Abstract
Individuals vary in their sociosexual behaviors and reactivity. How the organism interacts with the environment to produce this variation has been a focus in psychology since its inception as a scientific discipline. There is now no question that cumulative experiences throughout life history interact with genetic predispositions to shape the individual’s behavior. Recent evidence suggests that events in past generations may also influence how an individual responds to events in their own life history. Epigenetics is the study of how the environment can affect the genome of the individual during its development as well as the development of its descendants, all without changing the DNA sequence. Several distinctions must be made if this research is to become a staple in behavioral neuroendocrinology. The first distinction concerns perspective, and the need to distinguish and appreciate, the differences between Molecular versus Molar epigenetics. Each has its own lineage of investigation, yet both appear to be unaware of one another. Second, it is important to distinguish the difference between Context-Dependent versus Germline-Dependent epigenetic modifications. In essence the difference is one of the mechanism of heritability or transmission within, as apposed to across, generations. This review illustrates these distinctions while describing several rodent models that have shown particular promise for unraveling the contribution of genetics and the environment on sociosexual behavior. The first focuses on genetically-modified mice and makes the point that the early litter environment alters subsequent brain activity and behavior. This work emphasizes the need to understand behavioral development when doing research with such animals. The second focuses on a new rat model in which the epigenome is permanently imprinted, an effect that crosses generations to impact the descendants without further exposure to the precipitating agent. This work raises the question of how events in generations past can have consequences at both the mechanistic, behavioral, and ultimately evolutionary levels.
Keywords: Development, Genetically-modified mice, Knockout, Imprinting, Molar epigenetics, Context-Dependent epigenetic modification, Germline-Dependent epigenetic modification, Neural network, Cytochrome oxidase
“Genetic changes are stable and rarely reversed, whereas epigenetic changes are often reversed. A good example of that is genomic imprinting, where the changes imposed on DNA sequences may be lost during development, or if they persist, are erased and re-set during gametogenesis. Environmental influences do not change the genotype (leaving aside mutagens), and there is no inheritance of acquired characteristics. Epigenetics is quite different, because normal development depends on communication between cells. Thus, a hormone, morphogen or growth factor may induce an epigenetic change that may be heritable. This means that the environment of a cell may be all important in determining its properties or its fate in the developing organism. In this sense, epigenetics encompasses Lamarckian inheritance.” [43, pp. 78–79]
1. Introduction
The early stages of life, beginning before birth and, in mammals up to weaning, are the time of maximal neuronal plasticity. Although the individual’s capacity to respond to environmental change or insult with heritable phenotypic variation at a later stage is possible, it is during this early period that hormones and genotype predispose an individual’s responses to future experiences throughout the life cycle as well as the susceptibility to developing disorders (e.g., [8,9,32,38,48,59]). Obviously, suites of genes underlie the fundamental plasticity of an organism, particularly during development or life history transitions. How do these gene networks interact with the experiences that cumulate during an individual’s life history?
An important interface between the environment (either internal or external) and the genotype is that of epigenetic modifications (Fig. 1). Exactly how these modifications come about is still relatively unknown, but recent studies at both the molecular and organismal levels indicate that the origin of such effects may occur in previous generations. That is, experiences of earlier generations can modify regulatory factors affecting gene expression such that the DNA sequence itself is not changed but the individual’s physiology and behavior are substantially influenced not only as an effector but also as an affector system. And, as depicted in Fig. 1, further dimensions also apply in the form of both animal and human culture inheritance systems [57]. In a real sense, it may be said that Lamarck was right; he was simply born in the wrong century [57,83]. Thus, understanding how such modifications actually occur will increase our understanding of how the environment influences the relationship between genotype and behavior during sensitive developmental periods. Before reviewing this literature, it is important to first point out the dual origins of the study of epigenetic modification or what I term Molecular versus Molar epigenetics and, secondly, to distinguish between mitotic (non-germline) versus meiotic (germline) epigenetic imprints, or what I term Context-Dependent versus Germline-Dependent epigenetic modifications.
Fig. 1.
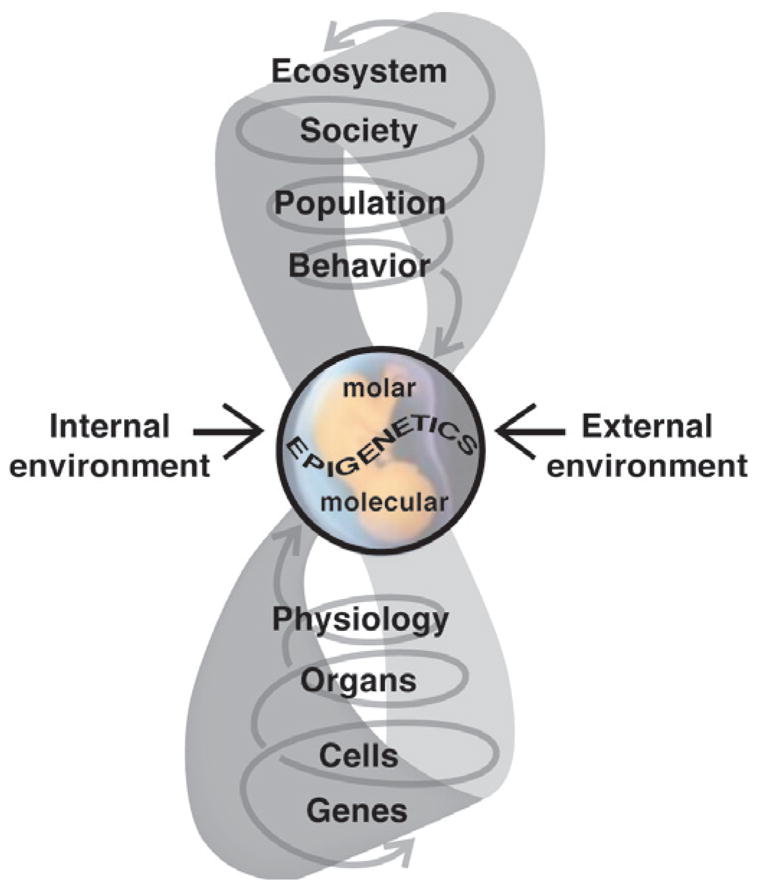
The external environment interacts with the internal environment to influence fetal development with both immediate and life-long consequences. Such environmentally-induced changes can occur at all levels of biological organization, from the molecular to the organism’s behavior, and tend to be amplified in their consequences as they ascend through these levels. Ultimately, these influences may be epigenetic in nature, inducing heritable alterations in gene expression without changing the DNA. Changes can occur at the physiological and morphological levels (Molar Epigenetics) as well as modification of normal patterns of gene expression (Molecular Epigenetics). These alterations can bring about functional differences in brain and behavior that result in changes in the phenotype. These then modify how individuals respond to conspecifics and their environment, bringing about changes at higher levels of biological organization. Whether these eventually can have an evolutionary impact is still open to question. What is known is that human society has changed the ecosystem in a manner that has had demonstrable impact on the health of humans and wildlife. Figure modified from Crews and McLachlan [21].
2. Molecular versus Molar epigenetics
There have been several reviews recently as to the origins of the field of epigenetics, all of which recognize the multiple roots of the current tree of research (e.g., [11,48,52,53,107,56]). Unfortunately, almost all have been written by molecular and developmental biologists who, being genocentric, are not versed in the history of the psychological or behavioral sciences (for exceptions, see [43,44,57]).
The debates in the natural sciences in the 16th–17th centuries pitted preformationism against epigenesis, with the central question being how a fully integrated multicellular organism develops from a single cell (the fertilized egg). The former camp believed that adult features were present fully formed in the egg and simply unfolded during growth, while the latter held that traits emerge as a consequence of the progressive interaction of the constituent parts of the zygote [36,37]. Spawned after the resolution of this conflict were two distinct research endeavors, one rooted in anatomy and geology, which ultimately became the broad study of biology, and the other focused on the study of sensation, perception and mind, which ultimately became the study of psychology. Thus, while both share a common origin, they evolved very differently both in perspective and substance. These I will label, respectively, “Molecular epigenetics” and “Molar epigenetics”. The former term is roughly equivalent to mitotic (non-germline) and meiotic (germline) epigenetic imprints. The latter term is from the historical literature in psychology, particularly functionalism, and re-introduced here because it encompasses the flavor of acting on emergent elements of behavior. Functionalism was a reaction of American psychologists such as William James and James Angell to the European Structuralist school of Hermann Helmholtz, Edward Tichenor, and Wilhelm Wundt, who started the first laboratory of experimental psychology. Rather than measuring sensory processes, the focus of functionalists was the organism itself and the organization of behavior in relation to its natural environment. As such it drew heavily from Darwin’s theory of natural selection and its adaptive consequences through evolution.
