Short abstract
Genomic analyses of human cells expressing dihydrofolate reductase provide insight into the effects of genome position on the propensity for a drug-resistance gene to amplify in human cells.
Abstract
Background
Amplifications, regions of focal high-level copy number change, lead to overexpression of oncogenes or drug resistance genes in tumors. Their presence is often associated with poor prognosis; however, the use of amplification as a mechanism for overexpression of a particular gene in tumors varies. To investigate the influence of genome position on propensity to amplify, we integrated a mutant form of the gene encoding dihydrofolate reductase into different positions in the human genome, challenged cells with methotrexate and then studied the genomic alterations arising in drug resistant cells.
Results
We observed site-specific differences in methotrexate sensitivity, amplicon organization and amplification frequency. One site was uniquely associated with a significantly enhanced propensity to amplify and recurrent amplicon boundaries, possibly implicating a rare folate-sensitive fragile site in initiating amplification. Hierarchical clustering of gene expression patterns and subsequent gene enrichment analysis revealed two clusters differing significantly in expression of MYC target genes independent of integration site.
Conclusion
These studies suggest that genome context together with the particular challenges to genome stability experienced during the progression to cancer contribute to the propensity to amplify a specific oncogene or drug resistance gene, whereas the overall functional response to drug (or other) challenge may be independent of the genomic location of an oncogene.
Background
Genetic instability resulting in chromosomal level alterations is frequent in solid tumors, which display a wide variety of types and frequencies of these aberrations. Amplifications, regions of focal high level copy number change, are likely to represent aberrations continuously under selection during tumor growth, since amplified DNA is unstable [1-4] and would otherwise disappear. They often harbor known oncogenes and thus are useful for identifying genes or pathways that foster tumor development. For ERBB2, amplification is the predominant method of its up-regulation and is the basis for FISH-based tests evaluating the ERBB2 status of breast tumors. On the other hand, change in DNA copy number is only one way to alter expression of a gene and expression of other oncogenes is much less tightly linked to DNA copy number or amplification. Other mechanisms for deregulation may be post-transcriptional, post-translational or involve alteration in expression of upstream genes. Tumor subtypes may also be distinguished by their propensity to amplify oncogenes, suggesting that the particular types of genomic instability present in a tumor are important determinants of how expression of an oncogene might be altered. Moreover, amplification is often associated with poor prognosis.
Amplification is a reiterative process, in which multiple copies of a genome region are accumulated. Studies in model systems indicate that amplification requires a DNA double strand break and progression through the cell cycle with this damaged DNA [5-9]. A role for genome context in promoting amplification has also been suggested, since introduction of a selectable gene into different genome positions in hamster and yeast cells resulted in site-dependent frequencies of resistant colonies following drug challenge [10,11]. Particular genome sequences prone to breakage have also been shown to set the boundaries of amplicons in rodent cells [6,12], further suggesting that genome position influences the propensity to amplify.
Common chromosomal fragile sites, of which there are approximately 90 in the human genome, have received the most attention as sites likely to promote amplification. Expression of fragile sites can be induced in cells in culture under conditions of replication stress and are visualized as gaps on metaphase chromosomes. Fragile sites can be divided into common sites (CFS), which are seen in all individuals and rare sites (RFS), which appear only in certain individuals. The sites are further distinguished by agents used to induce expression, which include aphidicolin, bromo-deoxyuridine (BrdU), 5-azacytidine and distamycin A. Folate stress caused by methotrexate exposure also induces a group of rare fragile sites. A small number of CFS have been molecularly identified and found to vary from hundreds of kilobases to over one megabase in size, to have some unusual sequence properties, but not to be conserved in sequence. Often they contain very large genes and are sites of viral integration in certain cancers [13]. Evidence supporting a role for fragile sites in promoting amplification in human cancer is provided by the MET oncogene, which is amplified in esophageal adenocarcinoma. The gene lies within FRA7G, and the amplicon boundaries in tumors also lie within this site [14]. Nevertheless, for many amplicons there is no obvious involvement of common chromosomal fragile sites.
Gene amplification has been studied in vitro in a variety of systems by selection for cells capable of growth in the presence of antimetabolites. To investigate the role of genome context on amplification in human cells, we chose methotrexate resistance as the model system, because clinical resistance to methotrexate targets a number of genes by a variety of mechanisms [15,16], thereby providing the opportunity to determine which types of aberration occur more frequently in different genetic backgrounds. We introduced a mutant copy of DHFR, which confers greater resistance to methotrexate than the endogenous wild-type DHFR, into random sites in the genome of chromosomally stable HCT116+chr3 cells and precisely determined the site of single copy integrations in the human genome sequence. We isolated colonies resistant to folate deprivation caused by methotrexate and characterized these cells with respect to site specific response to drug challenge. We used array comparative genomic hybridization (CGH) to identify and classify the types of genomic alterations in the drug resistant cells, fluorescent in situ hybridization (FISH) to study the organization and mechanism of amplicon formation, and expression profiling to investigate the functional consequences of amplification. These studies found site specific differences in the sensitivity to methotrexate, organization of amplicons and propensity to amplify. On the other hand, gene expression patterns of drug resistant cells were independent of integration site, with two major clusters revealed by hierarchical clustering of the expression profiles. The clusters differed significantly in expression of MYC target genes. Translated to human disease, these studies suggest that genome context together with the particular challenges to genome stability experienced during the progression to cancer contribute to the propensity to amplify a specific oncogene, whereas the overall functional response to drug (or other) challenge may be independent of the genomic location of an oncogene.
Results
Characteristics of clones with DHFR* integration
To study formation and structure of amplicons, we took advantage of the fact that resistance to methotrexate can be accomplished by a number of mechanisms, including copy number gain or amplification of DHFR and loss or down regulation of the folate transporter, SLC19A1 on chromosome 21 [15,16]. We have shown previously that HCT116+chr3 cells have stable karyotypes and that methotrexate resistant HCT116+chr3 cells frequently amplify DHFR on chromosome 5q [17]. The HCT116+chr3 cells are a variant of the mismatch repair deficient colorectal carcinoma cell line, HCT116; however, they are mismatch repair proficient due to the wild-type copy of MLH1 provided by an extra copy of chromosome 3p and proximal 3q [17,18]. Because these cells carry two wild-type copies of DHFR, we introduced a mutant form of DHFR (L22F), which confers greater resistance to methotrexate than the wild-type (endogenous) gene, into HCT116+chr3 cells by retroviral infection. The DHFR (L22F) variant also was fused to the gene encoding enhanced green fluorescence protein (EGFP) and is referred to here as DHFR*. We isolated 56 independent clones containing DHFR* at different positions in the genome and identified genome sequences flanking the integration site of DHFR* using inverse PCR (Additional data file 1). For further analysis, we selected only clones that were considered to have a single insertion of DHFR* by inverse PCR (13 independent insertion sites, Table 1).
Table 1.
Thirteen DHFR* insertion site clones and their response to methotrexate
| Name | Chr. | Chr. band | Sequence1 (bp) | Exp.2 | IC-503 (nM) | MTX4 (nM) | Expression5 (%) | No. of colonies6 | Colonies screened7 | DHFR* amplicon8 | DHFR* gain9 |
| 1M-34 | 9 | q34.3 | 137,580,684 | → | 43 | 120 | 4,172 | 26.5 (8.50) | 10 | 4 | 0 |
| 1M-39 | 13 | q32.1 | 96,707,451 | ← | 22 | 65 | 3,835 | 0.5 (1.00) | 0 | 0 | 0 |
| 1M-42 | 8 | q22.3 | 102,162,871 | ← | 21 | 75 | 2,660 | 5.5 (3.25) | 15 | 13 | 2 |
| 1M-43 | 11 | q13.1 | 65,526,684 | → | 22 | 65 | 1,828 | 13 (10.50) | 9 | 0 | 3 |
| 1M-45 | 11 | q23.3 | 118,293,388 | ← | 70 | 200 | 5,329 | 2.5 (3.00) | 8 | 1 | 6 |
| 1M-57 | 3 | q27.1 | 185,207,363 | → | 15 | 75 | 2,047 | 3.5 (3.75) | 12 | 3 | 8 |
| 1M-67 | 1 | p34.3 | 39,533,783 | → | 31 | 90 | 2,306 | 0.5 (1.00) | 3 | 1 | 0 |
| 1M-72 | 2 | q35 | 217,190,120 | ← | 16 | 65 | 5,077 | 1.0 (2.00) | 2 | 1 | 1 |
| 1M-73 | 12 | p13.2 | 12,767,229 | ← | 20 | 65 | 3,116 | 0.5 (1.00) | 1 | 0 | 0 |
| 1M-75 | 22 | q12.2 | 28,977,607 | → | 32 | 90 | 3,307 | 25.5 (12.75) | 3 | 1 | 1 |
| 1M-83 | 19 | q13.2 | 45,618,743 | → | 14 | 65 | 756 | 1.0 (1.75) | 6 | 0 | 1 |
| 1M-84 | 17 | q12 | 34,161,068 | → | 23 | 75 | 3,934 | 11.0 (11.00) | 4 | 0 | 0 |
| 1M-89 | 19 | q13.33 | 53,682,371 | → | 25 | 75 | 4,288 | 2.0 (1.00) | 9 | 3 | 0 |
1Position in the human genome sequence (UCSC Genome Browser, May 2004 freeze). 2Exp., Direction of DHFR* integration in the genome; arrow to the right indicates that the DHFR* coding sequence is integrated in the direction from the p-arm to the q-arm of the chromosome. 3Methotrexate concentration that causes 50% inhibition of the cell growth after 6 days. 4Methotrexate (MTX) concentration used for selection of resistant colonies. 5Expression of DHFR* measured indirectly by quantitative RT-PCR (EGFP expression normalized to GUSB expression, 2-(dCt) × 100). 6Median number (and interquartile range in parentheses) of resistant colonies per 10 cm plate after 28 days of methotrexate treatment. 7Number of independent resistant colonies screened by array CGH. 8Resistant colonies with amplification of the DHFR* integration region. 9Resistant colonies with low level copy number gain of the DHFR* integration region. Chr., chromosome.
