A number of techniques for the location of small and medium-sized model fragments in experimentally phased electron-density maps are explored. The application of one of these techniques to automated model building is discussed.
Keywords: model fragments, electron-density maps, model building
Abstract
Molecular replacement is a powerful tool for the location of large models using structure-factor magnitudes alone. When phase information is available, it becomes possible to locate smaller fragments of the structure ranging in size from a few atoms to a single domain. The calculation is demanding, requiring a six-dimensional rotation and translation search. A number of approaches have been developed to this problem and a selection of these are reviewed in this paper. The application of one of these techniques to the problem of automated model building is explored in more detail, with particular reference to the problem of sequencing a protein main-chain trace.
1. Introduction
Molecular replacement provides a powerful tool for phasing X-ray diffraction data by use of a known homologous atomic structure. The orientation of the atomic model in the unsolved crystal cell is determined by rotating it until a good match is obtained between the Patterson functions of the known and unknown structures. Historically, limitations of the method employed have required that the search model includes a substantial portion of the scattering density of the asymmetric unit of the unsolved structure and that the known and unknown structures be closely homologous. Recent developments in the use of maximum-likelihood techniques for molecular replacement (Read, 2001 ▶) have relaxed these constraints somewhat; however, current techniques do not allow the location of smaller models ranging from a few atoms to a few residues and in unfavourable cases fail to locate larger domains.
When molecular replacement cannot be used to solve and complete the structure readily, the alternative approach of experimental phasing using anomalous scattering or isomorphous derivatives is adopted. These approaches lead to a set of phase estimates from which a noisy electron-density map may be calculated. This map may then be interpreted in terms of atoms or polymer subunits by visual inspection of the electron density contoured in three dimensions, usually using a single contour level. This approach is effective when the phase estimates are sufficiently accurate.
However, there are problems associated with the visual inspection of electron-density maps. Firstly, it is a time-consuming process. As a result, a number of approaches to automatic electron-density interpretation have been developed over the last decade, largely focusing on features on a similar scale to that used by the human interpreter, i.e. polymer repeats. Secondly, the reduction of the electron-density map to a single contour level which can be visualized by the user throws away a great deal of the information contained in the features of the electron-density map. Thirdly, it is harder for the user to visualize electron density on a scale significantly different from a polymer repeat, thus a human interpreter would struggle to fit a whole missing domain into a noisy electron-density map, although it may be possible using a low-resolution difference map.
As a response to these problems, a number of approaches have been developed to allow the automatic fitting of molecular fragments varying in size from a few atoms to a complete domain into noisy electron-density maps. This problem involves at some level a six-dimensional search over possible orientations and translations of the search fragment in the target electron-density map, although performing the full six-dimensional search is computationally demanding and some form of optimization is usually adopted to improve performance. The use of a computational comparison between the model and the target electron density allows similarity to be measured without necessarily reducing the electron density to a single contour and allows the comparison of arbitrary-sized features.
A selection of fragment-fitting techniques are reviewed in this paper, followed by a discussion of the application of one of these techniques to the problem of automated interpretation of electron-density maps for protein structures. One common feature of all the methods described here is that the search is performed in an electron-density map. This represents a limitation, since the electron-density map does not contain all the information that is present in the structure-factor magnitudes and phase probability distributions. This is an area in which there is potential for future development.
1.1. Template convolution: ESSENS
The first approach to receive widespread application to the location of small fragments in an electron-density map was the ESSENS software of Kleywegt & Jones (1997 ▶). In this approach, a candidate position and orientation are chosen for the search model and the model is placed in the target electron-density map at the candidate position. A score function is calculated by consulting the electron density in the target map at the coordinates of each atom in the search model in turn (using the average of the eight density points surrounding the atom position). This gives rise to a list of N density values, where N is the number of values in the fragment. The list is scored using a minimum function following the approach of Nordman & Schilling (1986 ▶) using the mean of the lowest densities in the list, thus penalizing placings of the fragment which place more than a few atoms in low density.
The process is repeated over a sampling of all possible positions and orientations of the fragment and each placing is scored using the minimum function. The placings with the highest scores are candidates for possible locations of the fragment in the electron density. These may be refined using a finer search about the candidate solution. Correct solutions may be identified by scores which stand out from the rest of the list.