Molecular epigenetics originated from modern genetics and molecular biology about 30 years ago [51–53] and initially focused on gene regulation and its developmental significance. On the other hand, Molar epigenetics emerged soon after the re-discovery of Mendel’s studies and focuses on the organism. It has two independent roots; in the field of evolutionary biology/population genetics the emphasis has been on evolutionary and adaptive significance, whereas in psychology, the emphasis has been on behavioral development and the mechanisms underlying it. Thus, the object of study in Molecular epigenetics is gene expression usually during embryogenesis, while in Molar epigenetics, it is on the individual’s interactions with its biotic and physical environment usually after birth (what I term Molar epigenetics is similar to the “probabilistic epigenesis” [46] or “endophenotypes” [40,41]).
2.1. Molecular epigenetics
For most of the 20th century, the question of how the genotype produces different phenotypes in response to different environments fell out of favor among European and American biologists. Interestingly, it continued as a major field of study in Russia and was represented in small part in this country in the work of Theodosius Dobzhansky and his students, most notably Richard C. Lewontin [65]. Today, it has re-emerged as a vigorous area of research among evolutionary biologists and behavioral ecologists. This new work on the origins of polymorphisms and polyphenisms is now commonly called “Phenotypic Plasticity” and is one of the driving forces in the relatively new union of developmental biologists with evolutionary biologists (Evo-Devo, which I will not discuss further in this essay) and is making inroads into behavioral ecology [13,37].
Prior to the 1940s, the gene as the unit of heritable material was a theoretical concept without a physical identity. Conrad H. Waddington [109,110] proposed the term “epigenetics” from classical embryology, ascribing it to the study of the processes by which the genotype gives rise to the phenotype [36,37]. His concept of genetic assimilation was central to what became the modern era of epigenetics when Robin Holliday [51,54] used then modern genetic methods to propose a molecular model for heritability of gene activation (and inactivation) during development by DNA methylation (and demethylation). Since then the term epigenetics is commonly used in molecular and developmental genetics to connote the study of gene expression that does not involve change in the DNA. There are several mechanisms that can accomplish this end, such as DNA methylation and modification of histones by processes of methylation, deacetylation, and phosphorylation (see articles [60,87]). There is debate within the molecular community as to the efficacy of histone modification for change through time [82] but it is recognized that DNA methylation does qualify as a core process in epigenetics. Because manipulation of methylation patterns is often lethal, or at the least results in maladaptive traits or monsters, this method of research illuminates normal development by creating abnormalities or anomalies. While DNA methylation is clearly involved in genomic imprinting, the signal for the imprint is not yet known.
2.2. Molar epigenetics
Early comparative psychologists were also interested in epigenetics, but from the perspective of the interaction of the organism’s ‘heredity’ and the nature of species-typical behaviors or “instincts”. Work principally by Konrad Lorenz and Niko Tinbergen emphasized that such behaviors were products of natural selection, the result of gene(s) acting in the brain to generate behaviors that were unlearned and innate. This interpretation of behavioral organization was effectively countered by the work of Karl S. Lashley and his students [28] most notable for the purposes of this review Frank A. Beach (regarded as one of the founders of neuroendocrinology) and Theodore C. Schneirla [94,102]. Together they formulated an epigenetic approach to behavioral development that focused on the interaction of the various levels of biological organization, from the genetic to the environmental, that support the development and display of these species-typical behaviors. Examples of this integrative approach are now numerous, but two classic efforts were Daniel S. Lehrman’s elegant work on the elaborate interaction of parent and offspring as ringdoves learn to care for their young and Jay S. Rosenblatt’s exceptional research on the physiological and behavioral events that underlie the development of maternal behavior in cats and later rats. Both were students of Schneirla, but others also took this broad integrative approach (cf., [42–44,46,50,101,71]). Thus, the comparative approach in psychology emphasized the dynamic, interactive nature of a process thus occurs as two levels intersect, one from within the organism and the other that occurs between the organism and its environment, which must be defined broadly to include the behavior and physiology of socially important species members. In so doing these early comparative psychologists laid the foundation for psychobiology, a vibrant field that focuses on how experiences cumulate throughout life to shape the way in which the individual interacts with its social and physical environment (cf., [44,46,88]).
It is not the purpose of this essay to venture into the relatively unexplored frontier that lies in uniting the two sub disciplines of Molar epigenetics, namely that of evolutionary and developmental biology and psychobiology and behavioral neuroendocrinology. However, it is useful to be reminded of Ernst Mayr’s constant refrain that behavior is at the leading edge of evolution or Gottlieb’s adage that “changes in behavior create the new variants on which natural selection works” [45]; this important point, first made by Bateson [10] and Morgan [72] more than a century ago, never made it into mainstream biology until recently, although it has been occasionally commented on by endocrinologists [25,96,98]. It is necessary to emphasize here though that the individual is the unit of selection and that an approach that integrates both Molecular and Molar epigenetics will be necessary to reveal the mechanisms that underlie behavioral evolution. That is, the continuity of Molecular and Molar epigenetics will be revealed as the constituent elements of traits are accrued as genes and their products and regulators interact both positively and negatively in a temporal, spatial, and conditional (internal as well as the social and physical environments) context [62,74]. As adaptive responses emerge they, in turn, set the stage for future variation. Thus, evolution is a tandem process involving first development, with its built-in flexible responsiveness to both gene products and environment, followed by selection, which dictates which variants are spread and maintained [65,117]. In this sense the “genome learns from its experience” [58].
3. Context-Dependent versus Germline-Dependent epigenetic modifications
It is commonly thought that epigenetic modifications that occur within an individual’s own lifetime have the ability to become inherited. But unless one considers, for example, the cultural transmission of royalty an epigenetically modified state, this is not so. (That is, speaking of royalty in humans and not in honeybees [86]). At a molecular level, CpG sites however are often associated with 5′ promoter regions of genes and have a higher probability of undergoing mutation than other regions of the genome. Consequent changes in DNA methylation patterns at CpG islands would be persistent and if imprinted in the germline have the potential of becoming heritable. At issue then is what is meant by heritability. In Context-Dependent epigenetic modifications we are dealing with transmission within a generation (within an individual’s own lifetime, including the interaction of parent and young) while in Germline-Dependent epigenetic modifications we are dealing with transmission across generations.
3.1. Context-Dependent epigenetic modification
The best examples of Context-Dependent epigenetic modifications are those that either have an effect early in life, such as exposure to endocrine disrupting compounds in utero or smoking during childhood and adolescence (known collectively as the fetal basis of adult disease; see article [39]). In the first instance the onset of disease manifests later or the deleterious effects decline with time. However, the extent to which the modification is perpetuated is by simple persistence of the environmental factors that bring about the epigenetic modification; that is, in each generation individuals are exposed to the same conditions. For example, if the diet [112,29] or environmental toxicant such as lead continues to be present in the environment, then the epigenetic modification will be manifest each generation. This type of epigenetic modification lends itself to relatively straightforward therapeutic venues such as providing methyl donors to the diet [112] or removing the environmental toxicant (smoke or lead). Hence, the environment can induce epialleles, but that this environmentally induced epigenetic state can be reversed by a different environmental factor. This mitotically based transgenerational effect I term ‘Context-Dependent’ epigenetic change.
The best example of a Context-Dependent epigenetic modification on behavior is that of Meaney and colleagues [69,114,115,70] (see article [16]). In a long series of elegant studies this group has demonstrated that the nature and amount of care a pup receives from the mother modulates its reaction to stress later in life largely through effects on the glucocorticoid receptor (GR) in the hippocampus. This maternal effect can cross generations, but its heritability depends upon the pup’s experience in the first week of life. Recently this group has documented that being reared by a high quality mother results in the expression of the transcription factor A (NGFI-A), a nerve growth factor-inducible protein, that binds to the first exon of the GR gene, resulting in increased expression of GR. High quality maternal care during this critical period demethylates NGFI-A and the acetylation of histones. Just as cross-fostering can reverse these molecular and behavioral changes, infusion of methionine, a histone deacetylase inhibitor, into the hippocampus can also reverse these events [113]. It is important to point out, however, that selective breeding cannot stabilize these brain–behavior differences. That is, the effect of high and low quality mothering disappears after five generations, indicating that it is not a Germline-Dependent epigenetic modification but a Context-Dependent epigenetic modification. This of course does not detract from the implications of the work for the human condition. For example, in rhesus macaques, genotypic variation for the serotonin transporter gene influences how individuals respond to stress during early life [100] and in humans it has been reported that rearing environment can overcome the influence of a polymorphism in the gene encoding the neurotransmitter-metabolizing enzyme monoamine oxidase A in the etiology of violent behavior [14,15].
3.2. Germline-Dependent epigenetic modification
Germline-Dependent epigenetic modifications are fundamentally different than Context-Dependent epigenetic modification in that the epigenetic imprint has become independent of the original causative agent. Here the epigenetic modification is transferred to subsequent generations because the change in the epigenome has been incorporated into the germline. Thus, the effect is manifest each generation without the need for re-exposure. In such instances the DNA methylation of heritable epialleles are passed through to subsequent generations rather than being erased as occurs normally during gametogenesis and shortly after fertilization. Germline-Dependent epigenetic modifications tend to be influenced by parental sex (see contributions [26,60]). In addition such modifications can be associated with one sex, an important aspect as many behaviors and affective disorders show sex differences. Examples of this type of epigenetic modification are still relatively rare.