The individual insertion site clones were further characterized with respect to genome copy number profiles and expression of DHFR*. All clones with the exception of two (1M-39 and 1M-43) showed the same chromosomal changes by array CGH as HCT116+chr3 cells (Figure 1a). Clone 1M-39 gained part of the q-arm of chromosome 4 (between RP11-18D7 and the q-telomere). Clone 1M-43 contained one additional copy of chromosome 22 and had lost the q-arm of chromosome 18. We confirmed DHFR* expression in all clones by measuring the expression level of the fused EGFP portion of the gene using quantitative RT-PCR. Expression levels, as percentage of GUSB expression, showed an approximately seven-fold variation (Table 1).
Figure 1.
Parental DHFR* integration sites and copy number aberrations in methotrexate resistant colonies. (a) Copy number profile of cell line HCT116+chr3 and positions of 13 DHFR* integrations. This near-diploid cell line is characterized by partial chromosomal gains on chromosomes 3, 8, 10, 12, 16, losses on chromosomes 4, 16 and 10 and homozygous deletion on chromosome 16. Shown are the log2 ratios on BAC clones ordered according to genome position (UCSC Genome Browser, May 2004 freeze). Arrows indicate the positions of integration of one copy of DHFR* as mapped by inverse PCR to the human genome sequence. (b) Heatmap representation of copy number changes detected by array CGH in 82 methotrexate resistant colonies from 12 different insertion sites. Each column represents one resistant colony. Resistant colonies from each DHFR* integration site were clustered according to their copy number changes. Positions of the insertion sites are indicated by the arrowheads. Individual BAC clones are shown as rows and ordered according to their genome position (UCSC Genome Browser, May 2004 freeze). Copy number losses are indicated in red, gains in green and amplifications as yellow dots.
Frequency of DHFR* amplification at different genomic sites
Initially we measured the sensitivity of each clone to methotrexate by determining the methotrexate concentration that causes 50% reduction in cell number after six days exposure to varying concentrations of the drug (IC-50). The HCT116+chr3 cells showed the greatest sensitivity to the drug (IC-50 = ~7.6 nM). Clones with DHFR* showed 1.9- to 9.2-fold increase in methotrexate resistance (IC-50 ranged from 14.1 to 70.1 nM; Table 1). To select methotrexate resistant colonies, we exposed cells to a concentration of methotrexate that was three to four times the IC-50 for each integration site. We note that because DHFR is the target of methotrexate, exposure to the drug should inhibit synthesis of thymidylate and reduce levels of thymidine-based nucleotides. Such a reduction in nucleotide levels could cause DNA damage; however, the concentrations used here are not expected to do so [8]. Moreover, we determined that exposure of HCT116+chr3 cells to the range of concentrations used in these studies does not result in significant DNA damage as measured by the alkaline comet assay. The median number of resistant colonies obtained for each integration site is shown in Table 1.
Genomic copy number profiles were obtained for isolated resistant colonies that grew sufficiently well to be expanded to 5 × 106 cells (Figure 1; Additional data file 2). The retention of DHFR* at the original site of integration was confirmed in all resistant colonies using inverse PCR, as shown for untreated clone 1M-89 and its resistant colonies in Additional data file 1. Clones from different integration sites could be separated into four groups: The first group contained only one clone, 1M-39, which did not form any resistant colonies. The second group (1M-73 and 1M-84) formed resistant colonies that did not amplify DHFR*; however, partial gain of chromosome 5 and loss of chromosome 21 were among the copy number changes, suggesting that increased copies of the endogenous DHFR locus and loss of SLC19A1 contributed to resistance. Clones in the third group (1M-43 and 1M-83) showed low level copy number changes (partial or whole chromosome gains) of the region with DHFR* integration in at least one resistant colony. Finally, clones in the fourth group (1M-34, 1M-42, 1M-45, 1M-57, 1M-67 1M-72, 1M-75 and 1M-89) formed methotrexate resistant colonies with amplicons around the DHFR* integration site at varying frequencies (Table 1).
Structure of amplicons and mechanisms of formation
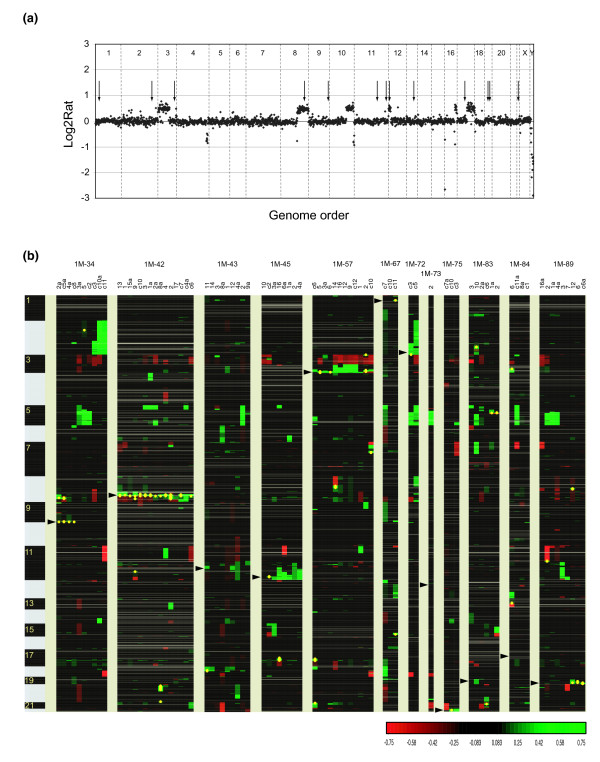
Amplified DNA can be present in various forms, including double minutes, amplified regions on a chromosome, which may be cytogenetically visible as a homogeneously staining region (HSR), or distributed across the genome [19]. The organization of 13 amplicons from five integration sites was investigated using FISH. All of the DHFR* amplicons in methotrexate resistant cells were present as amplified DNA on one or two chromosomes. In only one case (1M-89_6) was the amplified DNA present as double minutes in some cells rather than integrated into a chromosome. Hybridization of differentially labeled FISH probes from the amplicons revealed that eight of the chromosomal amplicons were organized as repeated units in inverted orientation (Figures 2 and 3). The organization of the remaining four was not determined. In five cases there were also copy number losses distal to DHFR* in the copy number profile. These observations are consistent with amplification being initiated by a double strand break distal to DHFR*, followed by breakage-fusion-bridge cycles. In two independent methotrexate resistant clones from 1M-57 in which DHFR* is integrated near the chromosome 3q telomere, we observed amplicons containing both the DHFR* integration site on 3q as well as amplification of 3pter. Cytogenetic analysis revealed the presence of ring chromosomes and patterns of hybridization of FISH probes on linear and ring chromosomes consistent with amplification occurring by a breakage-fusion-bridge process involving fusion of 3pter and 3qter (Figure 2).
Figure 2.