Kleywegt & Jones (1997 ▶) report very clear results for a 148-atom fragment in a 3 Å map, with the calculation taking a few hours, and the program has since been applied to a range of problems. Optimizations are possible by eliminating symmetry-related positions from the list of translations and by further restricting the positional search using packing constraints and a solvent boundary. A further optimization (Jones, 2004 ▶) is to combine the process with map skeletonization and to only test translations which correspond to points lying on the density ridges, leading to a calculation which is fast enough for interactive use.
The advantages of this approach are that it is very simple and also robust against the scaling of the map. However, it suffers from one problem somewhat similar to that of visual inspection of contoured maps: it is only sensitive to high density in positioning the model fragment. Thus, there is no penalty for placing a fragment in a position where there is significant additional density not accounted for by the atomic model. An additional limitation is that it takes no account of resolution: for example, at low resolutions the electron-density peaks may move away from the atomic centres (for example, in the case of α-helices at <5 Å resolution the highest electron density may run along the middle of the helix).
1.2. Fast Fourier feature recognition: FFFear
Cowtan (1998 ▶) was inspired by the work of Kleywegt & Jones (1997 ▶) to attempt an FFT-based implementation of the same ideas, although in the process some of the other limitations of the approach were also addressed. A new scoring function was developed that had two major differences from the approach of Kleywegt & Jones (1997 ▶).
(i) Instead of searching using an atomic model, an electron-density target function was constructed on a grid large enough to completely enclose the search model. The electron density was calculated over the whole of this grid, including regions where atoms were present in the fragment, regions where atoms were absent in the fragment and regions beyond the boundaries of the fragment.
(ii) Instead of looking for high density, the new target function looks for density matching the electron density of the search fragment. This allows the search function to identify placings of the search model where both high and low density match; however, it introduces a requirement that the map density and fragment density be correctly scaled.
Since the electron density of the target map is consulted over a region which may extend beyond the boundaries of the fragment, it is necessary to include a mask function along with the electron-density target to describe the volume over which the electron densities of the target map and search fragment should agree.
The resulting target function is therefore constructed as a weighted mean-squared difference between the target map and the search density placed in the map at a given position and orientation,
where x is a translation offset used for moving the search density through the unit cell, r describes a single rotation of the search fragment, t(x, r) is the score for a given translation and rotation (with low scores representing good matches between the map and the search model), ρ(x) is the electron density of the target map, ρf,r(x) is the rotated electron density of the search fragment and μf,r(x) is the rotated mask function of the search fragment. The rotation search is performed by rotating the search target density and performing a translation search for each orientation.
This approach may be performed in a computationally efficient manner by using fast Fourier transforms (FFTs) to perform the translation search, using five FFTs for a single translation search or three FFTs per orientation if multiple searches are being performed in the same map. The resulting calculation generally takes a few minutes on a current personal computer. (It is also possible to calculate a weighted correlation using this same approach, eliminating the need to scale the densities; however, additional FFTs are required and in some cases the local scale of the density is informative.)
In a subsequent refinement of the approach, Cowtan (2001 ▶) demonstrated that by using a modified electron-density target function and weight function, the same equation could be used to calculate an ‘electron-density likelihood’ score describing the probability of a particular piece of electron density being modelled by a specific placement of the search model. In this case, a nonbinary weighting function allows strongly conserved areas of the search model to be fitted more precisely than less conserved regions.
The advantages of this approach are that it is not very computationally demanding (although it is too slow for interactive use) and that it fits regions of both high and low electron density in the search model while allowing regions in which the model is uncertain. The time required for the computation of the translation function is also independent of the size of the search model (although ideally the rotation search becomes finer as the size of the model increases, in practice this is seldom necessary since a match to the core of the molecule may be found and refined). The main disadvantage is that method is dependent on the target-map density and the search-model density being correctly scaled. Another limitation is that the method is not susceptible to optimization by reducing the translation search region, unlike the ESSENS approach.
1.3. The phased rotation, conformation and translation function: PRCTF
The phased rotation, conformation and translation function (PRCTF) of Pavelcik (2006 ▶) is related in some ways to the FFFear approach in that it involves a six-dimensional search for the electron-density features associated with the search fragment and that it optimizes the six-dimensional search by using a fast method for one part of the rotation–translation search. The difference between the approaches is that instead of applying a form of fast translation search and slow orientation search, the PRCTF employs a fast rotation search and slow translation search.