4. Epigenetics, behavior and the brain
If the context in which the individual is nurtured influences its behavior as an adult, it is likely that the activity of the neural circuitry that underlies these behaviors must also be affected. This organizational principal should also apply to epigenetic modifications whether within the individual’s own life history (Context-Dependent) or inherited from previous generations (Germline-Dependent) (see articles [16,39]). What follows are two case studies, the first demonstrating how the genetic and litter environment influence later adult behavior and the neural circuitry underlying these behaviors. The second example is a new animal model system in which the epigenome of the male germline is permanently imprinted, an effect that crosses generations. This imprint results in altered patterns of behavior in males and females and is reflected in patterns of gene expression in critical brain areas known to be involved in the regulation of sociosexual behaviors.
4.1. Effects of sex ratio and genotype ratio of the litter influencing adult sexual behavior and the social behavior neural network in genetically-modified mice
The molecular modification of mice has been a major breakthrough in our understanding of the genetic basis of both simple and complex traits [12]. This includes behavior, in particular aggressive, sexual, and anxiety-related behaviors. While we have advanced from the simplistic ‘genes as adjectives’ interpretation for behavioral analyses, we have yet to explore the contribution of the dynamics within the natal nest. Not only is the sex ratio of the litter an issue [108,89,18], but the ratio of the various genotypes is equally important variable, particularly in model systems that are the result of the mating of heterozygotes (HTZ) to yield litters of varying numbers of wildtype (WT), HTZ, and knockout (KO) young of both sexes (Fig. 2).
Fig. 2.
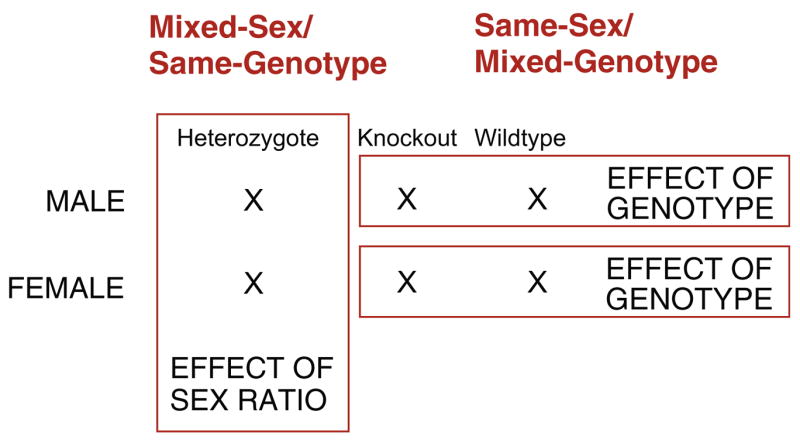
In most instances genetically-modified mice arise from the mating of individuals heterozygous for a null mutation. This results in litters that are a mixture of different numbers of male and female young of various genotypes. Thus, the sex ratio and genotype ratio of the litter can be a confound in interpreting the results of any phenotype measure, molecular or behavioral. However, by sexing and genotyping pups at birth and then re-constituting litters of equal numbers of specific young, it is possible to deconstruct the behavioral neural phenotype of the adult. The red-outline blocks represents groups that can be used to establish the effect of sex (e.g., Mixed-Sex/Same-Genotype groups) versus the effect of genotype (e.g., Same-Sex/Mixed-Genotype groups). Finally, by creating Mixed-Sex/Mixed-Genotype groups it is possible to study the precise interaction of sex and genotype in the development of the phenotype of interest. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Researchers using knockout (KO) mice do not control for the early social environment of their experimental animals. This is a mistake for this early social environment has a powerful effect on shaping the adult behavioral phenotype and brain. This now obvious point was first illustrated by the phenomenon of imprinting in birds. For example, if precocial birds such as ducks and geese are presented with an object (including a human) soon after hatching, they not only will fixate on that object and follow it, but they will attempt to mate with the object when sexually mature. This sexual imprinting also extends to primates as demonstrated by Harlow’s monkeys raised by “terricloth mothers” respond sexually only to terricloth dummies as adults [27]. Thus, both the social as well as endocrine experiences of the individual early in life can have a major influence on their adult behavior (cf., [6,20,49]).
Two laboratories have conducted extensive experimentation with genetically-modified mice with somewhat different results. For example, in their work on mice a null mutation of the estrogen receptor α (ERα), commonly known as ERKO mice, Sonoko Ogawa and Donald Pfaff routinely report that ♂ERKO mount stimulus females as much as males lacking the mutation or wildtype (WT), whereas in their initial work Emilie Rissman and colleagues claimed that ♂ERKO “fail to display sexual behavior” [116,95], although their data indicate that about one-half of intact ♂ERKO mounted receptive females compared to 100% of ♂WT. The distinction appears to lie in the different measurements used, the nature of the stimulus animal used (sex, hormonal condition, and genotype), testing paradigms, housing of animals after weaning, and, importantly, the genetic background of the knockouts; it is possible also that there are differences in how the females with their litters are maintained. The emphasis in the work of Ogawa/Pfaff is on mounting, while in the work of Rissman et al. it is intromission and ejaculatory behaviors, in essence a distinction between appetitive and consummatory behaviors [84,85]. In this context it is important to keep in mind that ♂ERKO can breed [66,33], indicating that they are capable of ejaculation. Similarly, there is a difference in reports of the level of aggression displayed by ♂ERKO; Ogawa et al. [77,78] report that ♂ERKO are less aggressive than ♂WT, whereas Skordalakes and Rissman [95] report the two genotypes are equally aggressive. The point of this discussion regarding the stark difference in behavior between laboratories is that the experience of the genetically-modified mouse is paramount in shaping its adult behavior.
In collaboration with Sonoko Ogawa I have been examining how the sex and genotype ratios of a litter might contribute to the development of behavior in knockout mice. We accomplish this by mating mice heterozygous (HTZ) for a null mutation of the ERα gene. The pups are sexed and genotyped within two days of birth [22]. Litters are then reconstituted to form same-sex litters of equal numbers of knockout (ERKO) and wildtype (WT) individuals such as (1) same-sex, same-genotype litters (e.g., ♀WT/♀WT), (2) same-sex, mixed-genotype (e.g., ♀WT/♀ERKO), (3) mixed-sex, same-genotype (♀WT/♂WT), or (4) mixed-sex, mixed-genotype (♂ERKO/♀WT) litters (Fig. 2). In this manner the effect of genotype is evident without the potential confound of the presence of the opposite sex in the litter and the effect of siblings of the opposite sex is evident without the potential confound of littermates with a different genotype.
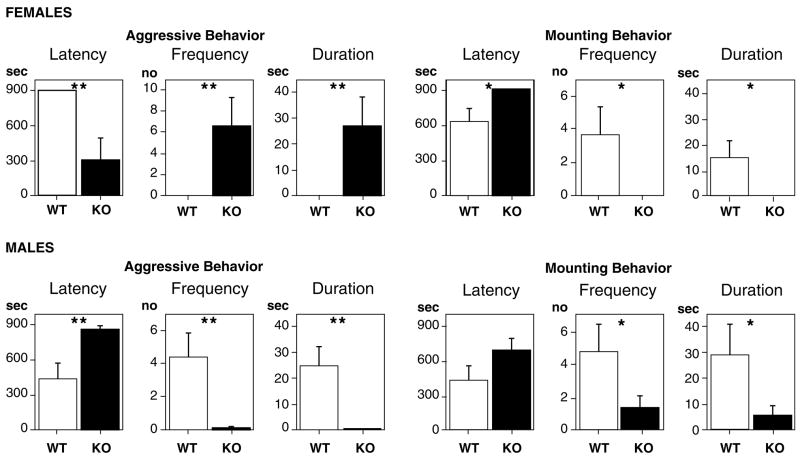
As adults individuals are tested in a standard resident-intruder paradigm. Our previous work [22] indicated that the behavioral differences between the genotypes are more sharply defined than reported previously in the literature where animals were raised in litters without concern for their composition (Table 1). For example, in our studies ♀WT display only aggressive behavior while ♀WT litter-mate’s display only mounting behavior (Fig. 3). In the case of ♂ERKO, both aggression and mounting behavior are greatly reduced. A replication and extension of this study revealed that ♀WT from both the ♀WT/♀ERKO and ♀WT/♂WT groups showed lower contact times compared to females raised in ♀WT/♀WT litters [78]. These data suggest that litter composition influences the development of sociosexual behaviors in ERKO mice of both sexes. Thus, in behavioral phenotyping studies of genetically manipulated animals it is important to understand and control for the potential role of the rearing environment in the development of the behavior of interest [97,34].
Table 1.