Mechanism of DHFR* amplification involving a ring chromosome intermediate. (a) Chromosome 3 copy number profile in untreated 1M-57 cells. Shown are the log2 ratios ordered according to position on the May 2004 freeze of the human genome sequence. The copy number gain extending from 3pter to RP11-233L3 reflects the presence of the two normal copies of chromosome 3 and the additional piece of chromosome 3 in HCT116+chr3 cells. The DHFR* insertion site is indicated by the arrowhead. (b) Chromosome 3 copy number profile in methotrexate resistant colony 1M-57_2. Two regions of amplification are evident at 3pter and near the distal end of 3q. Copy number losses include material from 3p and 3qter distal to DHFR*. (c) Proposed mechanism leading to amplification of the region around DHFR*. The ends of chromosome 3 fuse to create a ring chromosome with loss of material distal to DHFR* on 3q. Breakage at positions indicated by the arrowheads at anaphase results in the metacentric chromosome that now carries duplication of juxtaposed 3p and 3q sequences. The process can be repeated to generate additional copies of the 3p and 3q sequences. (d) FISH with RP11-107D22 (red), which contains sequences flanking the DHFR* integration site on 3q and RP11-28P14 (green), which maps to 3p. Shown are pseudocolor images showing hybridization signals on the chromosomes and the corresponding DAPI image (gray).
Figure 3.
Amplicons formed by two genomic regions, initially located on the different chromosomes. Chromosome 8 and 9 copy number profiles showing amplification on (a) 9q and (b) 8q. (c) Organization of the amplicon. CTD-3145B15 (red) maps to the 9q telomere near the DHFR* insertion site and RP11-237F24 (green) to the region of amplification on chromosome 8 shown in (b). The chromosome 9 signals appear to flank the chromosome 8 material on this chromosome. Amplification of the region around DHFR* is indicated by the large hybridization signal from CTD-3145B15. The amplified DNA was determined to be located on chromosome 9 by hybridization of RP11-62H18 to 9pter (not shown). Thus, material from 9qter appears to be amplified in situ on chromosome 9 and additional copies of material from the chromosome 9 amplicon are present on a separate chromosome together with amplified DNA from chromosome 8. (d, e) Copy number profiles of chromosomes 19 (d) and 8 (e). (f) Organization of the amplicon. RP11-691H23 (red) maps near the DHFR* integration site on chromosome 19 and RP11-175H20 (green) is one of the clones from the amplicon on chromosome 8 shown in (e). The chromosome 19 signals appear to flank a number of copies of chromosome 8, which could be as many as eight copies, since the CGH log2 ratio = ~2. Two additional copies of RP11-691H23, mapping near DHFR* on chromosome 19, were also present on chromosome 19 (data not shown). Thus, amplified DNA near the DHFR* integration site is present and independently amplified on two chromosomes. (g, h) 1M-42_9 CGH profile (chromosome 8) showing the DHFR* amplicon and its organization as determined by FISH. BAC clone RP11-91O11 (red) maps near the DHFR* integration site and is co-amplified with the distal part of chromosome 8 (RP11-237F24, green). The chromosomal region between the two co-amplified regions was lost. (i, j) 1M-42_2 CGH profile (chromosome 8) and FISH analysis showing that the two regions of chromosome 8 were amplified as two independent amplicons on different chromosomes.
Translocation prior to amplification also appears to have occurred in four resistant colonies in which the amplicons were formed from two separate genomic regions that were co-amplified in inverted repeats. In the two examples shown in Figures 3a–f, hybridization of FISH probes is consistent with different regions of chromosome 8 being translocated onto the chromosome carrying DHFR* followed by co-amplification. In both cases distal parts of chromosome 8 were lost. Amplicons containing two or more separate regions of the same chromosome organized as inverted repeats were also observed (Figure 3g,h). On the other hand, the contiguous genomic region of amplified DNA on 8q in 1M-42_2 was present on two different chromosomes (Figure 3i,j). In these cells ring chromosomes were also present.
Fragile sites and the propensity to amplify
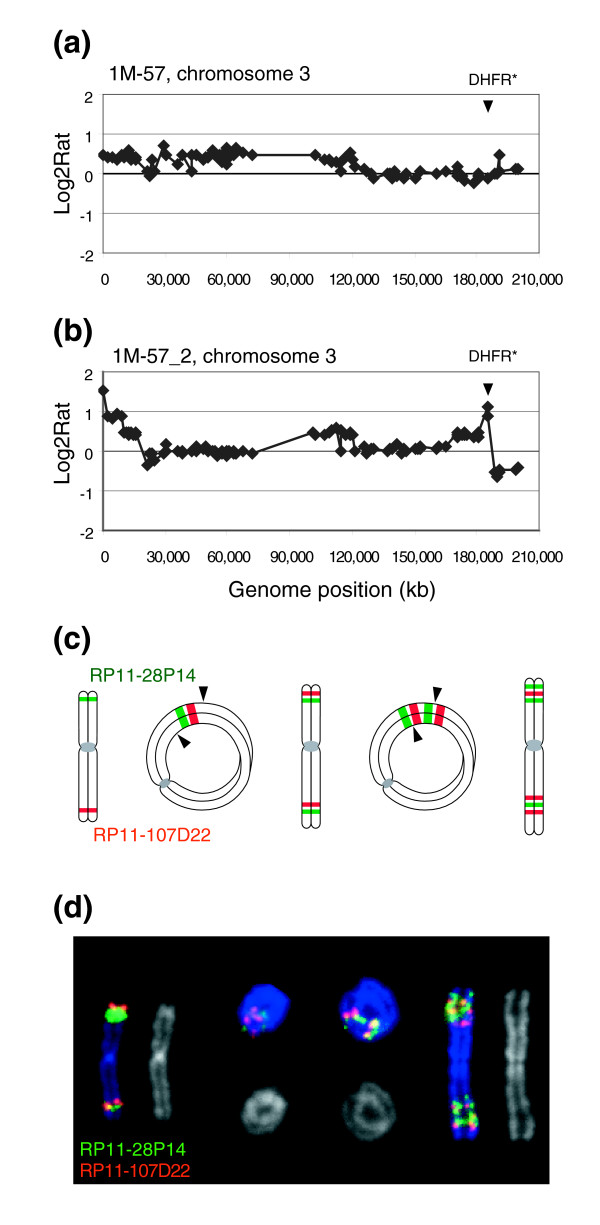
The integration sites varied in the frequency with which resistant clones amplified DHFR*. The 1M-42 integration site was unique in that DHFR* was amplified in almost all resistant clones (13/15), which was significantly more frequent than any other site (test for homogeneity of binomial proportion, p = 0.0002). Although the clones varied with respect to the regions of chromosome 8 that were amplified together with DHFR*, they all shared similar distal amplicon boundaries mapping between RP11-10G10 and CTD-2013D21. Higher resolution mapping on the 32K bacterial artificial chromosome (BAC) genome tiling path array [20] allowed the boundaries of five amplicons to be mapped more precisely (Figure 4). Four of the boundaries were positioned in a 1 Mb region between RP11-375I14 and RP11-97D1 (102322735 to 103386096 base-pairs (bp), May 2004 freeze, Additional data file 3).
Figure 4.
Recurrent amplicon boundaries in 1M-42 methotrexate resistant clones and fragile sites. (a) Chromosome 8 copy number profiles at approximately 1.4 Mb resolution. The vertical lines indicate the region from 99 to 105 Mb on chromosome 8 shown for (b) hybridization of these same DNAs to the 32K genome tiling array.
The consistent and recurrent location of amplicon boundaries to a limited region prompted us to investigate the possible involvement of fragile sites in initiating amplification. The integration site of DHFR* at 102,162,871 bp on chromosome 8 is close to several fragile sites, including the aphidicolin sensitive sites FRA8B, FRA8C and FRA8D and the distamycin A inducible site, FRA8E. A rare folate sensitive site, FRA8A, has also been localized to 8q22.3 (101,600-106,200 kb). As cells are being deprived of folates by challenge with methotrexate, a potential role for the folate sensitive fragile site, FRA8A seemed possible. Therefore, we sought evidence of a methotrexate induced fragile site in the region of the recurrent boundary of the 1M-42 amplicons in HCT116+chr3 cells. Metaphase spreads prepared from cells exposed to methotrexate for 24 hours were hybridized with FISH probes labeled with Cy3 (RP11-10G10) and fluoro-isothiocyanate (FITC; CTD-2013D21). Although rare metaphases were observed in which hybridization signals from these BACs appeared to bracket a fragile site, these experiments were inconclusive due to the very low frequency with which such patterns were seen.