The procedure involves calculating an expansion of the electron density of the search fragment in terms of spherical harmonics and the corresponding expansion of the electron density of the target map around each grid point in the map. This expansion is performed using FFTs (Pavelcik et al., 2002 ▶) and a list of coefficients is stored for every grid point in the map containing the l, m coefficients for the expansion of the electron density about that point. The data stored is substantial, running into multiple gigabytes.
However, once the expansion of the map is performed, it is comparatively efficient to search for a list of search fragments at the same time using a fast rotation function at each point in the grid. Therefore, a library of fragments prepared from the PDB, including if required multiple conformations of each fragment, are applied to each position in the map and the best orientation is chosen. The orientation may be refined to optimize the agreement between the fragment density and the map density at that point using a score based on either the overlap integral or correlation of the densities.
The method has been applied to the location of secondary-structure fragments in proteins and to the location of DNA and RNA fragments in nucleotide structures. The fragments located by this means can be connected to build a continuous model of the target structure (the automation of this completion step for nucleotides is novel). The calculation is more time-consuming than the other methods presented so far and is demanding in terms of storage; however, the ability to efficiently search against a whole library of fragments presents a significant opportunity for this approach in the interpretation of difficult maps.
1.4. The spherically averaged phased translation function: SAPTF
The spherically averaged phased translation function (SAPTF) is incorporated in the MOLREP program of Vagin & Isupov (2001 ▶). Unlike the previous methods, it reduces the six-dimensional search problem to a three-dimensional search only, which in turn is optimized using FFTs.
Unlike traditional molecular-replacement techniques, which eliminate the translation search by discarding the phases calculating a rotation search in the Patterson function, the SAPTF eliminates the rotation-dependent features by spherically averaging the search density to reduce the density to a list of radial Bessel function coefficients. The same expansion is applied to the electron-density map by expanding the spherically averaged density about each point in the map in terms of the same radial coefficients.
Calculating the fit between the search density and the target density at any position is then simply a matter of combining the coefficients of the model density and the map density at that point to determine the fit between the radial density distributions.
The calculation is by far the fastest of those discussed here, with only one FFT per radial Bessel coefficient and one search through the map. However, in spherically averaging the electron density, information about the shape of the fragment has been discarded. This process is therefore primarily applied as a filter for selecting possible sites at which a more exhaustive rotation search may then be applied.
2. Application of fragment-fitting techniques to model building
The techniques described here for fitting atomic model fragments into electron density have been applied to the problem of automated model building in a number of existing applications. Most notably, the highly successful RESOLVE software (Terwilliger, 2003a ▶) employs a search function similar to the FFFear search function to locate secondary-structure features in the electron-density map. These are subsequently extended by growing additional residues on each end by choosing the conformation which places the new atoms in the highest electron density.
Another approach, somewhat related to the methods described above, is employed in the CAPRA program from the TEXTAL package (Ioerger & Sacchettini, 2002 ▶) for locating likely α-carbon positions. Branch points in the skeleton are scored using a function which combines a range of electron-density feature scores using a neural network trained to select α-carbon-like features. This is somewhat related to the SAPTF method; however, while the feature scores are orientation-independent, they do not eliminate angular features of the density. However, the CAPRA approach is not a general-purpose density-fitting method, since it does not allow the user to choose an arbitrary search model for fitting.
A new method, in part inspired by ideas from both RESOLVE and CAPRA, is the Buccaneer model-building software. This approach applies a single feature-recognition technique, the FFFear search function, repeatedly in a number of different ways to build a protein model.
2.1. Model building in Buccaneer using fragment-fitting methods
The Buccaneer protein model-building calculation involves five successive applications of the FFFear target function to locate, extend and sequence protein chains. These applications are as follows.
Two types of electron-density target are used. The first, used to locate Cα atoms, is based on a 4 Å sphere of density centred on the Cα atom and the second, used for sequencing, is based on a 5.5 Å sphere of density centred on the Cβ atom. One α-carbon target is used and 20 β-carbon targets, one for each amino-acid type. The likelihood target and weight functions are constructed by calculating the mean and variance density for all features of that type from a known reference structure at a similar resolution to the unknown structure.
The FFFear search function is applied in the following ways in order to perform a complete model-building calculation.
(i) Use of the FFFear search function to locate probable α-carbon positions as starting points for chain building. For this stage alone, the search function is calculated using FFTs to perform a translation search over the whole map combined with an exhaustive search over all possible orientations of the search target.
(ii) Use of the FFFear search function to refine the most probable α-carbon positions. In this case, the search function is calculated by simple summation. A simplex search over the three rotational and three translational parameters is used to optimize the fragment location.