Litter composition affects in adult sociosexual behavior in genetically-modified mice
| Sex | Behavior | Litter compositon | Frequency | Study |
|---|---|---|---|---|
| Female | Aggression | Mixed-sex/mixed-genotype | ♀KO aggression higher than in ♀WT | [76] |
| Same-sex/mixed-genotype | Only ♀KO are aggressive | [22] | ||
| Mounting | Mixed-sex/mixed-genotype | Both ♀KO and ♀WT mount | [76,116] | |
| Same-sex/mixed-genotype | Only ♀WT mount | [22] | ||
| Male | Aggression | Mixed-sex/mixed-genotype | Only ♂WT are aggressive in some studies and in some ♂KO are as aggressive as ♂WT | [77,78,95] |
| Same-sex/mixed-genotype | Only ♂WT are aggressive | [22] | ||
| Mounting | Mixed-sex/mixed-genotype | ♂KO mount as much as ♂WT; however, in some studies ♂KO do not mount | [77,95] | |
| Same-sex/mixed-genotype | ♂KO mount less than ♂WT | [22] |
In mice wildtype (WT) or knockout (KO) for the estrogen receptor α gene, males and females exhibit different behavioral repertoires depending upon the sex and genotype ratio of the litter. Natural litters consist of a mixture of females and males of each of three possible genotypes (mixed-sex/mixed-genotype). If litters are reconstituted at birth such that they consist only of females or males, but equal numbers of WT and KO pups (same-sex/mixed-genotype), it becomes evident that the absence of the opposite sex in the litter influences these behaviors later when the young become adults.
Fig. 3.
Frequency of aggressive and mounting behavior in genetically-modified mice raised in single sex groups with equal numbers of wildtype (WT) or knockout (KO) female or male mice; top panel: ♀WT/♀KO; bottom panel: ♂WT/♂KO. Tests with female mice involved ovariectomized female intruders (top row) and tests with male mice involved olfactory-bulbectomized male intruders (bottom row). Values are group mean and standard errors. Statistical aralysis was computed on log-transformed data. (*p < 0,05, **p < 0.01). Reprinted with Permission from Crews et al. [22].
We have extended this work by examining the pattern of metabolic activity in the brains of animals raised in these controlled litter groups [23]. We use cytochrome oxidase (CO) histochemistry as a measure of metabolic activity because we are interested in the long-term effects of significant life-history events. This is a rate-limiting enzyme in oxidative phosphorylation, the major pathway in brain metabolism and, consequently, the abundance and activity of CO activity in a brain area is a measure of the metabolic capacity of that brain region. In other words, the CO abundance not only reflects the metabolic history of an area, it also determines the amount of ATP available in a neuron, thereby constraining the amount of activity a neuron can sustain [93]. Thus, CO histochemistry reveals long-term changes in brain activity while 2-DG autoradiography or immediate-early gene detection provide information on evoked or immediate activity. Indeed, the CO method can detect how metabolic activity has changed even several years after the event! For example, we find that in both mammals and reptile’s metabolic activity in limbic areas reflects the capacity to display sociosexual behaviors and, in turn, that differences in metabolic activity in these areas reflect individual differences in the propensity to display social behaviors [90–92].
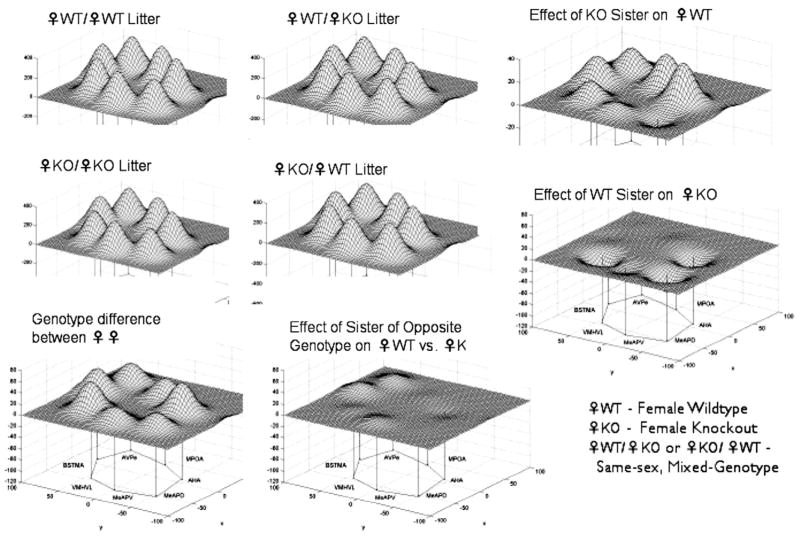
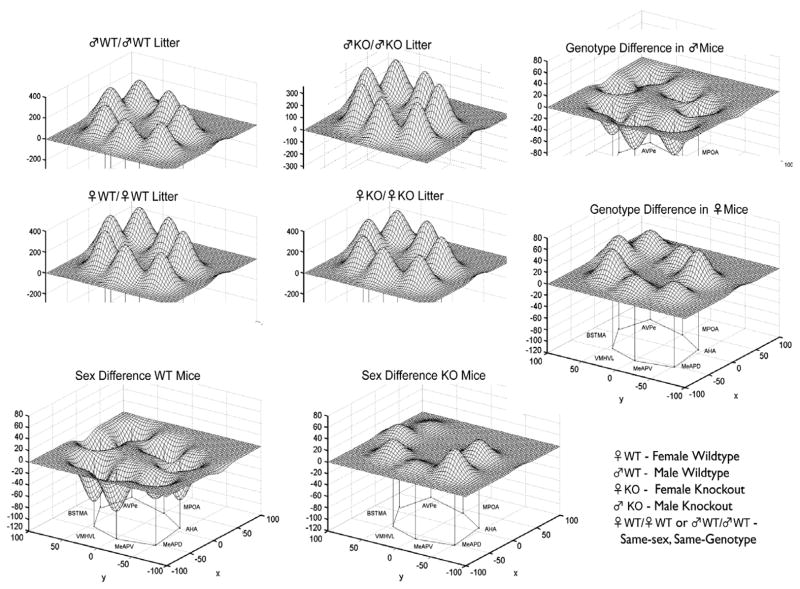
It is of interest that WT females raised in same-sex, same-genotype groups spend significantly more time in social contact in a Resident-Intruder test compared to ERKO females raised in same-sex, same-genotype groups (Fig. 3). Further, it appears that female WT siblings are able to compensate for this deficit, just as ERKO siblings cause a deficit in WT females. The neural network that subserves sociosexual behavior varies in different ways. First, as predicted, there is a significant genotype difference in the neural network of WT and ERKO females (Fig. 4). Further, the compensation/deficit in the behavioral scores (Fig. 3) are reflected in the metabolic activity of the neural circuit. The relative effects of sex independent of genotype, and of genotype independent of sex, are also striking (Fig. 5). Taken together these findings indicate that in studies with genetically-modified mice the litter composition during the preweaning period must be considered as it can effect the development of behavior and the neural network responsible for the regulation of emotional behaviors.
Fig. 4.
The genotype of siblings in the litter influences metabolic activity in the limbic landscape of adult mice wildtype (WT) or knockout (KO) for the estrogen receptor α gene. Limbic landscapes of ♀WT or ♀KO mice raised with sisters of the same or different genotype are depicted; genotype on left of diagonal of the caption (e.g., ♀WT/♀WT) is that of the experimental individual while the right of the diagonal indicates the genotype of the sisters it was raised with. Illustrated is the mean cytochrome oxidase (CO) abundance in specific brain nuclei. The limbic landscape map on the bottom left row is the genotype difference between the ♀WT and ♀KO mice, while the map on the bottom right reflects the effect of sisters of the opposite genotype on ♀WT vs. ♀KO. On the right column, the upper map indicates the effect of KO sisters on a WT female and below that is the effect of WT sisters on a KO female. The nuclei are presented in a clockwise fashion reflecting a rostral-caudal dimension: main bed nucleus of the stria terminalis (BNSTma); anteroventral periventricular nucleus (AVPe); medial preoptic area (MPOA); anterior hypothalamus, anterior (AHA); medial amygdaloid nucleus, posterodorsal (MeAPD); medial amygdaloid nucleus, posteroventral (MeAPV); ventromedial hypothalamic nucleus, ventrolateral (VMHVL).
Fig. 5.
The sex and genotype of siblings in the litter influences metabolic activity in the limbic landscape of adult mice wildtype (WT) or knockout (KO) for the estrogen receptor α gene. Limbic landscapes of ♂WT or ♂KO (or ♀WT or ♀KO) raised with brothers or sisters of the same or different genotype are depicted; sex/genotype on left of diagonal is the experimental individual while sex/genotype on right of diagonal is the type of sibling it was raised with. Illustrated is the mean cytochrome oxidase (CO) abundance in specific brain nuclei. The limbic landscape maps on the bottom row are the difference between the ♂WT/♂WT and ♀WT/♀WT and between ♂KO/♂KO and ♀KO/♀KO maps, respectively, indicating the effect of the sibling’s sex independent of genotype. The left column are maps indicating the difference between the effect of ♂WT/♂WT and ♂KO/♂KO and between ♀WT/♀WT and ♀KO/♀KO, respectively, indicating the effect of the siblings genotype independent of sex. See Fig. 4 for further details.