Seven of the 1M-42 amplicons contained more than one peak, indicating that breakage does not occur exclusively at one site on chromosome 8. Another frequent site of copy number transition occurred in the 5.8 Mb region between RP11-27I15 and RP11-238H10, which is included within the cytogenetically assigned position of the aphidicolin sensitive site FRA8B at 8q22.1. Nevertheless, we were unable to obtain evidence that induction of aphidicolin fragile sites near DHFR* in 1M-42 cells promotes amplification, since exposure of 1M-42 cells to aphidicolin for 24 hours prior to methotrexate challenge did not result in a statistically significant difference in the number of resistant clones compared to cells without aphidicolin pretreatment.
Amplification and gene expression
A primary reason for amplification of a gene under permanent selection pressure is its increased expression resulting from the copy number increase. A question frequently asked about amplified genes is whether they are the driver gene for amplification or are they simply passengers. In our model system, the presence of DHFR* significantly increases the IC-50 for cells challenged with methotrexate, suggesting that it is the driver gene for its amplicons. Moreover, expression of DHFR* as measured by quantitative RT-PCR is positively correlated with copy number determined by array CGH (p < 0.05), providing further support for DHFR* as the driver gene for amplification, regardless of site of integration. On the other hand, the amplicons always span regions much larger than DHFR* and include other genes (Figure 5); therefore, we asked which neighboring genes in the amplicons are also up-regulated by copy number. Twelve methotrexate resistant colonies (four different integration sites) were selected for microarray analysis of gene expression at the mRNA level (Additional data file 4). All of the resistant clones contained amplification of the region containing DHFR* and some also co-amplified additional regions. Considering only genes located within the 12 regions of amplification and with measured expression levels in both amplified and non-amplified samples (n = 370; Additional data file 5), we found that the mean expression levels of 139 were up-regulated when amplified compared to mean expression levels in samples without amplification (log2 fold change > 0.8), with the likelihood of up-regulation appearing to be independent of proximity to DHFR*. An additional 13 genes were highly expressed in methotrexate resistant samples without amplification (log2 ratio > 0.8) and 9/13 also showed additional modest increases in expression in samples with amplification. Although these genes could contribute to methotrexate resistance when amplified, we were unable to demonstrate any increase in IC-50 for methotrexate or growth advantage when two randomly selected amplified genes with positive correlation of expression with copy number (POLR2K and LOC157567) were overexpressed in HCT116+chr3 cells. Similarly, overexpression of MYC, which was co-amplified with DHFR* in a number of resistant cells from different integration sites, did not significantly alter the IC-50 for methotrexate or provide a proliferative advantage (data not shown). Thus, DHFR* appears to be the major driver gene for amplification, although we cannot rule out that one or more of the genes included in the amplicon could also provide a significant contribution to methotrexate resistance.
Figure 5.
Expression of genes mapping to the amplicon from methotrexate resistant colony 1M-89_6. (a) Chromosome 19 copy number profile at approximately 1.4 Mb resolution (HumArray3.0) shows four discrete regions of amplification. (b) Chromosome 19 copy number profile from the 32K BAC genome tiling path array in the amplified region showing the four regions of amplification detected on the lower resolution array and an additional small region distal to the others. The regions, ranging in size from 0.2 to 1.2 Mb, were amplified together in some cells as double minutes, while in others the amplified DNA was integrated into different chromosomes and present as a homogeneously staining region. Both copies of chromosome 19 were retained without rearrangement. As all regions are included together on the double minutes, their formation may have occurred by joining of broken pieces of DNA subsequent to resolution of stalled replication forks [63]. (c) Expression levels of genes mapping to the five amplified regions plotted according to position on the human genome sequence. Expression levels are shown as the log2 ratios of the signal intensities after hybridization of Cy3 labeled cDNA from the methotrexate resistant colony and Cy5 labeled cDNA from the untreated parent 1M-89 as reference. Shown is the list of genes in genomic order that map to the amplified regions according to the RefSeq database [64]. Genes that are expressed in this cell line with or without exposure to methotrexate are labeled in black and expression levels denoted by black dots. Genes present on the array, but not expressed in untreated or resistant cells, are colored by dark gray and represented by dots of the same color with zero change in expression. Genes not present on the array are listed in the upper line in light gray. Genes with log2 fold change >0.8 are indicated with an asterisk.
Response to methotrexate is independent of site of integration and amplification
To investigate further the question of the contribution of co-amplified genes to the overall response of cells to methotrexate, we asked whether expression profiles of methotrexate resistant colonies varied with respect to integration site. Unsupervised hierarchical clustering of the 12 samples with amplification revealed two major clusters, indicating at least two responses to methotrexate. Samples did not separate according to integration site, however, further suggesting that co-amplified genes do not contribute significantly to the overall response to methotrexate.
One notable difference between clusters was the presence of two samples with focal amplification of MYC in the right cluster (1M-34_c5 and 1M-42_9; Figure 6a,b). In general, samples in the right cluster with or without amplification of MYC showed higher MYC expression than those in the left cluster (log2 ratio change between clusters = 1.42). Using the MYC target gene database [21], we identified 380 human MYC target genes among the 3,931 variably expressed genes used for clustering. The median absolute log2 ratio change in expression of these target genes in the right cluster compared to the left cluster was significantly higher than a similar comparison using 1,000 sets of 380 randomly selected genes (p = 2.2e-16). Thus, differential expression of MYC and MYC target genes is one of the distinguishing features of the overall response of cells with amplified DHFR* to challenge with methotrexate, irrespective of integration site.
Figure 6.
Expression profiling of 12 methotrexate resistant colonies with amplification of DHFR*. (a) Unsupervised hierarchical clustering (Pearson) of genes with variable expression across the data set (SD ≥ 0.3) and present in >75% of samples (3,931 genes). Methotrexate resistant colonies represent four different integration sites, indicated by the shaded boxes below the dendrogram. (b) Comparison of expression of MYC target genes in the two clusters. The median absolute log2 ratio change in expression of MYC target genes in the right cluster compared to the left cluster is significantly higher than a similar comparison using 1,000 sets of 380 randomly selected genes (p = 2.2e-16).
Discussion
In order to investigate the effect of genome position on the propensity to amplify, we integrated a single copy of a mutant form of DHFR fused to the gene encoding EGFP (DHFR*) into different positions in the genome of HCT116+chr3 cells by retroviral transfer, challenged cells with methotrexate and then studied the genomic alterations arising in drug resistant cells. Since DHFR* confers greater resistance to methotrexate than the endogenous wild-type DHFR, we expected that increased copy number of this gene would be found in the methotrexate resistant cells, rather than the endogenous gene. This expectation was met, as the majority of the resistant colonies contained either gains or amplifications of the locus. Furthermore, DHFR* appeared to be the driver gene for the copy number change, since DHFR* mRNA levels were positively correlated with DHFR* copy number. Nevertheless, in this simple model system, we observed that at four different DHFR* insertion sites, approximately one-third of neighboring genes were also up-regulated when amplified along with DHFR*. It is unlikely that so many of these genes mapping to four different random locations would also be driver genes for amplification. Moreover, expression profiling divided methotrexate resistant cells into two groups independent of integration site, suggesting that neighboring genes in the amplicons, even though up-regulated by copy number increases, did not play a major role in the drug resistant phenotype. Taken together, these observations suggest that about one-third of genes can be regulated by copy number, which is consistent with global expression array profiling of tumors. If these four regions are representative of the genome as a whole, then a significant proportion of the amplified genes in tumors are also likely to be passengers. These observations have implications for studies of amplicons in tumors. Passenger genes will confound efforts to identify candidate oncogenes by expression analyses alone. For example, several candidate driver genes for amplification are thought to be present in amplicons in human cancers, including 8p11-p12 and 17q12 in breast cancer [22-24], 7p11.2 in glioblastoma [25] or 6p22 in bladder cancer [26], because gene expression is correlated with copy number. Further functional studies will be necessary to determine which overexpressed genes in the amplicons contribute to tumor development. On the other hand, BIRC2 and YAP1, two genes present in a narrow amplicon in oral squamous cell carcinomas [27], esophageal squamous cell carcinoma [28], and lung [29], pancreatic [30], and hepatocellular carcinomas, have recently been shown to collaboratively promote tumor formation in mice [31], indicating that the extent of some tumor amplicons may be determined by selection for multiple neighboring collaborating oncogenes.
