated activation of STAT3 as well as STAT1 (Croker et al. 2003; Lang et al. 2003). These observations are not artifacts confined to knockout cells in the mouse but have been found in humans as well. Notably, loss of STAT5B in patients resulted in an aberrant activation of STAT1 and STAT3 by GH and PRL (Kofoed et al. 2003), raising again the possibility that some of the symptoms in these patients are due to a gain of function.
The road ahead
The overt logic and cell specificity are the main attractions of the JAK–STAT pathway. We are far from understanding the algorithms used by receptors to pick and choose among seven STATs. Structural biologists will provide answers to some of these questions. STAT3 and STAT5 convey a wide range of cell-specific functions, which cannot simply be explained by receptor specificity. Cell-specific post-translational modifications and interactions with cofactors will be part of the picture. In addition to their transcriptional function, STATs also provide a means to regulate chromatin structure (Ye et al. 2001) and possibly act as cytoplasmic platforms for other proteins. The greatest challenge might lie in unlocking the mystery behind the multiplicity and diversity of transcriptional outputs elicited by STAT5, such as milk proteins in mammary tissue and FOXP3 in Tregs. The integration of genome-wide chromatin immunoprecipitation (ChIP) assays with massive parallel sequencing (Robertson et al. 2007) will provide a microscopic view of genetic areas occupied by STATs in different cells and physiological and pathophysiological states. Mouse genetics has provided insights into genuine functions of STATs, and now the time is ripe for a more comprehensive experimental approach across disciplinary borders.
Acknowledgments
We are supported by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. We apologize to all those whose work has not been cited due to space restrictions.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1643908.
References
- Azam M., Erdjument-Bromage H., Kreider B.L., Xia M., Quelle F., Basu R., Saris C., Tempst P., Ihle J.N., Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi A., Marino R., Ribas A., Rossi J., Ciaccio M., Oleastro M., Ornani A., Paz R., Rivarola M.A., Zelazko M., et al. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 2006;118:e1584–e1592. doi: 10.1542/peds.2005-2882. [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Cain J.A., Xiang Z., O’Neal J., Kreisel F., Colson A., Luo H., Hennighausen L., Tomasson M.H. Myeloproliferative disease induced by TEL-PDGFRB displays dynamic range sensitivity to Stat5 gene dosage. Blood. 2007;109:3906–3914. doi: 10.1182/blood-2006-07-036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wen R., Yang S., Schuman J., Zhang E.E., Yi T., Feng G.S., Wang D. Identification of Shp-2 as a Stat5A phosphatase. J. Biol. Chem. 2003;278:16520–16527. doi: 10.1074/jbc.M210572200. [DOI] [PubMed] [Google Scholar]
- Clark D.E., Williams C.C., Duplessis T.T., Moring K.L., Notwick A.R., Long W., Lane W.S., Beuvink I., Hynes N.E., Jones F.E. ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J. Biol. Chem. 2005;280:24175–24180. doi: 10.1074/jbc.M414044200. [DOI] [PubMed] [Google Scholar]
- Cohen A.C., Nadeau K.C., Tu W., Hwa V., Dionis K., Bezrodnik L., Teper A., Gaillard M., Heinrich J., Krensky A.M., et al. Cutting edge: Decreased accumulation and regulatory function of CD4+ CD25high T cells in human STAT5b deficiency. J. Immunol. 2006;177:2770–2774. doi: 10.4049/jimmunol.177.5.2770. [DOI] [PubMed] [Google Scholar]
- Costa-Pereira A.P., Tininini S., Strobl B., Alonzi T., Schlaak J.F., Is’harc H., Gesualdo I., Newman S.J., Kerr I.M., Poli V. Mutational switch of an IL-6 response to an interferon-γ-like response. Proc. Natl. Acad. Sci. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker B.A., Krebs D.L., Zhang J.G., Wormald S., Willson T.A., Stanley E.G., Robb L., Greenhalgh C.J., Forster I., Clausen B.E., et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Hosui A., Sun R., Shen K., Gavrilova O., Chen W., Cam M.C., Gao B., Robinson G.W., Hennighausen L. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology. 2007;46:504–513. doi: 10.1002/hep.21713. [DOI] [PubMed] [Google Scholar]