Finally, we have also examined the effect of litter composition on anxiety-related behaviors as measured by Open Field tests. It is perhaps significant that such behaviors are not affected. For example, litter composition has no substantial effect on open field activity (total distance, center distance, and center time); regardless of genotype, males consistently have higher scores than do females (D. Crews and S. Ogawa, unpublished data). Comparing three litter types tested (all-male/mixed-genotype, all-female/mixed-genotype, and mixed-sex/same-genotype litters), male and female mice differ significantly, but within each sex there is no difference between genotypes. This suggests that sex differences in open field behavior are organized early in life, perhaps prenatally, rather than environmentally determined.
The significance of the findings of litter type is evident when it is considered that the secondary sex ratio in rodents (gerbils, hamsters, mice, and rats) can be influenced by the birth mother’s own intrauterine position [18,89,106]. That is, a female that develops between two females in utero produces litters with a female-biased sex ratio whereas a female that developed between two males in utero produces litters with a male-biased sex ratio.
4.2. Transgenerational epigenetic imprint on the nuclear genome and its effect on brain and behavior
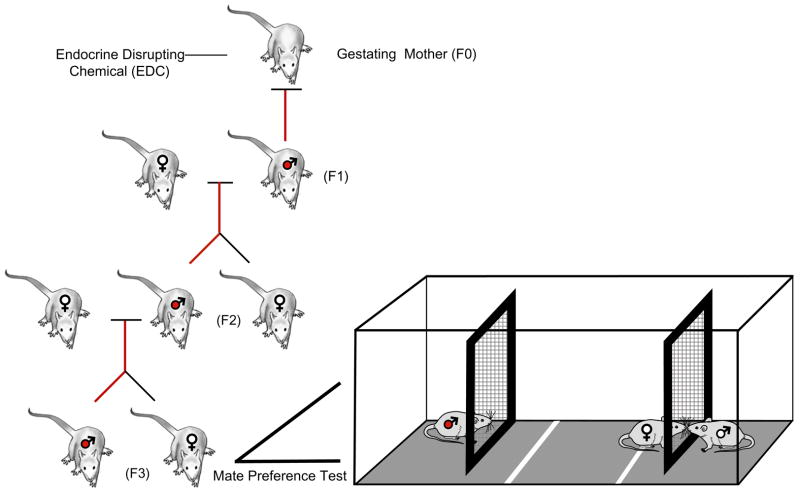
The two critical elements to demonstrating a Germline-Dependent or transgenerational epigenetic imprint are first, a single exposure that is never again repeated and second, the number of generations since that exposure (Fig. 6). Although there are studies of the “transgenerational” effects of endocrine disruptors on the behavior of animals treated as fetuses or as neonates, the tested individuals have body burden of the chemical. That is, the first generation is the exposed individual and even if continued to a second generation, these individuals have had their germline exposed. Hence, any effects must be considered as occurring within the individual’s own lifetime. Demonstration of a transgenerational epigenetic imprint, i.e., a meiotic (germline) versus a mitotic (non-germline) epigenetic imprint, requires at the minimum two generations if the male is exposed or three generations if the female is exposed. It is only then that these body burdens are absent and can be taken as evidence that the mechanism by which the imprint can continue is via the germline.
Fig. 6.
Determination of a transgenerational epigenetic imprint on mate preference behavior in the rat. The left panel shows that three generations separate the gestational exposure to vinclozolin, a common-use fungicide with endocrine-disrupting (EDC) properties. The right panel illustrates the testing apparatus for mate preference. Two groups of animals were tested. The control group was the F3 generation of a lineage (control-lineage) of animals in which the dams were exposed to vehicle (DMSO) three generations previously. The experimental group was the F3 generation of a lineage (EDC-lineage) of animals in which the dams were exposed to vinclozolin three generations previously. This EDC exposure epigenetically alters males to express early onset of various diseases states and this modification is transmitted via the germline. Third generation females from the EDC-lineage and the Control-lineage were tested with males from both lineages in simultaneous mate preference tests; males from the EDC-lineage (indicated by red-filled male symbols) and the Control-lineage (not shown) were similarly tested with females of both stimulus types. The trials are conducted under dim red light during the nocturnal (active) phase of the rats’ light cycle. The experimental animal (here a female from the Control-lineage) was placed in the center of the chamber; a stimulus male from each lineage type was at each end of the apparatus. The female could move freely in their chamber but separated from the stimulus males by a wire mesh. This enabled the animals to communicate by olfactory, pheromonal, or behavioral cues, but physical interaction was limited to touching across the wire mesh. Left portion of figure from Anway and Skinner [3]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To date there is but a single example of a Germline-Dependent epigenetic modification on behavior [24]. Working with Michael Skinner we took advantage of a new model system in which an endocrine disrupting chemical (EDC) reprograms methylation patterns that are then incorporated into the germline and, hence, transmitted to future generations [2,4,3,17]. The remarkable feature of this model system is that exposure of gestating female rats to the pesticide methoxychlor or the fungicide vinclozolin during the period of embryonic sex determination induces an epigenetic transgenerational phenotype through reprogramming the germline in a sex-specific manner. Specifically, in each generation males whose ancestor had been treated underwent progressive spermatogonial apoptosis, decreased sperm count and motility and, as the animals aged, adult onset disease is accelerated, including cancer, prostate disease, kidney disease, and immune cell defects.
We sought to determine if this altered epigenome also influences mate/partner preference behavior (Fig. 6) [24]. For these studies we used the F3 descendents of females that had been treated between E8 and E15 with dimethylsulfoxide buffer alone (Control-lineage) or with vinclozolin (= EDC-lineage) [2]. After habituation to the testing arena, each individual was tested individually (as the experimental subject) or in pairs (as the stimulus animal) in a round-robin design. To confirm that females were receptive, each female was placed with a sexually experienced male prior to the test; all females exhibited robust lordosis (arched back and lifted head posture) in response to mounting by the male.
Partner preference tests consisted of placing an individual (male or female) in the center of a large, three-chamber glass-testing arena. At either end was a small Plexiglas cage containing the stimulus rats separated by a wire-mesh barrier to allow exchange of olfactory, visual, and tactile cues. The area directly in front of the stimulus cage was marked by tape. All males were tested with both types of females as stimulus animals and all females were tested with both types of males as stimulus animals.
Behaviors directed to the stimulus animal included time spent in contact with the wire mesh separating the experimental and stimulus animal (Wire Mesh) during which the animals often touched noses through the wire mesh (Facial Investigation) and contacted the Plexiglas surface surrounding the front of the stimulus cage (Plexiglas); the cumulative total time in these behaviors directed toward each stimulus animal was also calculated (Total). Other activity measures included time spent: grooming (Grooming), aimless walking and sniffing (Walking), standing on hind paws and sniffing with nose pointed upwards (Standing/Sniffing), still with minimal head movement (Still), contacting the walls of the test cage (Glass), and time in the center compartment (Center).
The results were clear-cut and sex-specific (Fig. 7). The females discriminate and prefer males who do not have a history of exposure, while males do not exhibit such a preference. In social encounters in rodents and other animals, time is spent engaged in mutual facial investigation. This appears to be a critical aspect of the assessment process and may underlie our finding that males investigate females equally, while females show greater amount of time investigating males from the Control-lineage. It is known that pheromones from the vomeronasal organ [61] and urine are involved in mate recognition in rodents [68]. Methylation analysis revealed that the Major Urinary Protein 4 (MUP4) is one of the candidate imprinted-like genes induced in the vinclozolin generation males [17]. This MUP group of gene products binds and releases male-specific pheromones in rodents [5,19]. Finally, to rule out odor discrimination capacity as a potential explanation for their failure to show a preference in the mate preference test, we established that both males and females explored odors of the opposite sex much more than familiar (self) odors or novel odors of the same sex, and all animals explored novel odors of the same sex more than self odors.
Fig. 7.