A distinguishing feature of the expression profiles of methotrexate resistant cells was the expression of MYC target genes. Co-amplification of MYC irrespective of integration site also suggests that it played a role in the response to methotrexate; however, the exact mechanism whereby MYC contributes to resistance is not known. It does not appear that overexpression of MYC contributes to resistance simply by promoting progression through the cell cycle or genome instability, since overexpression of MYC prior to methotrexate challenge did not enhance drug resistance or proliferation. On the other hand, up-regulation of MYC expression may contribute to drug resistance by enhancing the capability of cells to evade checkpoints. For example, overexpression of MYC abrogates a p53-dependent cell cycle arrest in REF52 cells exposed to N-(phosphonacetyl)-L-aspartate (PALA), an inhibitor of pyrimidine nucleotide synthesis, allowing resistant cells to arise from these normally non-permissive cells [5].
The numbers or types of genomic alterations in resistant cells varied among the integration sites; however, they were not related to either expression levels or sensitivity to methotrexate (Table 1). Previous studies in hamster cells, yeast and protozoa have highlighted the association of genome position with the frequency of methotrexate resistant cells and repeated sequences have been implicated in promoting amplification in response to methotrexate challenge in yeast and Leishmania [10,32,33]. Moreover, in Leishmania the response to methotrexate differs among species and it was possible to show that not only was position important, but also the nature of the gene under selection; that is, amplification of the gene conferring the higher level of resistance was observed more frequently in resistant cells regardless of position [34] as observed here in human cells for DHFR*.
In spite of the fact that DHFR* confers greater resistance to methotrexate, DHFR* was not altered in copy number in resistant colonies from two integration sites, while no resistant colonies were recovered from a third integration site. Expression levels of DHFR* in these integrants, measured indirectly as EGFP expression, were in the middle of the range of expression levels for all integration sites as was sensitivity measured as IC-50. Furthermore, no variations in DHFR* were detected by sequencing DNA amplified from genomic DNA of the three clones. Thus, it appears that failure to observe copy number changes of DHFR* in these integration sites is related to genome position. We hypothesize that these regions harbor genes that, if amplified, would cause cell cycle arrest or cell death. The 1M-73 integration site is within <700 bp of CDKN1B (p27Kip1), a negative regulator of the cell cycle [35]. Overexpression of CDKN1B due to copy number increase of the locus could suppress growth and abrogate any advantage that might have been conferred by increased copy number and expression of DHFR*. Similarly, in 1M-84, DHFR* is integrated between PCGF2 (Mel-18) and PSMB3, approximately 1 Mb proximal to ERBB2, a gene frequently amplified in cancer. Nevertheless, PCGF2 and PSMB3 are rarely amplified with ERBB2 [36] in tumors, and PCGF2 has been reported to be a tumor suppressor [37,38]. Moreover, a recent report indicates that overexpression of PCGF2 leads to down regulation of MYC and senescence in human fibroblasts [39]. Thus, it is likely that amplification of DHFR* at the 1M-84 insertion site would be deleterious to cells exposed to methotrexate due to proximity to, and thus co-amplification with, PCGF2. These considerations suggest that there may be selection pressure from neighboring growth inhibitory genes on the positions of amplicon boundaries.
Copy number changes involving DHFR* encompassed regions much larger than the integrated DNA and, in most cases, the boundaries of the copy number changes were not recurrent as we had found previously for the endogenous locus on 5q13 [17]. At the 1M-42 integration site, however, we did observe a high frequency of amplification and recurrent copy number transitions or amplicon boundaries at RP11-238H10 and CTD-2013D21, which occurred in 50-70% of resistant colonies. A role for expression of fragile sites in promoting amplification and setting boundaries to the amplicons in tumor genomes was suggested more than 20 years ago [40,41]. Although several fragile sites, including the folate sensitive fragile site FRA8A, map near the 1M-42 integration site on 8q22 (Figure 7), we did not find that FRA8A was expressed at sufficiently high frequency in HCT116 cells for it to be mapped by FISH. Nevertheless, even low frequency expression of fragile sites in cells exposed to methotrexate leading to an increased rate of breakage in close proximity to a DHFR* integration site could be sufficient to enhance the probability of amplification and selection for cells with recurrent amplicon boundaries. Thus, in the 1M-42 resistant colonies, induction of FRA8A by exposure to methotrexate could have provided a double-strand DNA break in close proximity to DHFR*, which initiated amplification of this locus and subsequent survival advantage of these cells in the presence of methotrexate. Similarly, the association of folate deficiency with breast cancer suggests that in individuals who express folate sensitive fragile sites, breakage may occur at higher frequency at these loci, leading to chromosomal rearrangements promoting tumor development. We have, however, been unable to find evidence that fragile sites play a more general role in setting aberration boundaries in breast cancer (that is, using 1.5 Mb resolution array CGH data and looking over 5 Mb windows on chromosome arms, we found no evidence that fragile sites are associated with amplicon boundaries in breast tumors). We note that these analyses are currently hampered by the relatively low resolution mapping of both copy number aberration breakpoints and fragile sites. On the other hand, it is noteworthy that amplification was significantly more frequent at the 1M-42 integration site, suggesting that, if not fragile sites, then some, as yet undefined genomic features may have influenced the propensity to amplify. Similar genomic characteristics may also play a role in setting boundaries for amplicons in human tumors.
Figure 7.
Positions of DHFR* integration sites and fragile sites. The DHFR* integration sites were mapped onto the genome using BLAT [46] and are shown as a red bar on the chromosome ideogram from the UCSC genome browser [45]. Common fragile sites are shown under the chromosome and are mapped according to positions reported in NCBI Entrez and the literature [65-77]. Rare folate sensitive fragile sites are shown above the chromosome. Note that the 1M-45DHFR* integration site mapped just proximal to FRA11B, another folate sensitive fragile site; however, no copy number transitions or amplicon boundaries mapped to this region. Since FRA11B is a RFS, it may not be expressed in HCT116.
Conclusion
Altered expression of genes can occur by a number of mechanisms, of which copy number change is only one. In the simple system we have used to study genome position effects, cells are being challenged by a single agent and also offered a gene that can provide resistance, unlike the development of human tumors in which, as normal cells evolve to cancer cells, many barriers must be breached. There are many ways to alter signaling or checkpoint pathways, resulting in evasion of apoptosis and enhanced proliferation. Increasing expression of appropriate genes in these pathways by copy number changes is only one mechanism. The observations reported here on a model system translated to tumor development suggest that the nature of the gene under selection for growth advantage and genome context are, together, factors influencing the propensity to amplify. In addition, it is likely that the nature of the challenges experienced by the nascent cancer cell (for example, folate deprivation), individual variation (for example, frequency of expression of various fragile sites), and the types of genomic instability that may be present also influence which genes will be altered in expression by copy number changes.
Materials and methods
Cloning, cell culture, infection
The human colorectal HCT116+chr3 cell line (gift from Dr CR Boland) was maintained in DME H-21 media supplemented with 10% fetal bovine serum, 1 × non-essential amino acids, 100 U/ml penicillin and 100 μg/ml Streptomycin, and 400 μg/ml G418 (Invitrogen, Carlsbad, CA, USA). The MMLV vector containing the double mutant dihydrofolate reductase gene (L22F, F31S) fused to the gene encoding EGFP was a generous gift from Dr J Bertino, Cancer Institute, New Jersey. Using this vector, we prepared a DHFR variant with a single mutation (L22F) fused to the EGFP gene, which we refer to as DHFR*.
HCT116+chr3 genomic DNA was used as a template for amplifying the DHFR promoter and 5' segment of the gene (from -461 to +23 relative to the ATG translation initiation codon [42]) with primers containing Esp3I restriction sites at the 5' end. The 3'sequence of DHFR* (from +20 relative to the ATG translation initiation codon) was PCR amplified from the vector (primers also contained Esp3I restriction sites at the 5' end). Both PCR products were cleaved with Esp3I restriction enzyme, ligated together and cloned into pLPCX retroviral vector (Clontech, Mountain View, CA, USA), which had been linearized by digestion with Nco and HindIII restriction enzymes, resulting in removal of the CMV promoter from the vector.
To deliver DHFR* into HCT116+chr3 cells, we used the Phoenix retroviral system (Orbigen, San Diego, CA, USA). HCT116+chr3 cells were sparsely seeded into 6-well plates (103 cells per well) and infected with diluted viral particles the following day (MOI < 0.1). Two days after infection, resistant colonies were selected with puromycin (0.3 μg/ml media) for seven days. Individual colonies were expanded as separate integration site clones (1M-XX) without puromycin.