(iii) Use of the FFFear search function (by summation) to grow chains in each direction from the initial α-carbon positions by using an exhaustive search over allowed Ramachandran angles. A two-residue look-ahead is used in which two residues are built and scored by the combined FFFear score of their two α-carbon positions and the best results are stored for refinement.
(iv) Use of the FFFear search function (by summation) to refine the best attempts at chain growing at each step. The top five two-residue conformations determined at each step of chain growing are refined using the simplex algorithm with four Ramachandran angles as parameters. After refinement, the updated scores are used to select the best conformation from the list. The first of the two residues is added to the chain. The previous step is then repeated to grow the chain further.
(v) Use of the FFFear search function (by summation) with each of the 20 amino-acid side-chain functions to assign relative scores to each side-chain type for each α-carbon in the chain grown.
The final classification step is a new development and will be described in more detail.
2.2. Sequencing in Buccaneer using fragment-fitting methods
The Buccaneer electron-density target function is used as a test of how well a region of electron density matches a predetermined feature, taking into account possible variations in the electron density associated with that feature. These variations may arise from noise in the electron-density map arising from errors in the data or missing data or from variation within the feature itself. So, as an example, Cowtan (2006 ▶) described how to use this target function to identify likely α-carbon positions in a noisy electron-density map, despite the fact that the main-chain and side-chain conformations can vary and that the electron density associated with an α-carbon varies significantly with resolution. This approach has been demonstrated for both experimentally phased and molecular-replacement maps (Bahar et al., 2006 ▶).
The approach depends on simulating an electron-density map for a known whose resolution and noise levels match the unknown map. The electron-density features corresponding to any desired motif may be compiled from this map by combining the electron densities from every copy of that motif in the map to obtain a map of both mean density and the variation in the density as a function of position relative to the motif. The result is a function which describes for any position relative to the desired motif what the expected value of the electron density should be and how much it may be expected to vary from that value. As a result, the target function is able to identify the conserved features associated with an α-carbon while ignoring those features which are not conserved (such as the side chain).
This same approach may be applied to the identification of different residue types. The target function may be specialized to search for more specific features in the unknown structure by only using corresponding features from the known structure. Therefore, it is possible to search preferentially for alanine residues in the unknown map by only using alanine residues from the known structure in compiling the electron-density target function. If a target function is prepared for each type of amino-acid residue, then a residue in the unknown map may be tested against each target in turn to obtain an indication of the possible residue types which might explain the electron density.
One way of using this approach is to classify residues by both type and rotamer. This is the approach adopted by Terwilliger (2003b ▶) using a simpler target function. For an initial implementation, it was decided to test whether it is possible to identify side-chain types and thus assign sequence without attempting to identify their conformations. In other words, an electron-density target is prepared which is averaged over all side-chain rotamers for a given amino-acid type.
The likelihood density targets for classifying side-chain types are calculated over a sphere of density of radius 5.5 Å centred on the Cβ atom. This sphere is just large enough to enclose all the non-H atoms of tryptophan, the largest side chain with a high degree of rigidity. Only one target function is calculated per residue type using the pre-prepared simulated electron density from the known structure. The density target is therefore somewhat analogous to searching for a model which is an ensemble average over the possible side-chain conformations.
Each of the 20 side-chain targets is then applied to every α-carbon in the unsequenced model, using the positions of the N—Cα—C atoms to determine the position and orientation of the side chain and thus to position the target function in the map. This gives rise to a set of 20 scores, one for each amino-acid type, for each α-carbon in the model.
A problem arises from the fact that different side-chain types display different degrees of conformational variation and thus the scores for the different side-chain types show different ranges of variation. The raw scores are therefore converted to Z scores by subtracting the mean score for that amino-acid type and dividing by the standard deviation of the scores for that amino-acid type.
Finally, each chain is matched against the known sequence of the target structure, sliding the sequence along the chain fragment to try and locate of position where the sequence gives a good match against the electron-density features. The quality of the match at any sequence offset is scored by the sum of the Z scores for the corresponding amino-acid types at each position in the chain.
The best sequence match is that with the lowest Z score (since for the FFFear target function, low scores represent good agreements). A test is required to establish whether the resulting score is significant; for example, if the chain has been traced backwards then all attempts to sequence it must be wrong. The theoretical solution to this problem involves converting the scores to log-likelihood gains; however, in practice better results were obtained by the ad hoc method of examining the difference between the best and second best sequence matches: differences in the Z scores of greater than 3 provide a good indication that the match is correct. A similar approach is employed by Cohen et al. (2004 ▶).