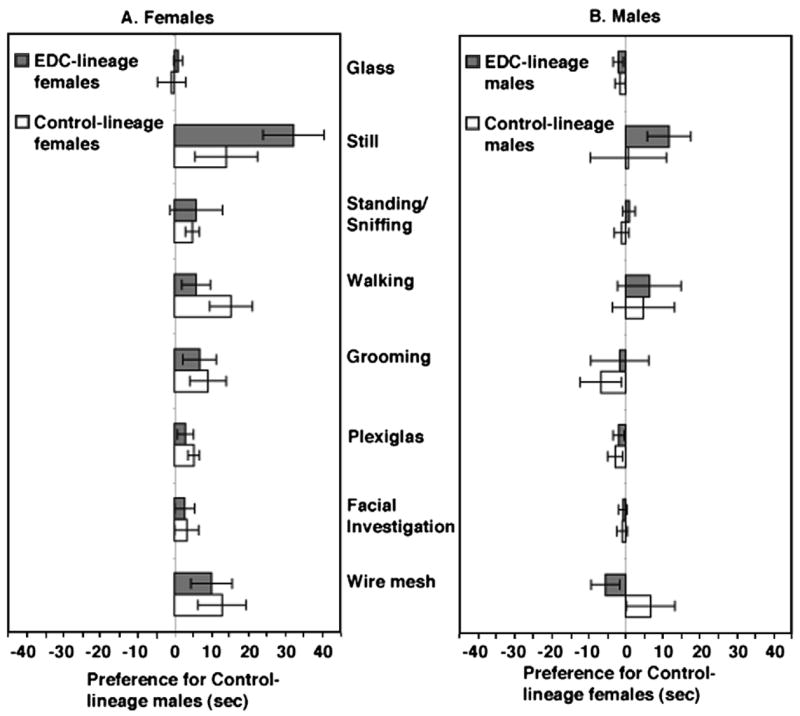
Third-generation female rats whose progenitors were exposed to vinclozolin, a common-use fungicide with endocrine-disrupting (EDC) properties, and hence epigenetically altered, prefer males from the unexposed Control-lineage. Males do not show this preference. See Fig. 6 for further details. Both females and males from Control- and EDC-lineages were tested with pairs of Control- and EDC-lineage stimulus partners. Presented are the mean (+1 standard error) differences in the time spent in each behavior. Left panel: Behaviors exhibited by females from Control- and EDC-lineages towards males from Control-lineage (positive, right side) and EDC-lineage (negative, left side). Right panel: Behaviors exhibited by males from Control- and EDC-lineages towards females from Control-lineage (positive, right side) and EDC-lineage (negative, left side). The various behavioral measures and test are described in Crews et al. (2007). Reprinted by permission from Crews et al. [24].
Are these behaviors reflected in differences in the pattern of gene expression in different brain areas? In pilot studies we have begun to analyze the extent to which this transgen-erational epigenetic imprint might influence patterns of gene expression in specific brain areas. Specifically, we have been using gene microarrays to analyze whole brain, amygdala and hippocampus transcriptomes in F3 generation Vinclozolin-lineage and Control-lineage males. Of the altered genes, only a limited number show similar changes in the whole brain, amygdala, and hippocampus (M. Skinner and D. Crews, unpublished data). Those genes common to all three include Senp5 (SUMO/sentrin specific protease 5); Nfix (Nuclear factor I/X), Akap5 (A kinase (PRKA) anchor protein 5), NTrkb (Neurotrophic tyrosine kinase, receptor) and COMT (catechol-o-methyltransferase); the latter three genes have been implicated in the etiology of schizophrenia [118,75] and other affective disorders including autism and depression [81,103,104]. As expected, Camk2a (calcium/calmodulin-dependent protein kinase II alpha subunit) was present, and regulated, in the amygdala and hippocampus; this gene has been implicated in both learning and memory [111] and stress-induced anxiety behavior [73].
A hallmark of epigenetically modifiable nuclear genes is their CpG-rich islands, a property shared, albeit little studied, by mitochondrial genes [1,99]. While epigenetic modification of traits encoded by nuclear genes has been established, it is only recently that evidence for a similar process for traits encoded by cytoplasmic genes. This is important as adaptive selection acts on traits and, as a consequence, on the frequency of genes (nuclear as well as cytoplasmic) that underlie these traits. Although it is commonly thought that the evolution of mitochondrial genes is due to neutral mutations [35,7], recent studies reveal that polymorphisms in mitochondrial genes play a significant role in adaptive evolution. For example, mitochondrial genes are important in determining male sperm quality, particularly motility [35,31] and there is some evidence that mitochondrial mutations result in decreases in human sperm motility [55]. In this regard, male rats of all five generations stemming from females treated during pregnancy with either methoxychlor or vinclozolin have increased spermatogenic cell apoptosis and decreased sperm number and motility [2]. The mitochondrial genome is also vitally involved in aging and, in particular, mutations in mitochondrial DNA are correlated with early onset of age-related phenotypes, including reduced fertility [105,64].
Selection may also act on the epistatic interactions between mitochondrial and nuclear genes, in other words, on co-evolved gene complexes. For example, oxidative metabolism and especially oxidative phosphorylation depend upon the coordinated and integrated action of the mitochondrial and nuclear genomes [79]. Using populations of the seed beetle fixed for their cytoplasmic and nuclear lineages, Dowling et al. [30] established that the effects of temperature, which dictates the rate of development in this species, and hence on fitness, are different in specific mito-nuclear genetic combinations. Studies similar to those with the seed beetle have been performed with Drosophila and demonstrate that the relative contributions of the mitochondrial and nuclear genomes in metabolic energy production can be manipulated; measurements of mito-nuclear fitness suggest both sex and environment specific effects due to mitochondrial polymorphisms [67,32].
Thus, it is particularly interesting to examine the genes coding for CO in transgenerationally imprinted male rates. It was surprising to find that none of the constituent genes of this mitochondrial-nuclear gene product appear to be altered in the brain, amygdala, or hippocampus of epigenetically imprinted male rats (M. Skinner and D. Crews, unpublished data). However, nuclear respiratory factor 2 (NRF-2), which modulates transcriptional levels of CO genes in mammalian cells [80] exhibits an increased expression in whole brain, suggesting that while the mito-nuclear complex is not altered, the gene regulating this functional system is over-expressed, perhaps accounting for the behavioral differences observed between mice of the two lineages. This finding that the genes coding for the individual CO subunits are unaffected, while NRF-2, the key gene regulating CO activity in the brain, is modified, is consistent with the interpretation that the relative fitness of specific mito-nuclear genotype combinations is dependent upon the modified DNA environment in which they persist. Thus, chemical agents such as EDCs could act via epigenetically modifying mitochondrial DNA as well as nuclear DNA and, as a consequence, influence the epistatic interactions between cytoplasmic and nuclear genes.
5. Conclusion
The future for epigenetic studies in neuroendocrinology is going to depend upon communication, principally between those schooled in what might be called collectively psychobiology (comparative and physiological psychology, behavioral neuroendocrinology, and behavioral biology) and molecular and developmental biology. Typically, scientific progress is in fits and starts, advancing only when practitioners in one discipline become aware of, and appreciate, other fields working on common problems [63]. This is the case when Waddington first proposed his theories of the role of gene action in phenotype development. It was not until geneticists and developmental biologists began to address common questions from their disparate perspectives that the field of molecular epigenetics blossomed to what it is today.
Ironically, the first suggestion that the process of DNA methylation might play a role in gene expression focused on the process of long-term memory [47]. This primacy for neuroscience was relinquished almost immediately to the union of genetics and developmental biology. Behavioral neuroendocrinologists are in a pivotal position to link Molecular with Molar epigenetics. The strong tradition of studies in behavioral development, our extensive knowledge of the role hormones play in the organization and activation of brain–behavior mechanisms, the predisposition in neuroscience research to use molecular methods to understand cellular function and development, and the identified goal of understanding social and emotional behaviors by integrating mechanistic with evolutionary analyses should facilitate the incorporation of epigenetics into neuroendocrinological research. While it is not essential that neuroendocrinologists actually employ molecular epigenetic tools if they wish to work on more molar questions, it is critical that they at least understand the basic distinctions within the field so that appropriate questions can be posed and the results obtained interpreted with validity.
Acknowledgments
The work reported herein was supported in part by grants from the NIMH (R21 MH068273 and ROl MH41770). I thank Gwen Cage for the illustration in Fig. 1, Vicky Huang for assistance with Figs. 4 and 5, Michael Skinner for permission in using a component of Fig. 6, and P. Bateson, G. Galef, E. Jablonka, M. McCarthy for critical comments on the manuscript.