To express POLR2K and LOC157567 in HCT116+chr3, cDNA clones (MSH1010-74358 and MHS1010-9205152, respectively) were purchased from Open Biosystems (Huntsville, AL, USA). The coding sequence was amplified with primers containing Esp3I restriction sites and cloned into the HindIII site of the pLHCX vector (Clontech). To overexpress MYC, the cDNA was cloned into the pBABE-hygro vector. The integrity of the constructs was verified by sequencing and the Phoenix retroviral system was used to deliver the genes into HCT116+chr3 cells. Two days after infection, cells were exposed to hygromycin (400 ng/ml media) for 7 days to select pools of cells with stable insertion.
Methotrexate sensitivity
Each clone was seeded into one 96-well plate (400 cells per well) leaving outer wells without cells, only with media. The next day different concentrations of methotrexate were added and cells were cultured for 72 hours. After three days, half of the media was replaced with fresh media with the same methotrexate concentration and cells cultured for another three days. Cells were washing gently with 1 × phosphate-buffered saline and plates were stored at 80°C prior to measuring cell proliferation using the CyQUANT Cell Proliferation Assay (Invitrogen, Carlsbad, CA, USA). The inhibition concentration (IC-50) was calculated for each integration site clone.
Selection of methotrexate resistant colonies
Individual clones were seeded into 6-well plates (103 cells per well) and grown until the population reached approximately 5 × 105 cells in each well. Then the cells from each well were counted and seeded into 100 mm dishes (4 × 105 cells per dish). The next day different concentrations of methotrexate (3-4 × IC-50, depending on clone sensitivity to methotrexate) were added to the cells. The medium was replaced every third day and after four weeks one colony from each dish was picked into a 12-well plate and then expanded into T75 culture flasks. DNA was isolated from a quarter of the cells from each T75 flask, and another quarter of the cells was expanded for isolation of RNA, proteins and metaphase spreads. The remaining half of the cells was frozen in liquid nitrogen. In other experiments, colonies were selected as above and then fixed and stained with crystal violet after four weeks to determine the number of resistant colonies.
Induction of fragile sites
Individual clones were seeded into 6-well plates (103 cells per well) and grown until the population reached approximately 106 cells in each well. Then the cells from each well were counted and seeded into two 100 mm dishes (4 × 105 cells per dish). After six hours, fragile site expression was induced in one of the two dishes by adding aphidicolin (at a final concentration of 0.15 μM). After 24 hours, aphidicolin was removed and methotrexate was added to all dishes (same concentration as used for selection of methotrexate resistant colonies). Colonies were fixed and stained with crystal violet after four weeks. Folate sensitive fragile sites were induced by adding methotrexate to HCT116+chr3 cells (25 nM) or the 1M-42 clone (75 nM). After 24 hours, metaphase spreads were prepared according to standard protocols.
Inverse PCR
Inverse PCR was performed as described previously [43,44] with slight modification as described in Additional data file 1. To map the integration sites on the genome sequence, the PCR products amplified in a second nested PCR were sequenced and mapped onto the human genome sequence using BLAT (UCSC Genome Browser, May 2004 freeze [45,46]). Methodological details and sequences obtained by inversion PCR for each insertion site are provided in Additional data file 1.
Array CGH
Array CGH was carried out as described previously [17]. Briefly, genomic DNA was isolated from one quarter of the cells from a T75 flask by incubation overnight at 55°C in 3 ml of 1 × TE buffer (pH 7.5) supplemented with 0.5 % SDS and 0.1 μg/μl proteinase K (Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN, USA), followed by ethanol precipitation after 24 hours. The DNA was dissolved in 100 μl of H2O. Genomic DNA (600 ng) was labeled by random priming to incorporate Cy3 or Cy5 dCTP in a 50 μl reaction and the labeled test and reference DNAs together with 100 μg human Cot-1 DNA were hybridized for approximately 48 hours at 37°C to arrays of 2,464 BAC clones, each printed in triplicate (HumArray2.0, 3.1, UCSF Comprehensive Cancer Center Microarray Core). Whole genome tiling path arrays containing 32,145 BAC clones [20] from the UCSF Comprehensive Cancer Center Microarray Core were used for more detailed analysis of some amplicons. A custom built CCD camera system was used to acquire 16 bit 1,024 × 1,024 pixel DAPI, Cy3 and Cy5 images [47,48]. Array data are available in Additional data files 2 and 3 and at NCBI Gene Expression Omnibus [49]. GSE6262 contains 95 CGH profiles of methotrexate resistant colonies and also untreated controls (HumArray2.0, 3.1). GSE6360 contains six CGH profiles (32K BAC tiling path array).
Expression microarrays
Test (20 μg) and reference (20 μg) RNA were reverse transcribed in the presence of aminoallyl-dUTP followed by coupling with Cy3 or Cy5 dyes. The untreated integration site clone was used as the reference (Cy5 labeled cDNA) for each of the hybridizations with its respective resistant colonies (Cy3 labeled cDNA). Hybridizations were carried out on arrays of printed long oligonucleotides (70 mers) containing 21,000 elements (Operon V2.0 [50]). Array images were analyzed using UCSF SPOT software [51]. Expression data were filtered to exclude all spots with foreground intensity below 500 units in the Cy3 or Cy5 channel and were subsequently median normalized and log2 transformed. Data are available in Additional data file 4 and at NCBI GEO (GSE6262).
Copy number analysis
Image and data analyses were carried out using UCSF SPOT [51] and SPROC software, as described previously [17]. We also made corrections for two well recognized sources of variation in ratios; the observed dependence on location of clones on the array (spatial or geometric variation) [52], and the dependence of ratios on GC content [53]. To correct for geometrical dependence of the log2 ratios over the array, we took advantage of the fact that neighboring loci in the genome are, for the most part, likely to have the same copy number [54]. We used backfitting [55] to iteratively obtain an estimate of the systematic spatial variation of the log2 ratios over the array, which was then subtracted from the original log2 ratios (Additional data file 6). The log2 ratios were corrected for GC content using a loess method [56]. That these procedures result in a reduction in the apparent 'noise' in the ratio profiles, but do not alter the ratios, is clearly seen by a side-by-side comparison with the array data prior to correction (Figures B and C in Additional data file 6).
Elucidation of copy number changes specific to methotrexate resistant cells
Although the copy number differences between a methotrexate resistant and untreated HCT116 cell line are easily distinguished by visible inspection of the copy number profiles, we developed an objective method to identify the copy number changes acquired by the drug resistant cells. Since the HCT116 cells have a small number of low level copy number changes (Figure 1a), it is necessary to exclude them from the analysis by 'subtracting out' aberrations that were present in the cell population prior to exposure to methotrexate. Therefore, to identify the copy number changes specific to methotrexate resistant cells, aberrations in the copy number profile of the untreated cells for each DHFR* insertion site (called 'parent') were subtracted from the resistant cell profiles for each colony from that insertion site (called 'colony'). However, since the amplitudes of the ratio changes in array CGH may vary among different measurements of related samples due, for example, to incomplete suppression of repeated sequences (Figure 1 in Additional data file 7), we implemented a correction procedure that took into account the differences in amplitude between the array CGH profile of a resistant colony relative to the parent. The correction was based on the assumption that when biases equally affect test and reference signals, a copy number change and the observed copy number linear ratio are linearly related, but the slope may be reduced as the bias increases [57]. Taking into account the known homozygous deletion on chromosome 16p in HCT116, a robust linear model was fit to the linear ratio of the colony and the parent with the colony as the response (Figure 2a in Additional data file 7). A weight of 500 was given to the homozygous deletion (using a cutoff of linear ratio <0.25). The large weight was necessary because of the small number of arrayed BACs reporting homozygous deletion (mean four BACs per sample) compared to the total number of arrayed BACs used for the analysis (2,056-2,282, before parent subtraction). The fitted value gave the parental ratio after correcting for bias. Thus, the ratio of the colony to the fitted parent ratio was considered the colony specific copy number change (Figure 2b in Additional data file 7). A comparison of this procedure with and without using the spatial correction procedure described in the previous section is shown in the upper panels in part (b) of Additional data file 6. Since we considered each insertion site separately and required that ratios on arrayed BACs be present in both the parent and the colony for each insertion site, the number of BACs remaining after this analysis varied among the different insertion site parents and their colonies (1705-2099).