- Engblom D., Kornfeld J.W., Schwake L., Tronche F., Reimann A., Beug H., Hennighausen L., Moriggl R., Schutz G. Direct glucocorticoid receptor–Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes & Dev. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Gavin M.A., Rudensky A.Y. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005a;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., Rudensky A.Y. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005b;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Germeshausen M., Ballmaier M., Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- Gil M.P., Bohn E., O’Guin A.K., Ramana C.V., Levine B., Stark G.R., Virgin H.W., Schreiber R.D. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh C.J., Rico-Bautista E., Lorentzon M., Thaus A.L., Morgan P.O., Willson T.A., Zervoudakis P., Metcalf D., Street I., Nicola N.A., et al. SOCS2 negatively regulates growth hormone action in vitro and in vivo. J. Clin. Invest. 2005;115:397–406. doi: 10.1172/JCI22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J., Stanford P.M., Sutherland K., Oakes S.R., Naylor M.J., Robertson F.G., Blazek K.D., Kazlauskas M., Hilton H.N., Wittlin S., et al. Socs2 and elf5 mediate prolactin-induced mammary gland development. Mol. Endocrinol. 2006;20:1177–1187. doi: 10.1210/me.2005-0473. [DOI] [PubMed] [Google Scholar]
- Hoelbl A., Kovacic B., Kerenyi M.A., Simma O., Warsch W., Cui Y., Beug H., Hennighausen L., Moriggl R., Sexl V. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Humphreys R.C., Hennighausen L. Signal transducer and activator of transcription 5a influences mammary epithelial cell survival and tumorigenesis. Cell Growth Differ. 1999;10:685–694. [PubMed] [Google Scholar]
- Hwa V., Little B., Adiyaman P., Kofoed E.M., Pratt K.L., Ocal G., Berberoglu M., Rosenfeld R.G. Severe growth hormone insensitivity resulting from total absence of signal transducer and activator of transcription 5b. J. Clin. Endocrinol. Metab. 2005;90:4260–4266. doi: 10.1210/jc.2005-0515. [DOI] [PubMed] [Google Scholar]
- Ilaria R.L., Van Etten R.A. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- Imada K., Bloom E.T., Nakajima H., Horvath-Arcidiacono J.A., Udy G.B., Davey H.W., Leonard W.J. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Iwama A., Tadokoro Y., Shimoda K., Minoguchi M., Akira S., Tanaka M., Miyajima A., Kitamura T., Nakauchi H. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J. Exp. Med. 2005;202:169–179. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Lesperance J., Zhang E.E., Bard-Chapeau E.A., Oshima R.G., Muller W.J., Feng G.S. Conditional deletion of Shp2 in the mammary gland leads to impaired lobulo-alveolar outgrowth and attenuated Stat5 activation. J. Biol. Chem. 2006;281:34374–34380. doi: 10.1074/jbc.M607325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klover P., Hennighausen L. Postnatal body growth is dependent on the transcription factors signal transducers and activators of transcription 5a/b in muscle: A role for autocrine/paracrine insulin-like growth factor I. Endocrinology. 2007;148:1489–1497. doi: 10.1210/en.2006-1431. [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., Hwa V., Little B., Woods K.A., Buckway C.K., Tsubaki J., Pratt K.L., Bezrodnik L., Jasper H., Tepper A., et al. Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med. 2003;349:1139–1147. doi: 10.1056/NEJMoa022926. [DOI] [PubMed] [Google Scholar]
- Lang R., Pauleau A.L., Parganas E., Takahashi Y., Mages J., Ihle J.N., Rutschman R., Murray P.J. SOCS3 regulates the plasticity of gp130 signaling. Nat. Immunol. 2003;4:546–550. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- Laron Z., Pertzelan A., Mannheimer S. Genetic pituitary dwarfism with high serum concentration of growth hormone—A new inborn error of metabolism? Isr. J. Med. Sci. 1966;2:152–155. [PubMed] [Google Scholar]
- Li G., Wang Z., Zhang Y., Kang Z., Haviernikova E., Cui Y., Hennighausen L., Moriggl R., Wang D., Tse W., et al. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output. Exp. Hematol. 2007;35:1684–1694. doi: 10.1016/j.exphem.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman G.J., Wittlin S., Lada H., Naylor M.J., Santamaria M., Zhang J.G., Starr R., Hilton D.J., Alexander W.S., Ormandy C.J., et al. SOCS1 deficiency results in accelerated mammary gland development and rescues lactation in prolactin receptor-deficient mice. Genes & Dev. 2001;15:1631–1636. doi: 10.1101/gad.880801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Robinson G.W., Gouilleux F., Groner B., Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Robinson G.W., Wagner K.U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes & Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Liu X., Kunter G., Krem M.M., Eades W.C., Cain J.A., Tomasson M.H., Hennighausen L., Link D.C. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J. Clin. Invest. 2008 doi: 10.1172/JCI32704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long W., Wagner K.U., Lloyd K.C., Binart N., Shillingford J.M., Hennighausen L., Jones F.E. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development. 2003;130:5257–5268. doi: 10.1242/dev.00715. [DOI] [PubMed] [Google Scholar]