One additional refinement is employed. Sometimes a chain trace will jump from one chain to another in the electron density. In this case, it is often possible to sequence segments on one or both sides of the jump, but not to sequence the whole chain fragment. To this end, attempts are also made to sequence subregions within the whole chain fragment, varying the start and end point within the fragment. To compare scores for sequences of different lengths, some form of weighting is required to account for the fact that longer chains will give rise to more extreme scores through random-walk statistics. Again, a log-likelihood gain should correct this problem, although in practice ad hoc methods give better results. Terwilliger (2003b ▶) uses random-walk statistics to suggest a weighting according to the inverse of the square root of the fragment length. In the current work, a formula which gives constant weight to the Z scores of shorter sequences and inverse square-root weight to the Z scores of longer sequences gave the best results,
where l is the length of the subfragment and l 0 is a constant controlling the changeover from constant to inverse square-root weighting. The value l 0 = 50 was found to be effective.
3. Results
The method described above was applied to two test structures from the Joint Center for Structural Genomics (JCSG) data archive (Joint Center For Structural Genomics, 2006 ▶). The chosen structures were solved using experimental phasing. For each structure, the JCSG software pursued multiple phasing paths using different software and parameters. A single initial phasing set was chosen for each structure by automatically selecting a structure on the basis of the statistics of the electron-density map.
Two structures were used for this test: 1vkn, for which the data extended to 2.45 Å resolution and the map correlation to the final map after density modification was 0.87, and 1vrb, for which the data extended to 3.2 Å resolution and the map correlation to the final map after density modification was 0.63.
For each structure, a large set of chain fragments of lengths between five and 30 residues were created by chopping up an initial model. Two starting models were used in each case: firstly the final model after refinement and secondly a model constructed by automatic chain tracing in Buccaneer. In the latter case, segments which were clearly incorrectly traced were removed, since these will not be sequenceable. However, a few cases remained where the chain trace jumped from one chain to another.
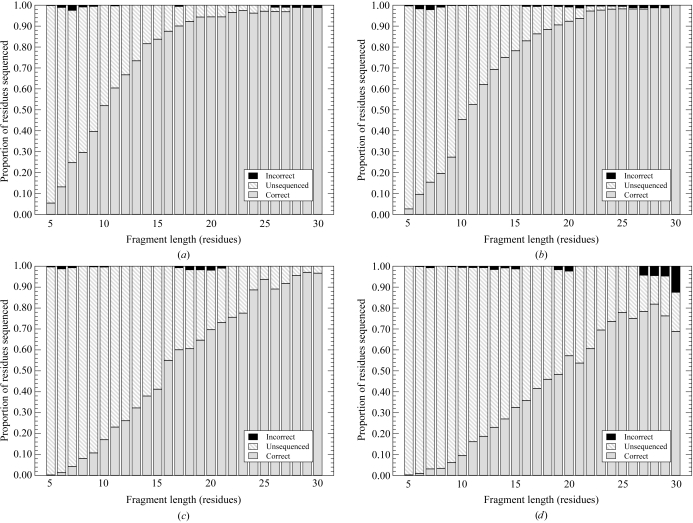
Each chain fragment was then sequenced using the algorithm described. For each sequencing attempt, the result was classified according to whether the chain fragment was sequenced correctly, sequenced incorrectly or not sequenced at all. The proportion of correct, unsequenced and incorrect results are plotted for each structure and for each starting model in Fig. 1 ▶.
Figure 1.
Success rates in sequencing fragments of different lengths for two structures using two different chain traces for generating the fragments. (a) 1vkn at 2.45 Å, using chain fragments taken from the true model. (b) 1vkn at 2.45 Å, using chain fragments taken from the Buccaneer auto-built model. (c) 1vrb at 3.2 Å, using chain fragments taken from the true model. (d) 1vrb at 3.2 Å, using chain fragments taken from the Buccaneer auto-built model.
For the better structure (1vkn), the map is easily interpretable and thus the chain tracing is very reliable. As a result, little difference is seen between using the true chain trace and the Buccaneer model as a starting point for sequencing. Using the true model, 50% of fragments of ten amino acids can be sequenced and 90% of fragments of 19 amino acids can be sequenced. Using the auto-traced model from Buccaneer, one or two extra residues are needed to achieve the same success rates. Wrongly sequenced fragments are very rare.