After subtraction of the linear parent ratios, the array CGH data were converted to log2 and analyzed as reported previously [58] using circular binary segmentation (CBS) [59] with default parameters to translate experimental intensity measurements into regions of equal copy number as implemented in the DNAcopy R/Bioconductor package [60]. Missing values for BACs mapping within segmented regions of equal copy number were imputed by using the value of the corresponding segment. A few clones with missing values were located between segmented regions and their values were imputed using the maximum value of the two flanking segments. Thus, each array clone was assigned a segment value referred to as its 'smoothed' value. The scaled median absolute deviation (MAD) of the difference between the observed and smoothed values was used to estimate the sample-specific experimental variation. All the samples had MAD less than 0.15. Due to additional random noise introduced when subtracting the parent from the resistant colonies, the smoothed values from CBS were further 'merged' using the MergeLevels procedure [61]. In this process, segmental values across the genome were merged to create a common set of copy number levels for each sample. The minimum and maximum criteria used in MergeLevels were 0.05 and 0.5, respectively.
To identify single technical or biological outliers, such as high level amplifications, the presence of the outliers within a segment was allowed by assigning the original observed log2 ratio to the clones for which the observed values were more than four tumor-specific MAD away from the smoothed values. The amplification status for a clone was then determined as described previously [58] by considering the width of the segment to which that clone belonged (0, if an outlier) and a minimum difference between the smoothed value of the clone (observed value, if an outlier) and the segment means of the neighboring segments. The clone was declared amplified if it belonged to the segment spanning less than 20 Mb and the minimum difference was greater than exp(-x3) where x is the final merged value for the clone.
Expression of genes in amplicons
For 12 samples the borders of the regions determined to have been amplified by array CGH were further refined by manually incorporating additional data from the whole genome tiling path arrays and FISH experiments. The border of each amplicon was set between the last not amplified BAC clone and the first amplified BAC clone. Expression probes were first filtered for ones with missing annotation, resulting in 19,777 probes, which were then mapped to the genome sequence. From these mapped probes, a list of expression probes included in all the amplicons within the samples was compiled manually, resulting in 685 expression probes, of which 370 provided measured expression in both amplified and non-amplified samples. To assess whether a gene was up-regulated when amplified, the mean expression levels were calculated in samples with and without amplification of the gene. Mean expression levels with and without amplification were compared and those with log2 fold change >0.8 were considered to be up-regulated when amplified.
Analysis of clustering of gene expression driven by MYC target genes
Genes with variable expression across the data set (standard deviation (SD) ≥ 0.3) and present in >75% of samples were identified and filtered for those with missing annotation. The samples were clustered based on the remaining clones (3,931) using the Pearson correlation distance metric and complete linkage. Human 'MYC target' genes (1,287) were identified by reference to the MYC database [21], and 380 were included in the set of 3,931 variably expressed genes. The fold change in expression of 380 MYC genes between the two sample clusters was computed and compared to the median log2 fold change for 1,000 selections of 380 random genes.
Quantitative RT-PCR
Real-time quantitative RT-PCR was performed on RNA isolated from methotrexate resistant colonies and untreated insertion site clones using Trizol reagent (Invitrogen) and treatment with DNase (Promega, Madison, WI, USA) as previously described [62] in the UCSF Comprehensive Cancer Center Genome Analysis Shared Resource Facility. Primers (forward, 5'CGTAAACGGCCACAAGTTCAG, reverse, 5'GGGTCAGCTTGCCGTAGGT) and probe (5'6FAM-CGCCCTCGCCCTCGC-BHQ_1) for EGFP were designed using GenScript Primer Design (GenScript Corp., Piscataway, NJ, USA). The ABI Assays-on-Demand were used to confirm overexpression of MYC, POLR2K and LOC157567 genes (ABI, Foster City, CA, USA).
Fluorescent in situ hybridization
Metaphase spreads were prepared from untreated insertion site clones and methotrexate resistant colonies at the time of RNA and protein isolation. Before hybridization slides were aged in 2 × SSC at 37°C for 30 minutes, dehydrated in an ethanol series (70%, 80% and 96%) and incubated in 70% formamide, 2 × SSC at 72°C for 4 minutes to denature the DNA. Fluorescent probes were prepared by labeling 20 ng of BAC DNA by random priming to incorporate Cy3- or FITC-coupled dCTP. The labeled DNA was precipitated, pre-annealed with 30 μg Cot-1 DNA and hybridized to metaphase spreads for 48 hours at 37°C in a humid chamber. Slides were washed three times with 50% formamide, 2 × SSC at 45°C for 15 minutes, 2 × SSC for 15 minutes, 0.1 × SSC for 5 minutes, counterstained with DAPI and mounted in Vectashield (Vector Labs, Burlingame, CA, USA).
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 shows the mapping and sequencing of DHFR* integration sites. Additional data file 2 lists array CGH log2 ratio data on clones for methotrexate resistant colonies and untreated DHFR* integrants. Additional data file 3 lists the Chromosome 8 array CGH dataset for five 1M-42 methotrexate resistant colonies hybridized on the 32K genome tiling path BAC array. DHFR* was integrated on chromosome 8 at 102,162,871 bp and this position is highlighted in red. All five colonies formed amplicons around the DHFR* integration site. Amplified regions are highlighted in yellow. Additional data file 4 lists Expression array data. Additional data file 5 shows the expression of genes mapping in amplicons in 12 methotrexate resistant cell lines. Additional data file 6 provides a Comparison of analysis of array data with and without spatial and GC correction. Additional data file 7 is an Example of elucidation of copy number changes specific to methotrexate resistant cells (results of the analyses for the 1M-34_c5a colony).
Supplementary Material
Mapping and sequencing of DHFR* integration sites
Array CGH log2 ratio data on clones for methotrexate resistant colonies and untreated DHFR* integrants
DHFR* was integrated on chromosome 8 at 102,162,871 bp and this position is highlighted in red. All five colonies formed amplicons around the DHFR* integration site. Amplified regions are highlighted in yellow.
Expression array data
Expression of genes mapping in amplicons in 12 methotrexate resistant cell lines
Comparison of analysis of array data with and without spatial and GC correction
Example of elucidation of copy number changes specific to methotrexate resistant cells (results of the analyses for the 1M-34_c5a colony)
Acknowledgments
Acknowledgements
We thank Richard Segraves for assistance with hybridizations to the 32K BAC human genome tiling arrays and Stewart MacArthur for mapping expression array probes onto the genome sequence. This work was supported by NIH grants CA90421 and CA94407.
Contributor Information
Pavla Gajduskova, Email: gajduskova@ibp.cz.
Antoine M Snijders, Email: Snijders@cc.ucsf.edu.
Serena Kwek, Email: skwek@cc.ucsf.edu.
Ritu Roydasgupta, Email: Roydasgupta@cc.ucsf.edu.
Jane Fridlyand, Email: fridlyand@cc.ucsf.edu.
Taku Tokuyasu, Email: tokuyasu@cc.ucsf.edu.
Daniel Pinkel, Email: Pinkel@cc.ucsf.edu.
Donna G Albertson, Email: albertson@cc.ucsf.edu.