References
- 1.Ames N, Gilley J, Fried M. The comparative genomic structure and sequence of the surfeit gene homologs in the puffer fish Fugu rubripes and their association with CpG-rich islands. Genome Res. 1997;7:1138–1152. doi: 10.1101/gr.7.12.1138. [DOI] [PubMed] [Google Scholar]
- 2.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147(Suppl):S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 4.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong SD, Robertson DH, Cheetham SA, Hurst JL, Beynon RJ. Structural and functional differences in isoforms of mouse major urinary proteins: a male-specific protein that preferentially binds a male pheromone. Biochem J. 2005;391:343–350. doi: 10.1042/BJ20050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakker J, van Ophemert J, Slob AK. Postweaning housing conditions and partner preference and sexual behavior of neonatally ATD-treated male rats. Psychoneuroendocrinology. 1995;20:99–310. doi: 10.1016/0306-4530(94)00061-e. [DOI] [PubMed] [Google Scholar]
- 7.Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Ann Rev Ecol Syst. 2005;36:621–642. [Google Scholar]
- 8.Barker DJ. The developmental origins of adult disease. Eur J Epidemiol. 2003;18:733–736. doi: 10.1023/a:1025388901248. [DOI] [PubMed] [Google Scholar]
- 9.Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Mirazo’n Lahr M, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE. Developmental plasticity and human health. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 10.Bateson W. Materials for the Study of Variation: Treated with Especial Regard to Discontinuity in the Origin of Species. Macmillan; London: 1894. [Google Scholar]
- 11.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 12.Blaustein JD, Erskine MS. Feminine sexual behavior: cellular integration of hormonal and afferent information in the rodent brain. In: Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, editors. Hormones, Brain and Behavior. Vol. 1. Academic Press; San Diego: 2002. pp. 139–214. [Google Scholar]
- 13.Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- 14.Caspi A, McClay J, Moffitt TE, Mill J, Martin C, Craig IW, Taylor A, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 15.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig I, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 16.Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol. doi: 10.1016/j.yfrne.2008.03.003. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HS, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147:5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- 18.Clark MM, Karpiuk P, Galef BG. Hormonally mediated inheritance of acquired characteristics in Mongolian gerbils. Nature. 1993;364:712. doi: 10.1038/364712a0. [DOI] [PubMed] [Google Scholar]
- 19.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 20.Cooke BM, Chowandadisai W, Breedlove SM. Post-weaning social isolation of male rats reduces the volume of the medial amygdala and leads to deficits in adult sexual behavior. Behav Brain Res. 2000;117:107–113. doi: 10.1016/s0166-4328(00)00301-6. [DOI] [PubMed] [Google Scholar]
- 21.Crews D, McLachlan JA. Epigenetics, evolution, endocrine disruptors, health and disease. Endocrinology. 2006;147(Suppl):S4–S10. doi: 10.1210/en.2005-1122. [DOI] [PubMed] [Google Scholar]
- 22.Crews D, Fuller T, Mirasol EG, Pfaff DW, Ogawa S. Postnatal environment affects behavior of adult transgenic mice. Exp Biol Med. 2004;229:935–939. doi: 10.1177/153537020422900910. [DOI] [PubMed] [Google Scholar]
- 23.Crews D, Lou W, Fleming A, Ogawa S. From gene networks underlying sex determination and gonadal differentiation to the development of neural networks regulating sociosexual behavior. Brain Res. 2006;1126:109–121. doi: 10.1016/j.brainres.2006.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crews D, Gore AC, Hsu T, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints and mate preference. Proc Natl Acad Sci USA. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham JT. Hormones and Heredity. A Discussion of the Evolution of Adaptations and the Evolution of Species. Constable and Co., Ltd; London: 1921. [Google Scholar]
- 26.Davies W, Phoebe MY, Relkovic D, Wilkinson LS. Imprinted genes and neuroendocrine function. Front Neuroendocrinol. doi: 10.1016/j.yfrne.2007.12.001. in this issue. [DOI] [PubMed] [Google Scholar]
- 27.Deutsch J, Larson K. Model-oriented sexual behavior in surrogate-reared rhesus monkeys. Brain Behav Evol. 1974;9:157–164. doi: 10.1159/000123662. [DOI] [PubMed] [Google Scholar]
- 28.Dewsbury DA. The Chicago Five: A family group of integrative psychobiologists. Hist Psychol. 2002;5:16–37. doi: 10.1037/1093-4510.5.1.16. [DOI] [PubMed] [Google Scholar]
- 29.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2006;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Dowling DK, Abiega KC, Arnqvist G. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution. 2006;61:194–201. doi: 10.1111/j.1558-5646.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 31.Dowling DK, Nowostawski AL, Arnqvist G. Effects of cytoplasmic genes on sperm viability and sperm morphology in a seed beetle: implications for sperm competition theory? J Evol Biol. 2007;20:358–368. doi: 10.1111/j.1420-9101.2006.01189.x. [DOI] [PubMed] [Google Scholar]
- 32.Dowling DK, Friberg U, Hailer F, Arnqvist G. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics. 2007;175:235–244. doi: 10.1534/genetics.105.052050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy EM, Washburn TF, Bunch DO, Goulding EH, Gladen BC, Lubahn DB, Korach KS. Targeted disruption of the estrogen receptor gene in male mice causes alteration of spermatogenesis and infertility. Endocrinology. 1996;137:4796–4805. doi: 10.1210/endo.137.11.8895349. [DOI] [PubMed] [Google Scholar]
- 34.Francis DD, Szegda K, Cambell G, Martin WD, Insell TR. Epigenetic sources of behavioral differences in mice. Nat Neurosci. 2003;6:445–446. doi: 10.1038/nn1038. [DOI] [PubMed] [Google Scholar]
- 35.Gemmell NJ, Metcalf VJ, Allendorf FW. Mother’s curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19:238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert SF. The genome in its ecological context philosophical perspectives on interspecies epigenesis. Ann NY Acad Sci. 2002;981:202–218. doi: 10.1111/j.1749-6632.2002.tb04919.x. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert SF, Sarkar S. Embracing complexity: organicism for the 21st century. Dev Dyn. 2000;219:1–9. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1036>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 38.Gluckman PD, Hanson MA. The Fetal Matrix: Evolution, Development and Disease. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 39.Gore AC. Fetal programming and endocrine disruptor effects on reproductive neuroendocrine systems. Front Neuroendocrinol. doi: 10.1016/j.yfrne.2008.02.002. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottesman II, Shields J. Schizophrenia and Genetics—A Twin Study Vantage Point. Academic Press; New York: 1972. [Google Scholar]
- 41.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Amer J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 42.Gottleib G. Development of Species Identification in Birds. University of Chicago Press; Chicago: 1971. [Google Scholar]
- 43.Gottleib G. Synthesizing Nature-Nurture. Prenatal Roots of Instinctive Behavior. Lawrence Erlbaum Associates; Mahwah: 1977. [Google Scholar]
- 44.Gottlieb G. Individual Development and Evolution: The Genesis of Novel Behavior. Lawrence Erlbaum Associates; Mahwah: 2002. [Google Scholar]
- 45.Gottlieb G. Developmental-behavioral initiation of evolutionary change. Psychol Rev. 2002;109:211–218. doi: 10.1037/0033-295x.109.2.211. [DOI] [PubMed] [Google Scholar]
- 46.Gottlieb G. Probabilistic epigenesis. Dev Sci. 2007;10:1–11. doi: 10.1111/j.1467-7687.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- 47.Griffith JS, Mahler HR. DNA ticketing theory of memory. Nature. 1969;223:580–582. doi: 10.1038/223580a0. [DOI] [PubMed] [Google Scholar]
- 48.Haig D. The (dual) origins of epigenetics. Cold Spring Harb Symp Quant Biol. 2004;69:67–70. doi: 10.1101/sqb.2004.69.67. [DOI] [PubMed] [Google Scholar]
- 49.Hard E, Larsson K. Dependence of adult mating behavior in male rats on the presence of littermates in infancy. Brain Behav Evol. 1968;1:405–419. [Google Scholar]
- 50.Hinde RA. Animal Behaviour: A Synthesis of Ethology and Comparative Psychology. McGraw-Hill; New York: 1970. [Google Scholar]
- 51.Holliday R. The inheritance of epigenetic effects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 52.Holliday R. Epigenetics comes of age in the twenty-first century. J Genet. 2002;81:1–4. doi: 10.1007/BF02715863. [DOI] [PubMed] [Google Scholar]
- 53.Holliday R. Epigenetics. A historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 54.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 55.Holyoake AJ, McHugh P, Wu M, O’Carroll S, Benny P, Lin IL, Sin FYT. High incidence of single nucleotide substitutions in the mitochondrial genome is associated with poor semen parameters in men. Int J Androl. 2001;24:175–182. doi: 10.1046/j.1365-2605.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- 56.Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann NY Acad Sci. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- 57.Jablonka E, Lamb MJ. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. MIT Press; Cambridge: 2005. [Google Scholar]
- 58.Jaenisch R, Bird B. Epigenetic regulation of gene expression, how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 59.Jirtle R, Skinner MK. Environmental epigenomics and diseases susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keverne EB, Curley JP. Epigenetics, brain evolution and behaviour. Front Neuroendocrinol. doi: 10.1016/j.yfrne.2008.03.001. in this issue. [DOI] [PubMed] [Google Scholar]