- Malek T.R., Porter B.O., Codias E.K., Scibelli P., Yu A. Normal lymphoid homeostasis and lack of lethal autoimmunity in mice containing mature T cells with severely impaired IL-2 receptors. J. Immunol. 2000;164:2905–2914. doi: 10.4049/jimmunol.164.6.2905. [DOI] [PubMed] [Google Scholar]
- Medina F.A., de Almeida C.J., Dew E., Li J., Bonuccelli G., Williams T.M., Cohen A.W., Pestell R.G., Frank P.G., Tanowitz H.B., et al. Caveolin-1-deficient mice show defects in innate immunity and inflammatory immune response during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2006;74:6665–6674. doi: 10.1128/IAI.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Shillingford J.M., Smith G.H., Grimm S.L., Wagner K.U., Oka T., Rosen J.M., Robinson G.W., Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R., Topham D.J., Teglund S., Sexl V., McKay C., Wang D., Hoffmeyer A., van Deursen J., Sangster M.Y., Bunting K.D., et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- Mui A.L., Wakao H., O’Farrell A.M., Harada N., Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. The JAK–STAT signaling pathway: Input and output integration. J. Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- Neculai D., Neculai A.M., Verrier S., Straub K., Klumpp K., Pfitzner E., Becker S. Structure of the unphosphorylated STAT5a dimer. J. Biol. Chem. 2005;280:40782–40787. doi: 10.1074/jbc.M507682200. [DOI] [PubMed] [Google Scholar]
- Neubauer H., Cumano A., Muller M., Wu H., Huffstadt U., Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Ormandy C.J., Camus A., Barra J., Damotte D., Lucas B., Buteau H., Edery M., Brousse N., Babinet C., Binart N., et al. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes & Dev. 1997;11:167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- Parganas E., Wang D., Stravopodis D., Topham D.J., Marine J.C., Teglund S., Vanin E.F., Bodner S., Colamonici O.R., van Deursen J.M., et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Park D.S., Lee H., Frank P.G., Razani B., Nguyen A.V., Parlow A.F., Russell R.G., Hulit J., Pestell R.G., Lisanti M.P. Caveolin-1-deficient mice show accelerated mammary gland development during pregnancy, premature lactation, and hyperactivation of the Jak-2/STAT5a signaling cascade. Mol. Biol. Cell. 2002;13:3416–3430. doi: 10.1091/mbc.02-05-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C.V., Gil M.P., Han Y., Ransohoff R.M., Schreiber R.D., Stark G.R. Stat1-independent regulation of gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt H.M., Kaestner K.H., Tuckermann J., Kretz O., Wessely O., Bock R., Gass P., Schmid W., Herrlich P., Angel P., et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Reichardt H.M., Tuckermann J.P., Gottlicher M., Vujic M., Weih F., Angel P., Herrlich P., Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Cai H.R., Li M., Furth P.A. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rosenfeld R.G., Belgorosky A., Camacho-Hubner C., Savage M.O., Wit J.M., Hwa V. Defects in growth hormone receptor signaling. Trends Endocrinol. Metab. 2007;18:134–141. doi: 10.1016/j.tem.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Rowland J.E., Lichanska A.M., Kerr L.M., White M., d’Aniello E.M., Maher S.L., Brown R., Teasdale R.D., Noakes P.G., Waters M.J. In vivo analysis of growth hormone receptor signaling domains and their associated transcripts. Mol. Cell. Biol. 2005;25:66–77. doi: 10.1128/MCB.25.1.66-77.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Creamer B.A., Triplett A.A., Wagner K.U. The Janus kinase 2 is required for expression and nuclear accumulation of cyclin D1 in proliferating mammary epithelial cells. Mol. Endocrinol. 2007;21:1877–1892. doi: 10.1210/me.2006-0316. [DOI] [PubMed] [Google Scholar]
- Schepers H., van Gosliga D., Wierenga A.T., Eggen B.J., Schuringa J.J., Vellenga E. STAT5 is required for long-term maintenance of normal and leukemic human stem/progenitor cells. Blood. 2007;110:2880–2888. doi: 10.1182/blood-2006-08-039073. [DOI] [PubMed] [Google Scholar]