References
- Miele M, Bonatti S, Menichini P, Ottaggio L, Abbondandolo A. The presence of amplified regions affects the stability of chromosomes in drug-resistant Chinese hamster cells. Mutat Res. 1989;219:171–178. doi: 10.1016/0921-8734(89)90012-x. [DOI] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays. 2004;26:1164–1174. doi: 10.1002/bies.20125. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Amplification-mutagenesis - how growth under selection contributes to the origin of genetic diversity and explains the phenomenon of adaptive mutation. Res Microbiol. 2004;155:342–351. doi: 10.1016/j.resmic.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Chernova OB, Chernov MV, Ishizaka Y, Agarwal ML, Stark GR. MYC abrogates p53-mediated cell cycle arrest in N-(phosphonacetyl)-L-aspartate-treated cells, permitting CAD gene amplification. Mol Cell Biol. 1998;18:536–545. doi: 10.1128/mcb.18.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/S0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Kuo MT, Sen S, Hittelman WN, Hsu TC. Chromosomal fragile sites and DNA amplification in drug-resistant cells. Biochem Pharmacol. 1998;56:7–13. doi: 10.1016/S0006-2952(98)00040-9. [DOI] [PubMed] [Google Scholar]
- Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipiras E, Coquelle A, Bieth A, Debatisse M. Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. Embo J. 1998;17:325–333. doi: 10.1093/emboj/17.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay BJ, Ondrusek NK, Wildenhain YD, Huang T, Carlone RL, Clement JM, Wahl GM. Effect of genomic position on amplification of the DFR1 gene in Saccharomyces cerevisiae. Adv Exp Med Biol. 1993;338:545–550. doi: 10.1007/978-1-4615-2960-6_111. [DOI] [PubMed] [Google Scholar]
- Wahl GM, Robert de Saint Vincent B, DeRose ML. Effect of chromosomal position on amplification of transfected genes in animal cells. Nature. 1984;307:516–520. doi: 10.1038/307516a0. [DOI] [PubMed] [Google Scholar]
- Kuo MT, Vyas RC, Jiang LX, Hittelman WN. Chromosome breakage at a major fragile site associated with P-glycoprotein gene amplification in multidrug-resistant CHO cells. Mol Cell Biol. 1994;14:5202–5211. doi: 10.1128/mcb.14.8.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Miller CT, Lin L, Casper AM, Lim J, Thomas DG, Orringer MB, Chang AC, Chambers AF, Giordano TJ, Glover TW, Beer DG. Genomic amplification of MET with boundaries within fragile site FRA7G and upregulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006;25:409–418. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR. Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim Biophys Acta. 2002;1587:164–173. doi: 10.1016/s0925-4439(02)00079-0. [DOI] [PubMed] [Google Scholar]
- Matherly LH. Molecular and cellular biology of the human reduced folate carrier. Prog Nucleic Acid Res Mol Biol. 2001;67:131–162. doi: 10.1016/s0079-6603(01)67027-2. [DOI] [PubMed] [Google Scholar]
- Snijders AN, Fridlyand J, Mans D, Segraves R, Jain AN, Pinkel D, Albertson DG. Shaping tumor and drug resistant genomes by instability and selection. Oncogene. 2003;22:4370–4379. doi: 10.1038/sj.onc.1206482. [DOI] [PubMed] [Google Scholar]
- Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N'-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Bosdet I, Smailus D, Chiu R, Mathewson C, Wye N, Barber S, Brown-John M, Chan S, Chand S, et al. A set of BAC clones spanning the human genome. Nucleic Acids Res. 2004;32:3651–3660. doi: 10.1093/nar/gkh700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, Ethier SP. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64:40–47. doi: 10.1158/0008-5472.CAN-03-1022. [DOI] [PubMed] [Google Scholar]
- Kauraniemi P, Barlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- Rossi MR, La Duca J, Matsui S, Nowak NJ, Hawthorn L, Cowell JK. Novel amplicons on the short arm of chromosome 7 identified using high resolution array CGH contain over expressed genes in addition to EGFR in glioblastoma multiforme. Genes Chromosomes Cancer. 2005;44:392–404. doi: 10.1002/gcc.20256. [DOI] [PubMed] [Google Scholar]
- Evans AJ, Gallie BL, Jewett MA, Pond GR, Vandezande K, Underwood J, Fradet Y, Lim G, Marrano P, Zielenska M, Squire JA. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am J Pathol. 2004;164:285–293. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin K, Roy G, Ouellette M. Formation of extrachromosomal circular amplicons with direct or inverted duplications in drug-resistant Leishmania tarentolae. Mol Cell Biol. 1996;16:3587–3595. doi: 10.1128/mcb.16.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin K, Papadopoulou B, Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundig C, Leblanc E, Papadopoulou B, Ouellette M. Role of the locus and of the resistance gene on gene amplification frequency in methotrexate resistant Leishmania tarentolae. Nucleic Acids Res. 1999;27:3653–3659. doi: 10.1093/nar/27.18.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- Kauraniemi P, Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer. 2006;13:39–49. doi: 10.1677/erc.1.01147. [DOI] [PubMed] [Google Scholar]
- Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. Embo J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, Ishihara H, Kanno R, Kamiyasu M, Inoue H, Tokuhisa T, Taniguchi M, Kanno M. mel-18 negatively regulates cell cycle progression upon B cell antigen receptor stimulation through a cascade leading to c-myc/cdc25. Immunity. 1998;9:439–448. doi: 10.1016/S1074-7613(00)80627-5. [DOI] [PubMed] [Google Scholar]
- Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536–546. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBeau MM, Rowley JD. Heritable fragile sites in cancer. Nature. 1984;308:607–608. doi: 10.1038/308607a0. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Urlaub G, Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986;6:1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol. 2003;77:2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT - the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Brown N, Oseroff V, Huey B, Segraves R, Sudar D, Kumler J, Albertson D, Pinkel D. A large field CCD system for quantitative imaging of microarrays. Nucleic Acids Res. 2006;34:e58. doi: 10.1093/nar/gkl160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- NCBI GEO http://www.ncbi.nlm.nih.gov/geo/
- Operon V2.0 http://www.gladstone.ucsf.edu/gladstone/site/genomicscore/section.php?id=1477
- Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–332. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce TE, Rozowsky JS, Luscombe NM, Emanuelsson O, Yu H, Zhu X, Snyder M, Gerstein MB. Extrapolating traditional DNA microarray statistics to tiling and protein microarray technologies. Methods Enzymol. 2006;411:282–311. doi: 10.1016/S0076-6879(06)11015-0. [DOI] [PubMed] [Google Scholar]
- Royce TE, Rozowsky JS, Gerstein MB. Assessing the need for sequence-based normalization in tiling microarray experiments. Bioinformatics. 2007;23:988–997. doi: 10.1093/bioinformatics/btm052. [DOI] [PubMed] [Google Scholar]
- Neuvial P, Hupe P, Brito I, Liva S, Manie E, Brennetot C, Radvanyi F, Aurias A, Barillot E. Spatial normalization of array-CGH data. BMC Bioinformatics. 2006;7:264. doi: 10.1186/1471-2105-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie TJ, Tibshirani RJ. Generalized additive models, volume 43 of Monographs on Statistics and Applied Probability. 1. New York Chapman and Hall; 1990. [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;(37 Suppl):S11–17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- http://bioconductor.org/packages/1.8/bioc/html/DNAcopy.html
- Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- Neve RM, Ylstra B, Chang CH, Albertson DG, Benz CC. ErbB2 activation of ESX gene expression. Oncogene. 2002;21:3934–3938. doi: 10.1038/sj.onc.1205503. [DOI] [PubMed] [Google Scholar]
- Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci U S A. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Reference Sequences (RefSeq) http://www.ncbi.nlm.nih.gov/RefSeq/
- Limongi MZ, Pelliccia F, Rocchi A. Characterization of the human common fragile site FRA2G. Genomics. 2003;81:93–97. doi: 10.1016/S0888-7543(03)00007-7. [DOI] [PubMed] [Google Scholar]
- Rozier L, El-Achkar E, Apiou F, Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: possible association with an inherited disease and cancer. Oncogene. 2004;23:6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli C, Karayianni E, Magnanini C, Mungall AJ, Thorland E, Negrini M, Smith DI, Barbanti-Brodano G. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene. 2002;21:7266–7276. doi: 10.1038/sj.onc.1205573. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R, Gostout BS, Cheung TH, Chung TK, Fu WY, Smith DI. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–7242. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- Ferber MJ, Montoya DP, Yu C, Aderca I, McGee A, Thorland EC, Nagorney DM, Gostout BS, Burgart LJ, Boix L, et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene. 2003;22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- Callahan G, Denison SR, Phillips LA, Shridhar V, Smith DI. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene. 2003;22:590–601. doi: 10.1038/sj.onc.1206171. [DOI] [PubMed] [Google Scholar]
- Sarafidou T, Kahl C, Martinez-Garay I, Mangelsdorf M, Gesk S, Baker E, Kokkinaki M, Talley P, Maltby EL, French L, et al. Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84:69–81. doi: 10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon WY, Sutherland GR, Richards RI, Tunnacliffe A. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature. 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- Savelyeva L, Sagulenko E, Schmitt JG, Schwab M. The neurobeachin gene spans the common fragile site FRA13A. Hum Genet. 2006;118:551–558. doi: 10.1007/s00439-005-0083-z. [DOI] [PubMed] [Google Scholar]
- Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre HJ, Lapsys N, Le Paslier D, Doggett NA, Sutherland GR, Richards RI. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/S0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mapping and sequencing of DHFR* integration sites
Array CGH log2 ratio data on clones for methotrexate resistant colonies and untreated DHFR* integrants
DHFR* was integrated on chromosome 8 at 102,162,871 bp and this position is highlighted in red. All five colonies formed amplicons around the DHFR* integration site. Amplified regions are highlighted in yellow.
Expression array data
Expression of genes mapping in amplicons in 12 methotrexate resistant cell lines
Comparison of analysis of array data with and without spatial and GC correction
Example of elucidation of copy number changes specific to methotrexate resistant cells (results of the analyses for the 1M-34_c5a colony)