- 61.Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- 62.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhn TS. The Structure of Scientific Revolutions. University of Chicago Press; Chicago: 1970. [Google Scholar]
- 64.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JE, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TAC. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 65.Lewontin RC. The Triple Helix: Gene, Organism and Environment. Harvard University Press; Cambridge: 2000. [Google Scholar]
- 66.Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci USA. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maklakov AA, Friberg U, Dowing DK, Arnqvist G. Within-population variation in cytoplasmic genes affects female life span and aging in Drosophila melanogaster. Evolution. 2006;60:2081–2086. [PubMed] [Google Scholar]
- 68.McClintock MK. Pheromones, odors, and vasanas: the neuroendocrinology of social chemosignals in humans and animals. In: Pfaff D, Arnold A, Etgen A, Faahrbach S, Rubin R, editors. Hormones, Brain and Behavior. Vol. 1. Academic Press; San Diego: 2002. pp. 797–870. [Google Scholar]
- 69.Meaney MJ. Maternal care gene expression and the transmission of individual differences in stress reactivity across generations. Ann Rev Neurosci. 2001;24:161–192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 70.Meaney MJ, Syzf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moltz H. Imprinting: an epigenetic approach. Psychol Rev. 1963;70:123–138. doi: 10.1037/h0044362. [DOI] [PubMed] [Google Scholar]
- 72.Morgan CL. Habit and Instinct. Edward Arnold; London: 1896. [Google Scholar]
- 73.Muller MB, Zimmermann S, Sillaber I, Hagemeyer TP, Deussing JM, Timpl P, Kormann MS, Droste SK, Kuhn R, Reul JM, Holsboer F, Wurst W. Limbic corticotropin-releasing hormone receptor 1 mediates anxiety-related behavior and hormonal adaptation to stress. Nat Neurosci. 2003;6:1100–1107. doi: 10.1038/nn1123. [DOI] [PubMed] [Google Scholar]
- 74.Nijhout HF. The importance of context in genetics. Amer Sci. 2004;91:416–418. [Google Scholar]
- 75.O’Donovan MC, Williams NM, Owen MJ. Recent advances in the genetics of schizophrenia. Hum Mol Genet. 2003;12:R125–R133. doi: 10.1093/hmg/ddg302. [DOI] [PubMed] [Google Scholar]
- 76.Ogawa S, Eng T, Taylor J, Lubahn DB, Korach KS, Pfaff DW. Roles of estrogen receptor-a gene expression in reproduction-related behaviors in female mice. Endocrinology. 1998;139:5070–5081. doi: 10.1210/endo.139.12.6357. [DOI] [PubMed] [Google Scholar]
- 77.Ogawa S, Washburn TF, Taylor J, Lubahn DB, Korach KS, Pfaff DW. Modifications of testosterone-dependent behaviors by estrogen receptor-a gene disruption in male mice. Endocrinology. 1998;139:5058–5069. doi: 10.1210/endo.139.12.6358. [DOI] [PubMed] [Google Scholar]
- 78.Ogawa S, Luk S, Murphy L, Matthews D, Pfaff DW, Tomihara K, Soga T, Crews D. Effects of the litter composition during pre-weaning period on the development of anxiety-related behaviors in mice. Soc Neurosci Abstr. 2005;31:892.14. [Google Scholar]
- 79.Ongwijitwat S, Wong-Riley MTT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 80.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Pezet S, Malcangio M. Brain-derived neurotrophic factor as a drug target for CNS disorders. Exp Opin Ther Targets. 2004;8:391–399. doi: 10.1517/14728222.8.5.391. [DOI] [PubMed] [Google Scholar]
- 82.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 83.Richards EJ. Inherited epigenetic variation-revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 84.Rissman EF, Wersinger SR, Taylor JA, Lubahn DB. Estrogen receptor function as revealed by knockout studies: neuroendocrine and behavioral aspects. Horm Behav. 1997;31:232–243. doi: 10.1006/hbeh.1997.1390. [DOI] [PubMed] [Google Scholar]
- 85.Rissman EF, Wersinger SR, Fugger HN, Foster TC. Sex with knockout models: behavioral studies of estrogen receptor a. Brain Res. 1999;835:80–90. doi: 10.1016/s0006-8993(99)01452-3. [DOI] [PubMed] [Google Scholar]
- 86.Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: social life in molecular terms. Nat Rev Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 87.Ruden DM, Jamison DC, Zeeberg BR, Garfinkel MD, Weinstein JN, Rasouli P, Lu X. The EDGE hypothesis: epigenetically directed genetic errors in repeat-containing proteins (rcps) involved in evolution, neuroendocrine signaling, and cancer. Front Neuroendocrinol. doi: 10.1016/j.yfrne.2007.12.004. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutter M. Gene-environment interdependence. Dev Sci. 2007;10:12–18. doi: 10.1111/j.1467-7687.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 89.Ryan BC, Vandenbergh JG. Intrauterine position effects. Neurosci Biobehav Rev. 2002;26:665–678. doi: 10.1016/s0149-7634(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 90.Sakata JT, Crews D. Developmental sculpting of social phenotype and plasticity. Neurosci BioBehav Rev. 2004;28:95–112. doi: 10.1016/j.neubiorev.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 91.Sakata JT, Gupta AJ, Crews D, Gonzalez-Lima F. Animal models of experiential effects on neural metabolism: plasticity in limbic circuits. In: Handa R, Hayashi S, Terasawa E, Kawata M, editors. Neuroplasticity, Development and Steroid Hormone Action. CRC Press; Boca Raton: 2001. pp. 257–272. [Google Scholar]
- 92.Sakata JT, Gonzalez-Lima F, Gupta AJ, Crews D. Repeated interactions with females elevate metabolic capacity in the limbic system of male rats. Brain Res. 2002;936:27–37. doi: 10.1016/s0006-8993(02)02491-5. [DOI] [PubMed] [Google Scholar]
- 93.Sakata JT, Crews D, Gonzalez-Lima F. Behavioral correlates of differences in neural metabolic activity. Brain Res Rev. 2005;48:1–15. doi: 10.1016/j.brainresrev.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 94.Schneirla TC. Behavioral development and comparative psychology. Quart Rev Biol. 1966;41:283–302. doi: 10.1086/405056. [DOI] [PubMed] [Google Scholar]
- 95.Scordalakes EM, Rissman EF. Aggression in male mice lacking functional estrogen receptor. Behav Neurosci. 2003;117:38–45. [PubMed] [Google Scholar]
- 96.Shire JG. The forms uses and significance of genetic variation in endocrine systems. Biol Rev Camb Philos Soc. 1976;51:105–141. doi: 10.1111/j.1469-185x.1976.tb01121.x. [DOI] [PubMed] [Google Scholar]
- 97.Smotherman WP, Robinson SR. Caveats in the study of perinatal behavioral development: utility of fetal study. Neurosci Biobehav Rev. 1994;18:347–354. doi: 10.1016/0149-7634(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 98.Spearow JL, Doemeny P, Sera R, Leffler R, Barkley M. Genetic variation in susceptibility to endocrine disruption by estrogen in mice. Science. 1999;285:1259–1261. doi: 10.1126/science.285.5431.1259. [DOI] [PubMed] [Google Scholar]
- 99.Staines DR. Does dysregulation of key epigenetic and biochemical pathways occur in postulated vasoactive neuropeptide autoimmune disorders? Med Hypotheses. 2005;65:1154–1160. doi: 10.1016/j.mehy.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 100.Suomi SJ. Genetic and environmental factors influencing the expression of impulsive aggression and serotonergic functioning in rhesus monkeys. In: Tremblay RM, Hartup WW, Archer J, editors. Developmental Origins of Aggression 63–82. Guilford Press; New York: 2004. pp. 63–82. [Google Scholar]
- 101.Tinbergen N. On aims and methods of ethology. Z Tierpsychol. 1963;20:410–433. [Google Scholar]
- 102.Tobach E. The identity of comparative psychology: its status and advances in evolutionary theory and genetics. Int J Comp Psychol. 2006;19:129–150. [Google Scholar]
- 103.Tsai SJ. Down-regulation of the Trk-B signal pathway: the possible pathogenesis of major depression. Med Hypotheses. 2004;62:215–218. doi: 10.1016/S0306-9877(03)00299-8. [DOI] [PubMed] [Google Scholar]
- 104.Tsai SJ. TrkB partial agonists: potential treatment strategy for epilepsy, mania, and autism. Med Hypotheses. 2006;66:173–175. doi: 10.1016/j.mehy.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 105.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly M-Y, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature aging in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 106.Vandenbergh JG, Huggett CL. Mother’s prior intrauterine position affects the sex ratio of her offspring in house mice. Proc Natl Acad Sci USA. 1995;91:1155–1159. doi: 10.1073/pnas.91.23.11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Speybroeck L. From epigenesis to epigenetics: the case of C.H. Waddington. Ann NY Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 108.Vom Saal FS, Bronson FH. Sexual characteristics of adult female mice are correlated with their blood testosterone levels during prenatal development. Science. 1980;208:597–599. doi: 10.1126/science.7367881. [DOI] [PubMed] [Google Scholar]
- 109.Waddington CH. Organisers and Gene. Cambridge University Press; Cambridge: 1940. [Google Scholar]
- 110.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Ferguson GD, Pineda VV, Cundiff PE, Storm DR. Overexpression of type-1 adenylyl cyclase in mouse forebrain enhances recognition memory and LTP. Nat Neurosci. 2004;7:635–642. doi: 10.1038/nn1248. [DOI] [PubMed] [Google Scholar]
- 112.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 113.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weaver IC, Cervoni N, Champagne FA, D’Allessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 115.Weaver IC, Champagne FA, Brown SE, Dymov S, Shama S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wersinger SR, Sannen K, Villalba C, Lubahn DB, Rissman EF, De Vries GJ. Masculine sexual behavior is disrupted in male and female mice lacking a functional estrogen receptor gene. Horm Behav. 1997;32:176–183. doi: 10.1006/hbeh.1997.1419. [DOI] [PubMed] [Google Scholar]
- 117.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford University Press; New York: 2003. [Google Scholar]
- 118.Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet. 2006;15:743–749. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]