- Schwaller J., Parganas E., Wang D., Cain D., Aster J.C., Williams I.R., Lee C.K., Gerthner R., Kitamura T., Frantsve J., et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Seita J., Ema H., Ooehara J., Yamazaki S., Tadokoro Y., Yamasaki A., Eto K., Takaki S., Takatsu K., Nakauchi H. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc. Natl. Acad. Sci. 2007;104:2349–2354. doi: 10.1073/pnas.0606238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexl V., Piekorz R., Moriggl R., Rohrer J., Brown M.P., Bunting K.D., Rothammer K., Roussel M.F., Ihle J.N. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of stat5. Blood. 2000;96:2277–2283. [PubMed] [Google Scholar]
- Shillingford J.M., Miyoshi K., Robinson G.W., Grimm S.L., Rosen J.M., Neubauer H., Pfeffer K., Hennighausen L. Jak2 is an essential tyrosine kinase involved in pregnancy-mediated development of mammary secretory epithelium. Mol. Endocrinol. 2002;16:563–570. doi: 10.1210/mend.16.3.0805. [DOI] [PubMed] [Google Scholar]
- Stocklin E., Wissler M., Gouilleux F., Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kundig T.M., Furlonger C., Wakeham A., Timms E., Matsuyama T., Schmits R., Simard J.J., Ohashi P.S., Griesser H., et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor β. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- Teglund S., McKay C., Schuetz E., van Deursen J.M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J.N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Tronche F., Opherk C., Moriggl R., Kellendonk C., Reimann A., Schwake L., Reichardt H.M., Stangl K., Gau D., Hoeflich A., et al. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes & Dev. 2004;18:492–497. doi: 10.1101/gad.284704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy G.B., Towers R.P., Snell R.G., Wilkins R.J., Park S.H., Ram P.A., Waxman D.J., Davey H.W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten R.A. Aberrant cytokine signaling in leukemia. Oncogene. 2007;26:6738–6749. doi: 10.1038/sj.onc.1210758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsdottir S., Walenkamp M.J., Pereira A.M., Karperien M., van Doorn J., van Duyvenvoorde H.A., White S., Breuning M.H., Roelfsema F., Kruithof M.F., et al. Clinical and biochemical characteristics of a male patient with a novel homozygous STAT5b mutation. J. Clin. Endocrinol. Metab. 2006;91:3482–3485. doi: 10.1210/jc.2006-0368. [DOI] [PubMed] [Google Scholar]
- Wagner K.U., Krempler A., Triplett A.A., Qi Y., George N.M., Zhu J., Rui H. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol. Cell. Biol. 2004;24:5510–5520. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao H., Gouilleux F., Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig G., Mercher T., Okabe R., Levine R.L., Lee B.H., Gilliland D.G. Expression of Jak2V617F causes a polycythemia vera-like disease with associated myelofibrosis in a murine bone marrow transplant model. Blood. 2006;107:4274–4281. doi: 10.1182/blood-2005-12-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel T.M., Bock D., Fleig V., Greiner E.F., Schutz G. The epithelial glucocorticoid receptor is required for the normal timing of cell proliferation during mammary lobuloalveolar development but is dispensable for milk production. Mol. Endocrinol. 2005;19:340–349. doi: 10.1210/me.2004-0068. [DOI] [PubMed] [Google Scholar]
- Woelfle J., Billiard J., Rotwein P. Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J. Biol. Chem. 2003;278:22696–22702. doi: 10.1074/jbc.M301362200. [DOI] [PubMed] [Google Scholar]
- Yakar S., Liu J.L., Stannard B., Butler A., Accili D., Sauer B., LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc. Natl. Acad. Sci. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S.K., Agata Y., Lee H.C., Kurooka H., Kitamura T., Shimizu A., Honjo T., Ikuta K. The IL-7 receptor controls the accessibility of the TCRγ locus by Stat5 and histone acetylation. Immunity. 2001;15:813–823. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- Ye D., Wolff N., Li L., Zhang S., Ilaria R.L. STAT5 signaling is required for the efficient induction and maintenance of CML in mice. Blood. 2006;107:4917–4925. doi: 10.1182/blood-2005-10-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang H.Y., Liu X., Wasik M.A. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat. Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Xu B.C., Maheshwari H.G., He L., Reed M., Lozykowski M., Okada S., Cataldo L., Coschigamo K., Wagner T.E., et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc. Natl. Acad. Sci. 1997;94:13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E., Nelson E.A., Mohseni M., Porcheray F., Kim H., Litsa D., Bellucci R., Raderschall E., Canning C., Soiffer R.J., et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]