For the lower resolution structure (1vrb), the automatic chain trace is somewhat poorer and this also affects the success rates for sequencing. Using the true model, 16 residues are now required to achieve a 50% success rate and 25 residues to achieve a 90% success rate. With the autotraced model, 19 residues are required to achieve a 50% success rate and a 90% success rate is not achieved. Note that in this last case the number of long fragments available is quite small and some contain jumps, so the results on the right of the graph show more errors and some statistical noise.
4. Discussion
The sequencing algorithm is clearly useful in both these cases, sequencing some of the autotraced model even at 3.2 Å resolution in a noisy map. The most interesting part of this result is that it is possible to sequence the chain fragments without determining side-chain rotamers at all: this is effectively an example of fitting a fragment to the electron density using an ensemble of models covering the possible side-chain conformations.
This surprising result may be explained by considering the types of conformational variation of the different side-chain types. Ala and Gly do not have rotamers and so are fully handled by this approach. Other small residues (e.g. Ser, Val, Thr, Pro) will readily be identified as small, even in low-resolution maps, by the absence of density at more than one bond distance from the β-carbon. In contrast, the longest side chains (e.g. Arg, Lys) have sufficient flexibility that their rotamers are not very clearly defined. Additionally, these side chains are often disordered in real structures. The remaining residue types are those for which the ensemble approach will be weaker than existing methods.
Including more targets to allow classification of side chains by both type and rotamer may well improve the results. The largest improvements will probably be obtained by introducing rotamer-specific targets for the large rigid side-chain groups, in particular Trp, Tyr and His.
5. Conclusions
Density-based fragment-fitting methods provide a useful tool in the interpretation of electron-density maps, firstly from the point of view of automating tasks which would previously be performed by the crystallographer in front of a graphics program, but also by allowing the fitting of larger fragments or rapid fitting of many small fragments as a means of interpreting larger regions of density. A range of techniques have been developed by different authors to address different problem domains. A particular driving force of these efforts has been the need to optimize the six-dimensional rotation–translation search required to locate a fragment in an electron-density map.
An example of the application of fragment-fitting techniques to the automatic interpretation of protein features in electron-density maps has been examined, demonstrating how a single technique can be employed in multiple ways to perform a complete model-building calculation. An interesting result of this work has been the discovery that the important sequence-assignment part of this calculation may be performed using a set of ‘ensemble density models’ combining all the possible side-chain conformations of each amino-acid type. The method is very successful when the resolution is better than 2.5 Å and parts of the chain may be sequenced when the resolution is worse than 3.0 Å.
Acknowledgments
The author would like to thank the JCSG data archive for providing a source of well curated test data. This work was supported by The Royal Society under a University Research Fellowship.
References
- Bahar, M. et al. (2006). Acta Cryst. D62, 1170–1183. [DOI] [PubMed]
- Cohen, S. X., Morris, R. J., Fernandez, F. J., Ben Jelloul, M., Kakaris, M., Parthasarathy, V., Lamzin, V. S., Kleywegt, G. J. & Perrakis, A. (2004). Acta Cryst. D60, 2222–2229. [DOI] [PubMed]
- Cowtan, K. (1998). Acta Cryst. D54, 750–756. [DOI] [PubMed]
- Cowtan, K. (2001). Acta Cryst. D57, 1435–1444. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Ioerger, T. R. & Sacchettini, J. C. (2002). Acta Cryst. D58, 2043–2054. [DOI] [PubMed]
- Joint Center for Structural Genomics (2006). JCSG Data Archive. http://www.jcsg.org/datasets-info.shtml.
- Jones, T. A. (2004). Acta Cryst. D60, 2115–2125. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1997). Acta Cryst. D53, 179–185. [DOI] [PubMed]
- Nordman, C. E. & Schilling, J. W. (1986). Crystallographic Computing, edited by F. R. Ahmed, pp. 110–114. Copenhagen: Munksgaard.
- Pavelcik, F. (2006). J. Appl. Cryst.39, 483–486.
- Pavelcik, F., Zelinka, J. & Otwinowski, Z. (2002). Acta Cryst. D58, 275–283. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Terwilliger, T. C. (2003a). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003b). Acta Cryst. D59, 45–49. [DOI] [PMC free article] [PubMed]
- Vagin, A. A. & Isupov, M. N. (2001). Acta Cryst. D57, 1451–1456. [DOI] [PubMed]